Abstract
Cataracts are a significant public health problem. Here, we describe the genetic alteration responsible for a progressive form of cataract, segregating as an autosomal dominant trait in a three-generation pedigree. Unlike most autosomal dominant cataracts, these are not clinically apparent at birth but are initially observed in the first year or two of life. The opacification evolves relatively slowly, generally necessitating removal of the lens in childhood or early adolescence. A genome-wide search in our kindred revealed linkage at 2q33–35 where the γ-crystallin gene cluster resides. A single base alteration resulting in an Arg- 14 → Cys (R14C) substitution in γD-crystallin was subsequently identified. Protein modeling suggests that the effect of this mutation is a subtle one, affecting the surface properties of the crystallin molecule rather than its tertiary structure, consistent with the fact that the patients’ lenses are normal at birth. This is the first gene defect shown to be responsible for a noncongenital progressive cataract, and studying the defective protein should teach us more about the mechanisms underlying cataract formation.
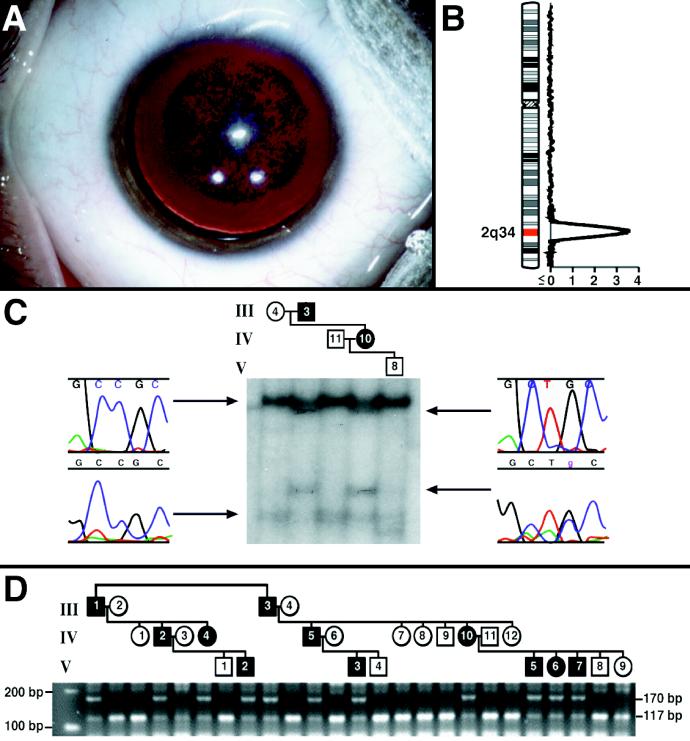
A number of inherited cataracts are known in both humans and animals (1–3). These cataracts are present at birth and generally static, suggesting a structural defect affecting lens development. Some, like the human cerulean (3) and Coppock-like (4) cataracts involve synthesis of grossly misfolded lens crystallins. Others, like the human zonular pulverulent (5) and autosomal dominant congenital cataract (2) cataracts, involve more subtle mutations in other lens proteins that cause congenital opacities. All of the nonsyndromal human cataracts known to be linked to a genetic locus or caused by a specific genetic mutation are congenital. We have identified a three-generation family with hereditary progressive cataracts transmitted as a fully penetrant autosomal dominant trait (Fig. 1). The lenses of affected individuals appear clear at birth, but with time focal opacities develop, suggesting some progressive alteration in protein structure or interaction. The cataract is always detectable by age 2.5 and consists of grayish-white punctate opacities in the nucleus and surrounding deep cortex, with intervening and peripheral clear lens (Fig. 1A). The cataracts are 5 to 6 mm in diameter, and the opacities become more dense over time such that lens extraction is ultimately required because of decreased vision. Once the opaque lens is removed and a synthetic lens inserted, vision returns to 20/20 and the eye functions normally.
Figure 1.
(A) Preoperative photograph of the right eye of the child V-3 (see D) representing the typical cataract in this pedigree. This child was initially examined at age 2.5 and was found to have focal opacities in the nuclear and perinuclear areas of both lenses causing minimal decreases in vision. By age 6 he complained of glare in bright light, but still had visual acuity of 20/50. At the time of surgery (age 10) his visual acuity was 20/70, and he had difficulty playing sports and performing in school because of blurring and glare. After the operation, his acuity returned to 20/20. His cousin (individual V-7, D) was examined at the earliest age (7 weeks). Faint “dust-like” opacities were seen when a careful slit lamp examination was performed. These were not visually significant. Other family members examined in early infancy (IV-4, V-6, and V-7) were found to have clear lenses, but developed obvious bilateral punctate opacities by age 2. Four of the five youngest members of the pedigree have already undergone cataract surgery (at ages 5, 7, 10, and 14). The remaining child is 4.5 years old and is nearing the point of needing surgery. At the other extreme, individual IV-5 did not have a lens removed until age 20. (B) Linkage analysis. Two-point lod scores greater than 2.00 were seen only with markers D2S325 and D2S2382 (see text) that gave scores of 3.68 and 2.53 at θ = 0, respectively. The sliding three-point analysis yielded a peak lod of 5.18 at markers D2S325 and D2S2382. (C) Detection of a nucleotide change by SSCP analysis in a subset of the pedigree members. The conformer illustrated was sequenced and found to contain a C->T nucleotide change at position 34 of exon 2 resulting in an R14C amino acid change. (D) The C->T nucleotide change in exon 2 segregates with the trait: restriction fragment length polymorphism analysis. The C->T missense mutation eliminates a HaeIII site in the second exon of the γD gene. PCR amplification of 50 ng of genomic DNA from each of the subjects studied followed by digestion for 1 hr at 37°C with HaeIII (New England Biolabs) and electrophoresis in 3% agarose (FMC) showed the expected 117-bp fragment in all the unaffected individuals and the uncut 170-bp fragment in all the affected members of the kindred.
To find the trait locus in the family, we performed a genome-wide search for linkage, obtained evidence for linkage at 2q33-q35 where the γ-crystallin genes are located, and looked for mutations in the two γ-crystallins that are expressed at high levels in the developing lens-γ-crystallins C and D. We discovered a mutation in the γD-crystallin (γD) gene that may alter its interaction with other proteins.
MATERIALS AND METHODS
These studies were performed in the context of an institutionally approved protocol. Members of the family were contacted, and after obtaining written informed consent, they were examined. Twenty milliters of blood were drawn from affected and selected unaffected individuals, and DNA was prepared from the blood samples by using a Puregene kit (Gentra Systems) according to the manufacturer’s instructions.
Genotyping and Linkage Analysis.
Genotyping was performed by using 396 fluorescent dye-labeled dinucleotide repeat markers (PE Applied Biosystems, Prism Linkage Mapping Set, Version 2). The PCR conditions used are given in detail at the following URL: http://www2.perkinelmer.com/ab/apply/dr/dra1a3.html. The PCR products were electrophoresed in 48-lane 5% denaturing polyacrylamide gels in Applied Biosystems 377 sequencers. The genescan and genotyper software packages (Applied Biosystems/Perkin–Elmer) were used to generate genotypes, and linkage analysis was performed by using fastlink for two-point and sliding three-point logarithm of odds (lod) scores. Allele frequencies were estimated from the founders of the pedigree for the genome scan. When evidence for linkage was obtained on chromosome 2, analyses of markers there were repeated by using maximum likelihood estimates of marker allele frequencies (obtained from the pedigree using the userm13 routine of mendel) and slight increases in the maximum multipoint lod score were observed.
Single-Strand Conformational Polymorphism (SSCP) and DNA Sequencing.
Two affecteds, two unrelated married-in individuals, and one unaffected sib of an affected child were chosen, to simplify the analysis while retaining power to eliminate out-of-phase polymorphisms (Fig. 1C). Primers flanking the exons of the γC-crystallin and γD isoforms were designed and used to PCR amplify 50 ng of subject genomic DNA in the presence of [α-32P]dCTP radioisotope. An equal amount of loading buffer (95% formamide/10 mM NaOH/0.25% bromphenol blue) was added to each PCR after cycling, and the mixture heated to 96°C for 5 min followed by snap cooling on ice. Denatured amplicons were loaded onto two different types of mutation-detection enhancement SSCP gels, prepared with or without 1% glycerol as recommended by FMC. The amplicons were electrophoresed at 5 W for 16 hr at room temperature. Gels were dried, stapled to film (Kodak XAR5), and exposed for 4 hr at room temperature. Developed film was realigned with the dried gel, and a conformer in exon 2 of γD with altered mobility compared with wild type and inherited with the phenotype was excised and eluted in 50 μl of TE buffer (10 mM tris/1mM EDTA, pH 7.6) overnight. The conformer was reamplified with the same primers that were initially used, the 287-bp product was gel purified (Qiagen, Chatsworth, CA), and it was sequenced with the same primers (5 μM final concentration) by using the FS Dye Terminator kit (PE Applied Biosystems). The sequencing reactions were amplified with an MJ-Tetrad Thermocycler (MJ Research, Cambridge, MA) as follows: 93°C, 3 min; 94°C, 30 sec; 55°C, 5 sec; 60°C, 4 min. The last three steps were repeated 24 times. Subsequently, the products were purified on Centri-sep columns according to the manufacturer’s instructions (Princeton Separations, Princeton, NJ), dried in a Speed Vac (Savant), redissolved/denatured in 3 μl of loading buffer (95% formamide/50 mM EDTA) at 90°C for 3 min, and analyzed with a Model 377 PE Applied Biosystems Fluorescent Sequencer with a 48-well comb by using the conditions in the user’s manual. Sequences were then aligned with those of the wild-type genes (Sequencher, Ann Arbor, MI).
Restriction Fragment Length Polymorphism Analysis.
The C->T missense mutation eliminates a HaeIII site in the second exon of the γD gene. PCR amplification of 50 ng of genomic DNA from each of the subjects studied followed by digestion for 1 hr at 37°C with HaeIII (New England Biolabs) and electrophoresis in 3% agarose (FMC) yielded the expected 117-bp fragment in unaffected individuals. The mutation was predicted to result in amplification of an uncut 170-bp fragment in all the affected members of the kindred. This technique was used to study members of the kindred and 400 additional unaffected control chromosomes.
Molecular Modeling.
Several γ-crystallin structures are known and all have extremely highly conserved tertiary structures. Bovine γD is 86% identical in amino acid sequence to its human homologue. Threading experiments were performed by using the method of Bryant and Lawrence (6). All possible alignments of the wild-type and mutant γD sequences against the three-dimensional coordinates of bovine γD (PDB ID code 1ELP; ref. 7) were examined. For each possible alignment, individual pairwise residue interactions were determined based on chemical type and distance intervals with no use of arbitrary gap penalties (6). By using these values, a conformational energy (ΔR/M), defined as the expected work for substitution of a specific sequence R for a random sequence with the same composition in the context of folding motif M, was then calculated for each alignment. Z-scores and chance occurrence probabilities (ER/M) were calculated to compare conformational energies for different alignments. Chance occurrence probabilities give the odds that a random sequence of the same length and amino acid composition would yield threading energy as low as that of the query sequence R. A significant match of sequence to structure results when ER/M for that thread is <0.05 (5%). Calculation of energies and statistical values was performed by using C and S-PLUS subroutines. All energy scaffolds were visualized by using the grasp software package (8).
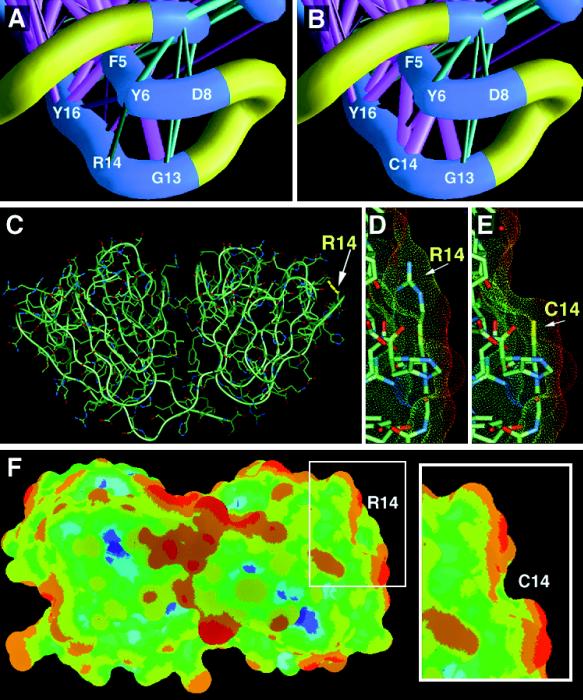
Human γD and the Arg-14 → Cys (R14C) mutant were also modeled from the experimentally determined coordinates of the bovine protein by side chain replacement and regularization using quanta (Molecular Simulations, San Diego, CA), courtesy of the National Institutes of Health Molecular Modeling Center. Identical residues were left in the experimentally determined conformation. Solvent-accessible areas, indicated in Fig. 2 D–F by dotted surfaces, were also calculated and colored by using quanta.
Figure 2.
Human γD and the R14C mutant were modeled from the coordinates of the bovine homologue (PDB ID code 1ELP). (A and B) Energy scaffolds for wild-type and mutant γD sequences modeled against the three-dimensional coordinates of bovine γD (1ELP). The portion of the structure containing the C-terminal end of strand a and the N-terminal end of strand b of the β-hairpin of motif 1 are shown. The α-carbon backbone of the protein is depicted as a curving worm. Strands a and b are shown in blue, while the characteristic folded loop is shown in yellow. Pairwise residue interaction energies between core residues are indicated by the thickness and coloring of the connecting α-carbon positions in the protein backbone. Thick magenta-colored cylinders are the most favorable interactions; thick cyan-colored cylinders indicate the least favorable interactions. Intermediate colors and cylinder thicknesses represent interactions falling between these extremes. (C) The overall structure of human γD with the position of residue 14 marked. The path of the polypeptide backbone is shown and the symmetrical two-domain structure of the protein can be seen. (D, E, and F) Solvent-accessible areas for wild-type and mutant proteins. These views are rotated by approximately 90° from the view in C for greater clarity. Solvent-accessible areas are indicated by dotted surfaces. D and E show close-up views of position 14 while F shows the overall surface accessible area for human γD and the panel (Inset) shows the change in the R14C mutant. Surfaces were colored according to electrostatic energy, with red representing least polar and blue, most polar regions. The R14C change causes a clear local change in surface shape and reduces polarity in that region.
RESULTS AND DISCUSSION
The genome-wide scan revealed linkage at 2q33–35 where the γ-crystallin gene cluster is located (Fig. 1B). This cluster is comprised of six genes, γA through γF, plus a quarter gene fragment, γG. Only two of these genes, γC-crystallin and γD, encode abundant lens proteins; γE and γF are pseudogenes with in-frame stop codons. For this reason we focused our attention on the γC-crystallin and γD genes. We used SSCP to screen for mutations in these genes and detected only an alteration in exon 2 of the γD gene (Fig. 1C). The altered conformer was sequenced and a C->T missense mutation was discovered, resulting in a nonconservative R14C substitution that segregated with the trait in the kindred studied (Fig. 1 C and D) but was absent from 400 control chromosomes. Twenty-two γ-crystallin isoforms have been cloned from human, rat, mouse, cow, carp, and Xenopus, and in 21 of 22 of these arginine is the fourteenth residue. In the only isoform without arginine (bovine γB), histidine is the fourteenth residue. Thus, we feel that R14C must be a mutation because it was the only alteration detected in the γC and γD crystallin genes, it segregates with the disease, it was not seen in unaffected individuals, and it is not found in any γD crystallin isoform known.
Lens transparency is maintained through a critical biophysical balance between proteins and water in a regular array of cells. Insults that disrupt this balance can cause opacity. In particular, modifications that reduce the effective solubility of lens proteins can lead to progressive aggregation and the formation of light-scattering centers in the lens fiber cells (9, 10). γD is one of only two γ-crystallins expressed at high concentrations in the fiber cells of the embryonic human lens (11, 12). These cells later form the lens nucleus, the site of the punctate cataract. As they mature, lens nuclear fibers lose their nuclei and other organelles (1). Consequently, their proteins must survive without turnover throughout life. γ-Crystallins are highly stable proteins, composed of four repeated “Greek key” structural motifs (13–15). They are particularly associated with high protein concentrations and high refractive index in vertebrate lenses. As such, they play a key role in maintaining the clarity and the gradient of refractive index on which the optical properties of the lens depend.
Several γ-crystallin structures are known, including that of bovine γD (PDB ID code 1ELP). This protein is 86% identical in sequence to the human γD and was used to model both wild-type and mutant γD (Fig. 2). In all γ-crystallins, residue 14 is conserved as a basic residue (arginine or histidine). In Greek key motifs of γ-crystallins and related β-crystallins other than the first one, however, residues including cysteine are found in the topologically equivalent position. In spite of this, the main chain fold of all crystallin Greek keys is essentially identical (see, for example, ref. 14). Thus, the R14C change would not necessarily be expected to have a significant effect on the tertiary fold of the protein, and the clarity of the lens at birth in affected individuals is consistent with an essentially normal structure in the R14C mutant. Furthermore, our model calculations show that replacing R14 with C may only cause a subtle change in the molecule — an increase in side chain interactions between C14 on strand b of the β hairpin and both the aromatics F5 and Y6 on strand a (Fig. 2 A and B). If anything, these interactions may serve to increase the stability of the characteristic folded hairpin structure of the γ-crystallin Greek key motif.
The greatest effect of the R14C mutation can be seen on the surface of γD. R14 is an exposed surface residue that contributes significantly to the solvent-accessible surface area of the protein. The views presented in Fig. 2 C–F illustrate the effect of this mutation: The change to cysteine produces a groove on the surface. The overall polarity of the surface in the region of the mutation also changes; the color shift indicates a reduction in polarity as would be expected when a basic polar residue is replaced by a nonpolar one.
The changes in the surface of the protein could affect the way the mutant γD interacts with other proteins. Residue 14 is involved in lattice contacts in several γ-crystallin crystallographic structures (16). In the case of human γD, surface properties may be particularly important. This protein belongs to the high critical temperature class of γ-crystallins (17). High critical temperature proteins are cryoproteins that undergo a reversible phase transition at temperatures above freezing, resulting in opacities known as “cold cataracts.” This phenomenon has often been used as a model system for lens opacification (2). Because the R14C mutation decreases the charge and increases the local hydrophobicity of γD, it is very likely that the critical temperature of the protein also rises, increasing its tendency toward self-association and entry into a protein-rich phase, even under normal physiological conditions. Furthermore, the presence of a new exposed cysteine residue would allow for the formation of intermolecular disulfide bonds under oxidizing conditions. Protein aggregation, particularly linked with disulfide formation, is a major feature of many cataracts (1), and while the lens of affected patients may be clear at birth, small seeds promoting the aggregation of γD may already be present. In this way the effect of the mutation may resemble what occurs through age-related insults in an older normal lens.
Future experiments will examine the biophysical effect of the R14C (and other mutations that it suggests) on the surface properties of γ-crystallins. Since the cataract is dominant, it should also be possible to generate a transgenic animal model of this human cataract, which may provide a useful system for testing strategies for cataract prevention. Because protein aggregation is a feature of several neurological diseases (18), insights gained from studies of cataracts such as this one may have wide applicability.
Acknowledgments
We are grateful to family members for their participation. We thank Darryl Leja for his assistance in making the figures and David Walton for his help with patient histories.
ABBREVIATIONS
- γD
γD-crystallin
- lod
logarithm of odds
- SSCP
single-strand conformational polymorphism
References
- 1.Hejtmancik J F. Am J Hum Genet. 1998;62:520–525. doi: 10.1086/301774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Litt M, Kramer P, LaMorticella D M, Murphey W, Lovrein E W, Weleber R G. Hum Mol Genet. 1998;7:471–474. doi: 10.1093/hmg/7.3.471. [DOI] [PubMed] [Google Scholar]
- 3.Litt M, Carrero-Valenzuela R, La Morticella D M, Schultz D W, Mitchell T N, Kramer P, Maumenee I H. Hum Mol Genet. 1997;6:665–668. doi: 10.1093/hmg/6.5.665. [DOI] [PubMed] [Google Scholar]
- 4.Brakenhoff R H, Henskens H A, van Rossum M W, Lubsen N H, Schoenmakers J G. Hum Mol Genet. 1994;3:279–283. doi: 10.1093/hmg/3.2.279. [DOI] [PubMed] [Google Scholar]
- 5.Shiels A, Mackay D, Ionides A, Berry V, Moore A, Bhattacharya S. Am J Hum Genet. 1998;62:526–532. doi: 10.1086/301762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryant S H, Lawrence C E. Proteins. 1993;16:92–113. doi: 10.1002/prot.340160110. [DOI] [PubMed] [Google Scholar]
- 7.Chirgadze Y, Nevskaya N, Vernoslava E, Nikonov S, Sergeev Y, Brazhnikov E, Fomenkova N, Lunin V, Urzhumtsev A. Exp Eye Res. 1991;53:295–304. doi: 10.1016/0014-4835(91)90233-5. [DOI] [PubMed] [Google Scholar]
- 8.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 9.Harding J J, Crabbe M J C. In: The Eye. Davson H, editor. 1B. New York: Academic; 1984. pp. 207–492. [Google Scholar]
- 10.Benedek G B. Invest Ophthalmol Visual Sci. 1997;38:1911–1921. [PubMed] [Google Scholar]
- 11.Brakenhoff R H, Aarts H J, Reek F H, Lubsen N H, Schoenmakers J G. J Mol Biol. 1990;216:519–532. doi: 10.1016/0022-2836(90)90380-5. [DOI] [PubMed] [Google Scholar]
- 12.Russell P, Meakin S O, Hohman T C, Tsui L C, Breitman M L. Mol Cell Biol. 1987;7:3320–3323. doi: 10.1128/mcb.7.9.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blundell T, Lindley P, Miller L, Moss D, Slingsby C, Tickle I, Trunell B, Wistow G. Nature (London) 1981;289:771–777. doi: 10.1038/289771a0. [DOI] [PubMed] [Google Scholar]
- 14.Wistow G, Turnell B, Summers L, Slingsby C, Moss D, Miller L, Lindley P, Blundell T. J Mol Biol. 1983;170:175–202. doi: 10.1016/s0022-2836(83)80232-0. [DOI] [PubMed] [Google Scholar]
- 15.Wistow G, editor. Molecular Biology and Evolution of Crystallins: Gene Recruitment and Multifunctional Proteins in the Eye Lens. Austin, TX: R. G. Landes; 1995. [Google Scholar]
- 16.Sergeev Y V, Chirgadze Y N, Mylvaganam S E, Driessen H, Slingsby C, Blundell T L. Proteins. 1988;4:137–147. doi: 10.1002/prot.340040207. [DOI] [PubMed] [Google Scholar]
- 17.Siezen R J, Thomson J A, Kaplan E D, Benedek G B. Proc Natl Acad Sci USA. 1987;84:6088–6092. doi: 10.1073/pnas.84.17.6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mezey E, Dehejia A, Harta G, Papp M I, Polymeropoulos M H, Brownstein M J. Nat Med. 1998;4:755–757. doi: 10.1038/nm0798-755. [DOI] [PubMed] [Google Scholar]