Abstract
Holocentric chromosomes assemble kinetochores along their length instead of at a focused spot. The elongated expanse of an individual holocentric kinetochore and its potential flexibility heighten the risk of stable attachment to microtubules from both poles of the mitotic spindle (merotelic attachment), and hence aberrant segregation of chromosomes. Little is known about the mechanisms that holocentric species have evolved to avoid this type of error. Our studies of the influence of KLP-19, an essential microtubule motor, on the behavior of holocentric Caenorhabditis elegans chromosomes suggest that it has a major role in combating merotelic attachments. Depletion of KLP-19, which associates with nonkinetochore chromatin, allows aberrant poleward chromosome motion during prometaphase, misalignment of holocentric kinetochores, and multiple anaphase chromosome bridges in all mitotic divisions. Time-lapse movies of GFP-labeled mono- and bipolar spindles demonstrate that KLP-19 generates a force on relatively stiff holocentric chromosomes that pushes them away from poles. We hypothesize that this polar ejection force minimizes merotelic misattachment by maintaining a constant tension on pole–kinetochore connections throughout prometaphase, tension that compels sister kinetochores to face directly toward opposite poles.
Keywords: chromokinesin; kinetochore; merotelic; congression; kinesin
Introduction
Equal distribution of genetic material during nuclear division in eukaryotic cells depends on the precise organization of chromosomes in the mitotic spindle before the actual act of chromatid separation and segregation (for reviews see Nicklas, 1997; Rieder and Salmon, 1998; McIntosh et al., 2002). At the beginning of prometaphase, spindle microtubules interact with chromosomes to initiate congression movements that accomplish that organization. In mitotic animal cells, spindle microtubules emanate in aster-shaped arrays from two microtubule-organizing centers that comprise the poles of the spindle. The minus-ends of microtubules are at or near the poles, whereas dynamic plus-ends extend outward, away from the poles. Plus-ends and microtubule walls can interact with chromosomes to move them toward or away from each pole via polymer-based ratcheting or by the action of plus- and minus-end–directed motor proteins. During prometaphase, such radial movements, biased away from the poles, move chromosomes to a plane at the equator known as the metaphase plate. Accompanying this “congression,” duplicated chromosomes become “oriented” such that each chromatid of a back-to-back sister chromatid pair makes stable attachments with the plus-ends of microtubules from just one spindle pole. Those plus-end interactions are mediated by kinetochores, complex protein structures that assemble on centromeric DNA. Proper orientation (amphitelic) ensures that when sister chromatids release one another at the start of anaphase, they are pulled by their kinetochore microtubules in opposite directions, segregating to form two identical nuclei.
Three forms of misorientation are known to occur. First, a failure of plus-end attachment to one or both kinetochores of a pair prevents the generation of opposing pulling forces, and thus segregation cannot occur. However, the kinetochore-dependent spindle checkpoint, sensing a lack of plus-end occupancy and/or bipolar tension across the kinetochore pair, commonly delays anaphase until the problem is corrected (Li and Nicklas, 1995; Waters et al., 1998; for reviews see Nicklas, 1997; Rieder and Salmon, 1998; McIntosh et al., 2002). Second, sister kinetochores can capture microtubules from the same pole (syntelic orientation), causing them to segregate together. This form of misorientation too is detected by the checkpoint, perhaps because of a lack of bipolar tension across the sister kinetochores. In the third case, merotelic orientation, a single kinetochore captures microtubules from both poles, resulting in a tug-of-war that often leaves the chromosome lagging at the equator during anaphase. Merotelic orientation causes little or no delay in the onset of anaphase, perhaps because the spindle checkpoint cannot distinguish aberrant bipolar tension on a single kinetochore from correct bipolar tension across a back-to-back kinetochore pair. Hence, merotelism is a serious source of chromosome segregation errors (Cimini et al., 2001, 2002; Kline-Smith et al., 2004).
To gain insight into mechanisms of chromosome movement and their relationship to orientation, we have used various function disruption and light microscopy approaches to study mitosis in Caenorhabditis elegans. Among the commonly studied model systems, the chromosomes of C. elegans are unique in that they are holocentric and thus have kinetochores that extend along the entire length of each chromatid (Albertson and Thomson, 1982; Dernburg, 2001; Moore and Roth, 2001). Holocentric chromosomes are also found in many less commonly studied organisms (for review see Lima-de-Faria, 1949). As discussed eloquently by Nicklas (1997), monocentric chromosomes reduce the probability of misorientation by assembling discrete disc-shaped sister kinetochores back-to-back, each in a pit-shaped depression. Chromatin surrounding the depression restricts access by microtubules, favoring contact and stable attachment only with those that approach from the front. After initial random attachment of one kinetochore to a pole, tension on that connection encourages it to face that pole and thus rotates its sister to face the other. This minimizes the likelihood that either kinetochore will make or maintain aberrant microtubule contacts (Nicklas, 1997).
In principle, the elongated kinetochores of holocentric chromosomes should have a high risk of misorientation. They present a large target for microtubule plus ends. Also, twisting or bending of chromatids could allow distant parts of individual kinetochores to face in opposite directions. C. elegans appears to have solved the twisting problem by making its chromosomes relatively stiff, via HCP-6–dependent condensation before kinetochores interact with microtubules (Stear and Roth, 2002). However, the stiffness does not solve the oversized plus-end target. Compounding this problem in C. elegans is the fact that the elongated kinetochore actually protrudes from the chromosome surface rather than being recessed (Albertson and Thomson, 1982; O'Toole et al., 2003). Perhaps the elongated, exposed architecture evolved to speed the capture of microtubules and thus facilitate a fast pace of mitosis. However, the range of angles for microtubule–kinetochore attachment should be quite wide, being blocked only from the rear. Even modest rotation of the sister kinetochore axis away from alignment with the pole–pole axis would invite merotelic microtubule attachments. Clearly, holocentric chromosomes in general and C. elegans chromosomes in particular face kinetochore orientation challenges that are exaggerated relative to monocentric chromosomes, suggesting that holocentrics offer new insights into mechanisms designed to prevent misorientation.
Here, we report an analysis of C. elegans mitotic chromosome behavior centered around KLP-19, whose sequence is related to plus-end microtubule motors of the kinesin-4 family. KLP-19 has a dynamic relationship with the spindle during mitosis. It accumulates around chromosomes in prometaphase and metaphase and becomes concentrated in the spindle interzone during anaphase. Depletion of KLP-19 allows aberrant poleward (P) chromosome motions during prometaphase, misorientation of kinetochores, and dramatic anaphase chromatin bridges. Analysis of chromosome movements in bipolar and monopolar spindles suggests that there are two stages of prometaphase chromosome congression: an early stage during which a polar ejection force immediately pushes chromosomes antipoleward (AP) and toward the equator, and then a second stage in which a KLP-19–dependent polar exclusion force competes with kinetochore-driven P forces to hold chromosomes near the metaphase plate. We suggest that the KLP-19 polar ejection force maintains constant tension on pole–kinetochore connections to rotate the sister kinetochore axis onto the pole–pole axis, minimizing merotelic connections by forcing sister kinetochores to always face directly toward opposite spindle poles.
Results
KLP-19 sequence and predicted structure
Polar exclusion force generation has been demonstrated most directly for vertebrate Kid (Levesque and Compton, 2001), a member of the kinesin-10 family (Lawrence et al., 2004). Similar forces may be produced by some members of the kinesin-4/chromokinesin family (e.g., Vernos et al., 1995; Kwon et al., 2004; and references therein). A clear Kid homologue has not been identified in C. elegans. However, there are two chromokinesin-like genes: klp-12 (GenBank/EMBL/DDBJ accession no. Z92811) and klp-19 (GenBank/EMBL/DDBJ accession no. AL021481). KLP-19 has an NH2-terminal motor domain, whereas KLP-12 has an internal motor domain. Both have conserved neck residues consistent with plus-end–directed motion (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200403036/DC1). One recent phylogenetic analysis of kinesin motor domains (∼340 amino acids) placed KLP-19 in a divergent chromokinesin clade with Kid (Lawrence et al., 2002) and placed KLP-12 with the classic chromokinesins (kinesin-4 family; e.g., mammalian Kif4, Xenopus laevis Xklp1, and Drosophila melanogaster KLP3A). Another analysis grouped both KLP-19 and KLP-12 with the classic chromokinesins and left Kid as an orphan kinesin (Dagenbach and Endow, 2004). The sequence of the neck region of kinesins (∼40 amino acids), which is a key element in force transduction, may provide insights into class-specific relatedness (Vale and Fletterick, 1997; Vale, 2003). Within the neck region, KLP-19 has 41% identity with Kif4 and 44% identity with KLP3A; KLP-12 shows 29 and 38% identity with Kif4 and KLP3A, respectively. The KLP-19 and -12 neck regions are less similar to that of Kid (18% identical; Fig. S1). This finding reinforces the idea that KLP-19 is a Kif4-like plus-end motor.
Studies of various chromokinesins using a variety of approaches have produced a surprising mix of function predictions, including roles in microtubule dynamics, spindle assembly, metaphase chromosome alignment, spindle pole separation, cytokinesis, and nonmitotic neuronal vesicle transport (e.g., Theurkauf and Hawley, 1992; Sekine et al., 1994; Vernos et al., 1995; Williams et al., 1995; Heald, 2000; Antonio et al., 2000; Funabiki and Murray, 2000; Peretti et al., 2000; Levesque and Compton, 2001; Goshima and Vale, 2003; Bringmann et al., 2004; Kwon et al., 2004). A genome-wide RNAi screen for microtubule motor functions (Powers et al., 1998; Segbert et al., 2003) suggested that KLP-19 was essential whereas KLP-12 was not. Therefore, our analysis of chromosome movement and orientation focused mainly on KLP-19.
KLP-19 concentrates around chromosomes and in the spindle
Some kinesins have been shown to interact with chromosomes via DNA-binding motifs in their stalk regions (Afshar et al., 1995; Wang and Adler, 1995; Tokai et al., 1996). Others have stalk sequences consistent with DNA binding, but direct binding has not yet been demonstrated (Vernos and Karsenti, 1995). Analysis of KLP-19 cDNAs and relevant genomic sequences failed to reveal a recognizable DNA-binding motif. To gain insight into possible chromosome and other associations, KLP-19 distribution was studied with antibodies raised against a nonconserved COOH-terminal peptide. Strong antibody staining was seen primarily in areas with dividing cells; i.e., the gonad and embryos (Fig. 1). In the gonad, staining was bright in germline nuclei of the distal mitotic zone, dim in early meiotic prophase nuclei, and then bright again in late prophase nucleoplasm. Just before fertilization, KLP-19 concentrated on prophase chromosomes. During metaphase of meiosis I, staining concentrated slightly in the body of the spindle, more around the periphery of chromosomes, and most between homologous chromosomes (Fig. 1 D). A similar pattern was seen in meiosis II. In embryos, during most of mitotic prophase, KLP-19 concentrated in nucleoplasm (Fig. 2 A). In prometaphase, it concentrated slightly in the body of the spindle and more strongly around the periphery of chromosomes (Fig. 2, B and C). In anaphase, it left chromosomes and concentrated in the spindle interzone (Fig. 2, D and E). These dynamic patterns predict that KLP-19 functions in meiotic and mitotic spindles and suggest that, despite the lack of a recognizable DNA-binding motif, it can associate with chromosomes during prometaphase congression.
Figure 1.
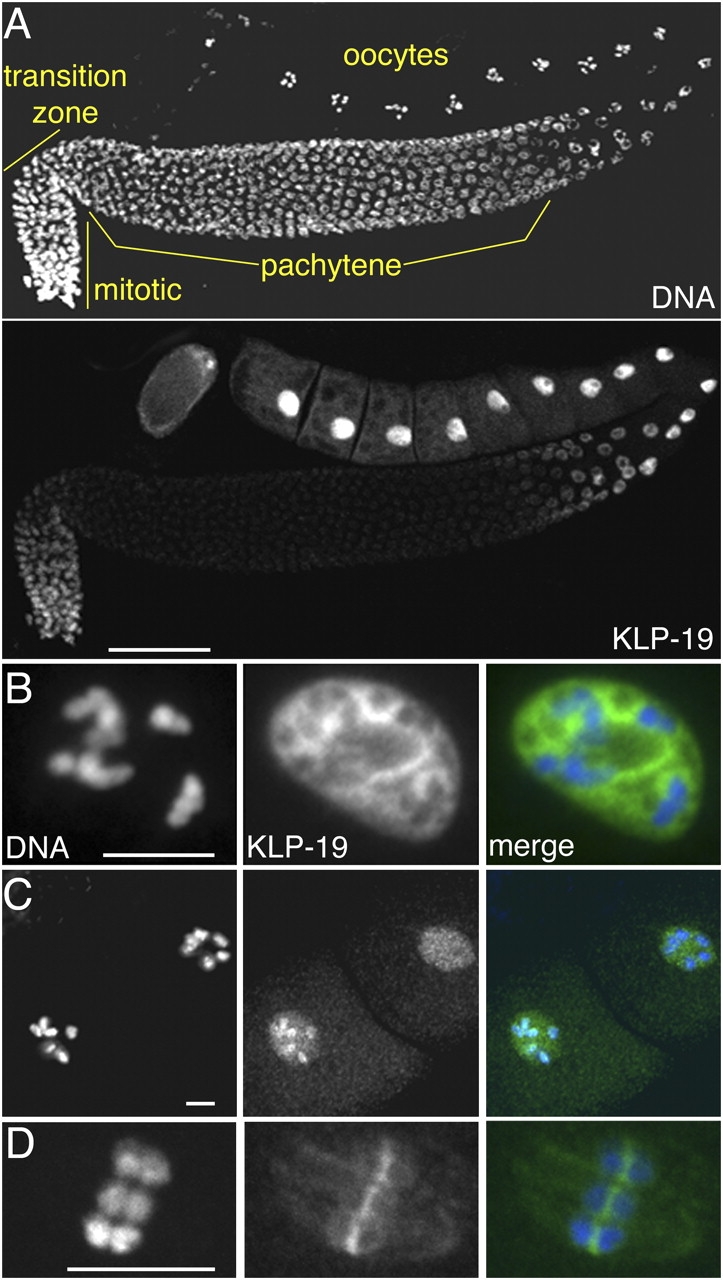
KLP-19 localization in the germline. (A) Hermaphrodite gonad arm, showing DNA (top) and anti–KLP-19 staining (bottom). Germ nuclei undergo mitosis in the distal tip of the gonad, exit mitosis and enter meiosis in the transition zone, and then progress through prophase I in the remainder of the gonad. KLP-19 accumulates in distal and late prophase nuclei. (B) High magnification image of the nucleus of a single immature oocyte, showing KLP-19 in the nucleoplasm. (C) Two oocytes approaching the spermatheca (to the left), where fertilization occurs. KLP-19 became concentrated on chromosomes just before fertilization. (D) Female meiosis I metaphase spindle in a newly fertilized embryo. KLP-19 is concentrated most between homologues. In merged panels, DAPI is blue and anti–KLP-19 is green. Bars: (A) 50 μm; (B–D) 5 μm.
Figure 2.
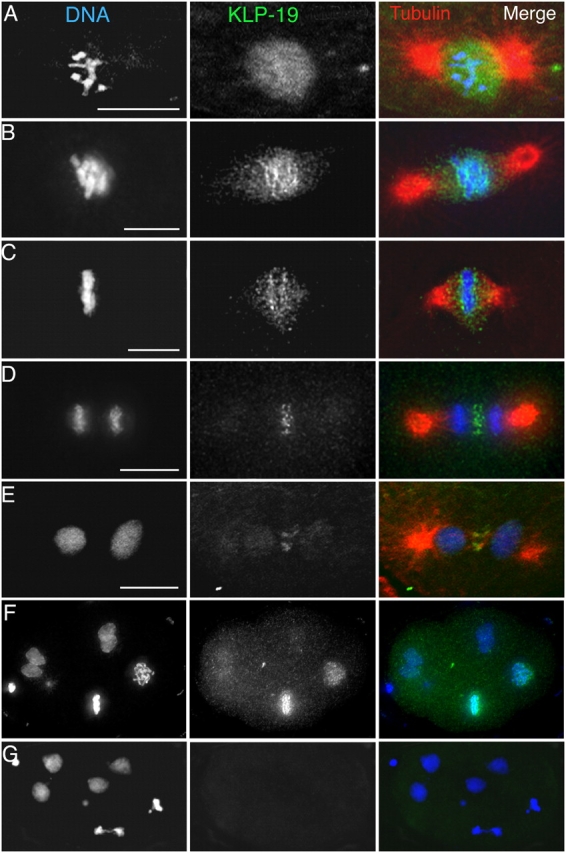
KLP-19 localization during mitosis in early embryonic blastomeres. Embryos isolated from adult worms were fixed and stained with DAPI (DNA, blue), antitubulin (red), and anti–KLP-19 (green). (A) Prophase. (B) Prometaphase. (C) Metaphase. (D) Anaphase. (E) Telophase. KLP-19 in the nucleoplasm (A) became concentrated along the edges of chromosomes during prometaphase and metaphase (B and C) and in the spindle interzone during anaphase and telophase (D and E). Bars, 5 μm. (F) A multicellular wild-type embryo. (G) A multicellular klp-19(RNAi) embryo. The absence of detectable KLP-19 staining after RNAi affirmed the specificity of the anti–KLP-19 antibody and the effectiveness of RNAi. In panels F and G and for whole embryos in other figures, C. elegans embryos are ∼50 μm in length.
Chromosome association of KLP-19 is independent of kinetochores
The concentration of KLP-19 at the edges of condensed chromosomes was similar to that of MCAK (Fig. 3), a protein known to associate with the holocentric C. elegans kinetochore as well as with spindle poles (Oegema et al., 2001). To determine if KLP-19 chromosome localization depends on kinetochores, the effects of kinetochore disruption were studied. The hcp-3 gene encodes C. elegans CENP-A, a histone H3-like protein required for recruiting C. elegans kinetochore proteins (Oegema et al., 2001; Desai et al., 2003). Depletion of HCP-3 by RNAi causes a “kinetochore null” phenotype: mitotic chromosomes appear normal during prophase, but in prometaphase they form two or occasionally more spherical clusters near the spindle equator and do not segregate during anaphase (Oegema et al., 2001). In hcp-3(RNAi) embryos, MCAK is present at poles but is absent from chromosomes (Fig. 3 C; Oegema et al., 2001). In contrast, KLP-19 remained concentrated around individual chromosomes (not depicted) and the spherical chromosome clusters (Fig. 3 D) as well as between chromosomes and poles. These results indicate that KLP-19 spindle localization is independent of kinetochores and suggest that its association with chromosomes is via nonkinetochore chromatin.
Figure 3.

KLP-19 relationship to C. elegans kinetochores. (A) In a metaphase blastomere, MCAK, a kinetochore protein, and KLP-19 are seen concentrated on the P edges of chromosomes. (B) In an anaphase blastomere, MCAK remained on chromosomes, whereas KLP-19 concentrated in the spindle interzone. Bar, 5 μm. (C) After disruption of kinetochores by RNAi depletion of HCP-3 (CENP-A), association of MCAK with chromosomes was not detected. (D) KLP-19 localization around chromosomes was not prevented by HCP-3 depletion. Color panels show merged images with colors as indicated by labeling.
KLP-19 is required for proper mitotic chromosome behavior
To study the effects of a loss of KLP-19 function, both genetic and RNAi approaches were used. A recessive lethal mutation, klp-19(bn126), was identified in a PCR-deletion screen. The mutation is an in-frame deletion beginning in α-helix 4 of the motor domain and ending near the carboxyl limit of the neck region (Fig. S1). Homozygous mutant klp-19 progeny from heterozygous hermaphrodite parents arrested as L1 larvae, verifying that KLP-19 is essential and suggesting that inherited maternal gene products are sufficient for embryogenesis.
To test for function in embryos, we used gene-specific RNAi to deplete KLP-19 from wild-type hermaphrodite germlines. Embryos from klp-19(RNAi) worms lacked detectable KLP-19 immunofluorescence (Fig. 2 G), indicating that the RNAi approach was effective. RNAi embryos underwent early mitotic divisions but later displayed aberrant patterns of nuclei and arrested before morphogenesis. To test for defects in spindle architecture like those caused by inhibition of KLP3A and Xklp1 (Vernos et al., 1995; Kwon et al., 2004), microtubule patterns were studied with antitubulin immunofluorescence in fixed embryos (Fig. 4 A) or with GFP::β-tubulin fluorescence in live embryos (not depicted). The bipolar organization of spindle microtubules and their dynamic progression through mitosis appeared normal. Spindle pole separation in anaphase was normal, and no failures in cytokinesis or aberrant cleavage patterns were observed.
Figure 4.
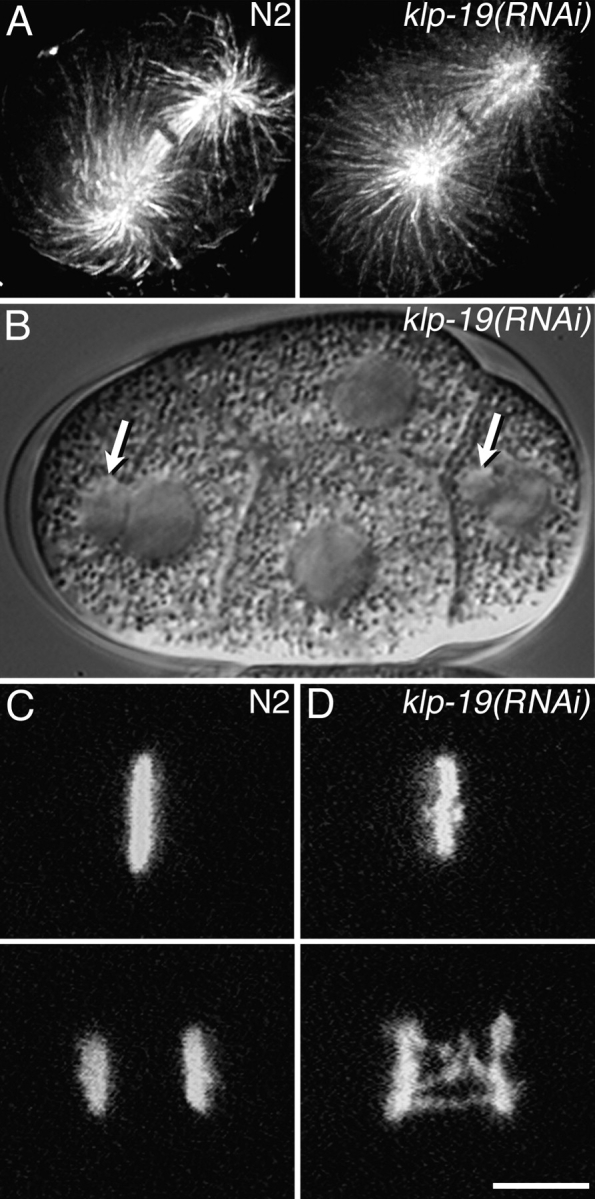
KLP-19 depletion causes chromosome segregation defects. (A) Antitubulin staining of fixed wild-type (N2) and klp-19(RNAi) one-cell embryos. Depletion of KLP-19 did not noticeably alter mitotic spindle structure. (B) A Nomarski DIC image of a live klp-19(RNAi) four-cell embryo. Micronuclei (arrows) are visible in two cells. (C and D) Time-lapse confocal images of chromosomes in live one-cell embryos expressing GFP::histone. (C) A wild-type (N2) embryo in metaphase (top) and then in anaphase (bottom; see Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200403036/DC1). (D) A klp-19(RNAi) embryo showing a disordered metaphase plate (top) and then lagging anaphase chromatin (bottom; see Video 2). Bar, 5 μm.
The formation of micronuclei in some klp-19(RNAi) embryos (Fig. 4 B) suggested defects in chromosome segregation. To investigate mitotic chromosome behavior directly, embryos expressing a GFP::histone fusion protein were imaged by time-lapse confocal microscopy (Fig. 4, C and D; see Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200403036/DC1). In prophase, during condensation, chromosomes often appeared bent. However, after prophase, bending was slight and rarely seen, consistent with the findings of Stear and Roth (2002), which suggested that proper condensation limits chromosome flexibility. Stiff rod-like behavior was observed for chromosomes in klp-19(RNAi) as well as wild-type embryos. In every klp-19(RNAi) mitosis observed, chromosomes formed a slightly disordered metaphase plate, and then multiple chromosomes lagged, forming bridges during anaphase. The chromosome bridges stretched, often appeared to break, and sometimes formed micronuclei. These anaphase defects are similar to those reported for vertebrate cells that enter anaphase with individual kinetochores pulled in two directions because of merotelic misorientation (Cimini et al., 2003).
To address the question of kinetochore orientation, fixed embryos were stained with anti-MCAK and observed by deconvolution microscopy (Fig. 5). In wild-type anaphase spindles, holocentric kinetochores were on the P sides of the chromosomes, aligned perpendicular to the spindle axis. In klp-19(RNAi) anaphase, kinetochores were not well aligned and some were stretched across the interzone, parallel to the pole–pole axis, suggesting that single chromatids were subjected to opposing anaphase forces. This finding is consistent with merotelic kinetochore misattachment.
Figure 5.
Kinetochore misorientation with lagging chromatin. (A) Wild-type (N2) and B) klp-19(RNAi) multicellular embryos were fixed and stained with anti-MCAK, which binds kinetochores and spindle poles, and with DAPI for DNA. Single optical sections generated by deconvolution microscopy of anaphase blastomeres are shown. The arrow in B indicates a kinetochore that lies along lagging chromatin that was stretched between two recently separated groups of anaphase chromosomes. Other kinetochores in B are also misoriented. Colors in the merge panels reflect the colors of the labels. Bar, 5 μm.
We considered several alternative explanations for disordered metaphase plates and anaphase bridges. Chromatids might fail to disjoin properly because of aberrant chromosome condensation, as seen in studies of SMC-1, a C. elegans condensin (Hagstrom et al., 2002), and of HCP-6 (Stear and Roth, 2002). SMC-1 depletion allows normal homologous chromosome segregation in meiosis I and causes defects in sister chromatid segregation in meiosis II. KLP-19 depletion causes anaphase bridges in both meiosis I and II. SMC-1 and HCP-6 depletion both cause a striking impairment of chromosome condensation during prophase. KLP-19 depletion does not noticeably alter chromatin condensation during prophase nor chromosome flexibility during the rest of mitosis (compare Videos 1 and 2).
We also considered and tested the possibility that chromosome bridges in embryos were due to earlier chromatin damage from missegregation in the germline during the RNAi treatment. In mei-1 mutant embryos, female meiosis fails, occasionally resulting in the complete absence of an oocyte-derived pronucleus and a subsequent haploid mitosis involving solely the paternal chromosomes (Mains et al., 1990). By subjecting mei-1 hermaphrodites to klp-19 RNAi after sperm formation was complete, we were able to study the mitotic behavior of “naive” paternal chromosomes in KLP-19–depleted embryos. In three embryos that excluded all oocyte-derived chromatin, the male pronucleus entered mitosis, produced disordered metaphase plates, and then showed lagging/stretched chromosomes in anaphase (Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200403036/DC1). This result confirms that KLP-19 is needed directly in the embryo to prevent chromosome misorientation and anaphase bridges.
Work with cultured mammalian cells has shown that bridging can be caused by premature entry into anaphase, before random merotelic attachments are corrected (Cimini et al., 2003). To determine if this was the basis of klp-19(RNAi) chromatin bridges, the start of anaphase was timed relative to the first clear prometaphase chromosome movement (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200403036/DC1). In wild-type embryos, the prometaphase–anaphase period was remarkably short (P0 = 189 ± 16 s, P1 = 148 ± 18 s). KLP-19 depletion caused no significant change (P0 = 228 ± 36 s, P1 = 142 ± 11 s), indicating that precocious anaphase was not the cause of merotelism and chromosome bridging.
KLP-19 resists P forces on prometaphase chromosomes
To gain insight into how KLP-19 influences chromosome behavior, chromosome movement relative to spindle poles was analyzed in embryos expressing GFP::histone and GFP::γ-tubulin (Fig. 6 and Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200403036/DC1). In wild-type embryos, the end of prophase and start of prometaphase was marked by immediate movement of chromosomes AP and toward the equator (Fig. 6 A, 0–81 s). This initial polar ejection, or “early congression,” which lasted an average of 30 s, often moved chromosomes to the periphery of the equator where they briefly formed a loose ring. This is consistent with AP polar ejection forces radiating outward from each pole. When opposing AP forces from two poles combine on a chromosome that is off the direct spindle axis, a resultant force vector will be directed away from the axis and toward the periphery (Ostergren et al., 1960; Rieder et al., 1986).
Figure 6.
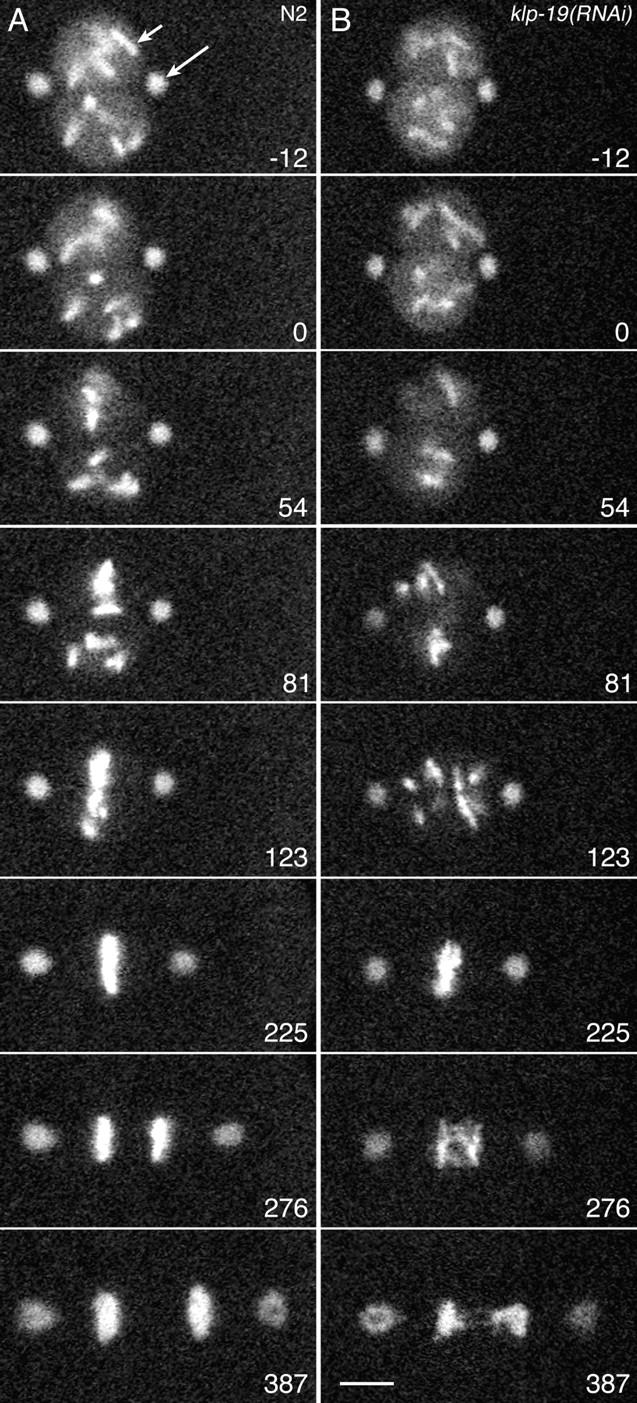
KLP-19 influences prometaphase congression and anaphase segregation of chromosomes. Images are single confocal optical sections from time-lapse movies of GFP::γ-tubulin (spindle poles, long arrow) and GFP::histone (chromosomes, short arrow) in the first mitotic divisions of (A) wild-type (N2) and (B) KLP-19–depleted embryos. Numbers represent seconds before or after the end of prophase (0), selected as the last frame before directed movement of at least one chromosome was observed. Early AP chromosome movements toward the equator were not substantially affected by KLP-19 depletion (A and B, 0–81 s). In wild type, short P and AP movements completed congression (A, 81–225 s). In contrast, the first P movements in KLP-19–depleted spindles were usually substantial, scattering chromosomes widely between the poles (B, 81–123 s). However, subsequent bidirectional oscillations accomplished congression to a disordered metaphase plate, and then anaphase resulted in multiple lagging strands of chromatin (B, 276–387 s). See Videos 4 and 5, available at http://www.jcb.org/cgi/content/full/jcb.200403036/DC1. Bar, 5 μm.
The first evidence of P force generation was either a short, fast chromosome movement directly toward one centrosome, which suggested microtubule attachment and P force generation by one kinetochore (mono-orientation), or slower movement of chromosomes from the periphery of the equator toward the spindle axis, which suggested microtubule attachment and opposing P force generation by both kinetochores (bi-orientation). Chromosomes remained fairly close to the equator, and short-range oscillations completed congression to a thin, well-defined metaphase plate (Fig. 6 A, 225 s). Segregation was accomplished by anaphase B pole separation with no sign of chromosome-to-pole movement, confirming a previous report that anaphase A does not occur in C. elegans embryos (Oegema et al., 2001).
After klp-19(RNAi) by feeding, chromosomes in most embryos showed normal early AP movement (Fig. 6 B, 0–81 s; and Video 5, available at http://www.jcb.org/cgi/content/full/jcb.200403036/DC1), although it occasionally appeared less robust. After that early congression, multiple chromosomes scattered dramatically back toward the poles (Fig. 6 B, 123 s), while a few remained near the equator and displayed normal, short-range bipolar oscillations. Shortly after scattering, the displaced chromosomes engaged in bipolar oscillations, recongressed to a loose metaphase plate, and engaged in anaphase B with normal timing (Table S1). The P scattering suggests that KLP-19 creates an AP force that resists the P movement of chromosomes that first attach to microtubules from just one pole. The post-scattering recongression may reflect attachment to microtubules from the second pole, followed by kinetochore-driven P movements in both directions. The recongression, although imperfect, suggests that chromosomes can find the equator with little input from the putative KLP-19 polar ejection force.
Early AP movement requires microtubules but only minor chromokinesin contributions
The minimal effects of klp-19(RNAi) by feeding on early AP chromosome movement raised questions about the mechanism of early AP force generation. To test the assumption that microtubules are involved, partially flattened wild-type P0 embryos were treated with 20 μg/ml nocodazole (Encalada, S., and B. Bowerman, personal communication; Strome and Wood, 1983) and imaged by time-lapse microscopy. At that concentration, prophase chromosome condensation and centrosome separation appeared normal and centrosomes maintained their separation after nuclear envelope breakdown (Video 6, available at http://www.jcb.org/cgi/content/full/jcb.200403036/DC1). Chromosomes did show small oscillations and a gradual drift toward the equator, but early AP congression and subsequent active movements were not evident. This confirms that microtubules are critical for early congression as well as the rest of mitosis.
To determine if kinetochores contribute force for the early movement, kinetochore assembly was disrupted by RNAi depletion of HCP-3. Chromosomes showed normal early AP motion, congressing into two or more clusters, usually near the periphery of the equator. However, subsequent chromosome movements were rare (Video 7, available at http://www.jcb.org/cgi/content/full/jcb.200403036/DC1), and chromosomes were left at the equator during anaphase, as expected for kinetochore disruption (Oegema et al., 2001; Desai et al., 2003). This result indicates that, although kinetochores are critical for P oscillation movements later in prometaphase, they are not needed for early AP chromosome movement.
To determine if KLP-12, the other chromokinesin, might contribute to early chromosome congression, either alone or redundantly with KLP-19, we studied the effects of single and double RNAi. To maximize depletion, RNAi was done by injecting young hermaphrodites with high-concentration double-strand RNA. Effects on early AP congression were quantified by measuring chromosome-to-pole distances in half-spindles at 24 s after nuclear envelope breakdown (wild type = 5.96 ± 0.18 μm, n = 48). Injection of klp-12 dsRNA alone did not have a significant effect on the early congression (5.64 ± 0.24 μm, n = 29) or on subsequent chromosome behavior. Injecting klp-12 and klp-19 RNA together (1/2 concentration of each) generated phenotypes similar to those seen with klp-19 RNAi by feeding, but had no significant effect on early congression (5.61 ± 0.23 μm, n = 26). Injection of klp-19 dsRNA alone (full concentration) caused a small reduction in early chromosome-to-pole distance (5.28 ± 0.16 μm, n = 59) that was significant (wild-type vs. klp-19(RNAi), P = 0.011). In summary, our results suggest that microtubules are critical, that kinetochores and KLP-12 are not required, and that KLP-19 serves a minor role in early AP chromosome motion, perhaps redundant with forces generated by microtubule polymerization-based ratcheting.
Chromosome movement in monopolar spindles
In bipolar spindles, a rigorous determination of whether a given chromosome movement is driven by AP or P forces is confounded by the presence of two poles; movements that are AP relative to one pole are P relative to the other. Interpretations that predict AP force production by chromokinesins have usually relied on observation of defective metaphase chromosome positioning in bipolar spindles after inhibition of motor function. However, similar positioning/congression defects can be caused by inhibition of a variety of proteins that are not microtubule motors, including EAST (a “nucleoskeletal” component), Nup358 (a nuclear pore component), and centromeric MCAK (Chang et al., 2003; Salina et al., 2003; Wasser and Chia, 2003; Kline-Smith et al., 2004). This uncertainty, in light of the diverse functions reported for Kif4-like chromokinesins, highlighted the need for a more critical test of whether or not KLP-19 produces AP forces in our system.
To distinguish AP versus P forces, we studied chromosome movements in embryos with monopolar spindles, a strategy based on tests of Kid function in cultured human cells by Levesque and Compton (2001). Embryos from zyg-1 mutant hermaphrodites are defective in centrosome duplication (O'Connell et al., 2001). We used a conditional allele of zyg-1 and a temperature-shift regime that produced a bipolar spindle in P0 and a monopolar spindle in P1, AB, and later blastomeres. Using time-lapse microscopy of zyg-1 mutant embryos containing GFP::γ-tubulin and GFP::histone, we determined that chromosome behavior and anaphase timing in P0 were similar to wild type (compare Videos 4 and 8, available at http://www.jcb.org/cgi/content/full/jcb.200403036/DC1; Table S1). In P1 and AB, chromosomes congressed to a normal-appearing metaphase plate on one side of the single spindle pole and then failed in anaphase segregation (Fig. 7 A and Videos 8 and 9, available at http://www.jcb.org/cgi/content/full/jcb.200403036/DC1). The pole–chromosome axis in AB was parallel to the plane of view and remained relatively stationary. In P1, the spindle axis often shifted and rotated unpredictably, so most monopolar observations were focused on AB. At the prophase–prometaphase transition, early AP motion cleared chromosomes away from the single pole, pushing them into the distal half of the remnant nucleus (Fig. 7 A, 0–70 s). Subsequently, chromosomes oscillated toward and away from the single pole for an extended period, eventually producing a metaphase plate with a mean pole–chromosome distance of 6.66 ± 0.43 μm (Table S1). The start of anaphase, which could be recognized by a small split in the metaphase plate, was delayed approximately twofold (Table S1). The delay may reflect prolonged activation of the spindle checkpoint by distal kinetochores that lack microtubules and tension (for reviews see Nicklas, 1997; Rieder and Salmon, 1998; McIntosh et al., 2002). No substantial P or AP movement of anaphase chromosomes was observed after the split. The lack of P migration by the proximal chromosomes is consistent with a lack of anaphase A in C. elegans, as noted above for bipolar spindles.
Figure 7.
KLP-19 generates polar exclusion forces in monopolar spindles. Single optical sections of monopolar AB spindles in zyg-1 mutant embryos containing GFP::γ-tubulin (centrosomes) and GFP::histone (chromosomes). Spindles in a control zyg-1 embryo (A) and a zyg-1 embryo depleted of KLP-19 (B) are shown from prophase to metaphase with times after the end of prophase in seconds. In both spindles, chromosomes initially moved AP (0–70 s). In the control, subsequent oscillations toward and away from the pole (70–290 s) drove congression to a well-ordered metaphase plate. In the KLP-19–depleted spindle, after the initial AP movement, chromosomes moved back toward the pole (180 s) and failed to form an ordered metaphase plate (290 s). See Videos 8 and 9, available at http://www.jcb.org/cgi/content/full/jcb.200403036/DC1. Bar, 5 μm.
To determine if KLP-19 generates AP forces on chromosomes, zyg-1 hermaphrodites were subjected to klp-19(RNAi) by feeding. The formation of extensive anaphase chromatin bridges in P0 (bipolar) confirmed that depletion had been effective in each embryo studied (Video 8). In the next mitotic cycle, early AP forces cleared chromosomes away from the single pole (Fig. 7 B, 0–70 s; and Videos 8 and 9), but then chromosomes immediately moved back P to form a disordered group spaced 4.17 ± 0.65 μm from the pole (Table S1). They remained there, and additional movements were insubstantial. These results confirm that KLP-19 produces an AP force on chromosomes, which is needed to resist P forces that are produced by kinetochores following the early AP congression movement. The fact that chromosomes did not move all the way to the pole may be due to steric hindrance by a high density of microtubules near the centrosome (Rieder and Salmon, 1998; O'Toole et al., 2003) or it may reflect a down-regulation of P force production as kinetochores approach the pole.
Discussion
Microtubule-based movements of chromosomes have been studied intensively for many years, and complete sets of microtubule motors have been identified in several organisms through genome sequencing. Nevertheless, our understanding of how individual motors contribute to spindle function remains sparse, particularly for higher eukaryotes. We have focused here on the contributions of C. elegans KLP-19 to the movement and orientation of holocentric prometaphase chromosomes. Depletion of KLP-19 from wild-type embryos leads to abnormal congression movements, slightly disordered metaphase plates, and anaphase segregation defects, evident as chromatin bridges. The bridges and the appearance of holocentric kinetochores aligned parallel to the spindle axis, rather than perpendicular, are consistent with merotelic attachment. Analysis of chromosome movements in bipolar and monopolar spindles suggests two periods of AP force generation: a brief early period that has a strong dependence on microtubules and a minor dependence on KLP-19, and then a second period persisting until anaphase that has a major dependence on KLP-19. We suggest below that the AP polar ejection forces provided by KLP-19 combat merotelic chromatid attachments by compelling sister kinetochores to face opposite poles.
Given that chromosome movement is dependent on microtubules, AP pushing forces might be generated by tubulin polymerization at microtubule ends, by plus-end motors walking along microtubule walls, or by a combination of the two (Rieder and Salmon, 1998; McIntosh et al., 2002). Clear support for the idea that a plus-end motor can contribute to AP forces on chromosomes, poetically termed “polar winds” (Carpenter, 1991), has been provided by studies of monocentric chromosomes in human cultured cells by Levesque and Compton (2001): Kid pushes chromosome arms away from the pole in monopolar spindles. However, inhibition of Kid in bipolar spindles had surprisingly mild effects. Although chromosome arms did not align at the equator, kinetochores did, showing that polar ejection force is not necessary for their congression. Furthermore, anaphase chromosome segregation appeared normal, suggesting that kinetochore orientation was normal (Antonio et al., 2000; Funabiki and Murray, 2000; Levesque and Compton, 2001). Those observations and the lack of compelling evidence that polar ejection force is essential has left the biological purpose of polar winds uncertain (Carpenter, 1991; Marshall, 2002).
Our studies of C. elegans embryos indicate that KLP-19 generates Kid-like AP force on chromosomes, in this case with an essential purpose in combating merotelic misorientation of kinetochores. This role for polar ejection force may be highlighted in C. elegans because each holocentric, exposed kinetochore can interact with microtubules from wide angles (Albertson and Thomson, 1982; O'Toole et al., 2003). In contrast, microtubule access to a vertebrate monocentric kinetochore is thought to be restricted to a more face-on, narrow range of angles because it is shielded by surrounding nonkinetochore chromatin (Nicklas, 1997; Rieder and Salmon, 1998). The elongated, exposed kinetochore of a C. elegans chromatid may facilitate fast mitosis by efficient capture of microtubules, but the trade-off is a high probability of merotelic attachment to microtubules from both poles. One way the spindle could minimize such misattachments is to maintain a constant AP force on nonkinetochore chromatin (Fig. 8). Then, as soon as one kinetochore captures microtubules from a pole, regardless of whether or not the kinetochore is in an active P force–generating state, tension on the connection would compel that kinetochore to face directly toward that pole and its sister to face the opposite pole. Exposure of individual kinetochores to both poles would be minimized, and any merotelic misattachment of a microtubule would be strained by a bend that might make it unstable (Nicklas, 1997). Thus, rather than polar wind blowing on a chromosome as on a sailboat (Ostergren et al., 1960), with the purpose of pushing it toward the spindle equator (Rieder et al., 1986), a better analogy for chromokinesins like KLP-19 might be that of polar wind blowing on a chromosome as on a kite, with the purpose of proper orientation. By keeping constant tension on the string, wind forces a kite to face directly toward the hand of the flyer.
Figure 8.

A model for the roles of prometaphase polar exclusion forces in C. elegans embryos. Only half-spindles are shown, for simplicity. (A) At the beginning of prometaphase, kinetochores are inactive (shaded gray) and a polar exclusion force that requires microtubules, pushes chromosomes AP. That early force may be generated primarily by microtubule plus-end pushing with some help from KLP-19. (B and C) Kinetochores become active, capture microtubules, and alternate between active P force generation (short black arrows) and a neutral state. KLP-19 on chromosomes generates AP force on nonkinetochore chromatin that creates tension on the microtubule– kinetochore connection. The constant tension forces the attached kinetochore to face directly toward its pole, even when the kinetochore is not generating P forces (noted by the absence of black arrows in C). This minimizes the probability that microtubules from opposite poles will attach to the same kinetochore. A KLP-19–generated torque could also increase the probability that incorrect microtubule–kinetochore attachments are bent and thus unstable. The centrosome diameter and the pole–chromosome distances in B and C were derived from measurements of GFP-tagged centrosomes and chromosomes in bipolar spindles. The sizes and geometry for chromosomes and kinetochores were derived from the electron micrographs of Albertson and Thomson (1982).
This orientation role may also be important for polar ejection force in spindles with monocentric chromosomes. In Drosophila mitosis, which is monocentric, the chromokinesin KLP3A is known to contribute to pole–pole separation, organization of the anaphase spindle interzone, and cytokinesis (Williams et al., 1995; Kwon et al., 2004). However, it has also been noted that inhibition of KLP3A can cause congression defects and lagging anaphase chromosomes (Goshima and Vale, 2003; Kwon et al., 2004). This finding is consistent with a KLP-19–like role for KLP3A in producing AP forces that help prevent merotelic kinetochore attachment of monocentric chromosomes. As discussed above, lagging chromosomes and other anaphase defects have not been noted in reports of the effects of Kid inhibition in a cultured human cell line (Levesque and Compton, 2001). However, the duration of prometaphase–metaphase in those cells is ∼30 min, compared with 2–3 min for the early mitotic divisions of Drosophila and C. elegans (Table S1; Sharp et al., 2000; Levesque and Compton, 2001). The less exposed monocentric kinetochores of human chromatids should make them less prone to initial merotelic attachment, and the leisurely prometaphase could allow intermittent bipolar tension generated solely by kinetochores to bend, and so correct aberrant microtubule connections. Thus, inhibition of a polar ejection force in slow, monocentric spindles might cause only a low incidence of merotelism-associated aneuploidy. Although not noticeable in cultured cells, a small increase in aneuploidy would place whole organisms at a selective disadvantage, which could explain the evolutionary retention of polar ejection force generation mechanisms by species that have monocentric, slow mitotic divisions.
The study of mitosis in early C. elegans embryos promises continued insights. A unique combination of features, including holocentric chromosomes, a fast mitotic cycle, lack of anaphase A, and the importance of cleavage plane orientation, places special demands on the mitotic machinery and offers exciting opportunities to study the mechanisms that have evolved to meet those demands. The availability of expressed fluorescent markers for chromosomes, spindle poles, and microtubules (Oegema et al., 2001; Strome et al., 2001); continued improvements in the speed and resolution of live cell imaging; and the ability to inhibit the functions of most proteins through mutation or RNAi (e.g., http://celeganskoconsortium.omrf.org/; Kamath and Ahringer, 2003) provide powerful tools to address questions about the mechanisms that ensure accurate chromosome segregation.
Materials and methods
Worm strains
C. elegans N2 variety Bristol was used for RNAi analysis except where noted. GFP strains used were WH204 pie-1::GFP::tbb-2 (Strome et al., 2001); AZ212 pie-1::GFP::histone H2B (Praitis et al., 2001; Strome et al., 2001); and TH32 pie-1::GFP::histone H2B; pie-1::GFP::tbg-1 (Desai et al., 2003). Strain HR75 mei-1(b284)unc-29(e1072)/mei-1(cf46)unc-13(e1091) I was crossed to AZ212 to create a mei-1 strain expressing GFP::histone. Strain OC0010 zyg-1(b1) II (O'Connell et al., 2001) was crossed to stain TH32 to create the zyg-1 strain expressing GFP::histone and GFP::γ-tubulin. To generate embryos with bipolar P0 spindles and monopolar spindles thereafter, zyg-1 larvae were grown to L3 stage at 16°C and then transferred to 25°C and grown to adulthood.
Isolation of the klp-19(bn126) deletion mutant
Worm libraries mutagenized with trimethylpsoralen and UV irradiation were screened for a deletion mutation in klp-19 (Y43F4B.6) according to the protocols of M. Koelle et al. (http://info.med.yale.edu/mbb/koelle/protocols/protocol_Gene_knockouts.html) and the C. elegans Knock-Out Consortium (http://celeganskoconsortium.omrf.org/). PCR primers used in the screen were as follows: external primer set, forward 5′-ATTGTGCGTGAACTCTGACG-3′ and reverse 5′-GCGATCTGCTTCTCCAAGTC-3′; poison primer, 5′-ATCCGAGAGGCTGAAGAAGAC-3′; and internal primer set, forward 5′-GTCCGTAAATACACTCGCGG-3′ and reverse 5′-TCATCTTGTCCACCAAGTGC-3′.
RNA interference
For klp-19 RNAi, the cDNA clones yk105a12, yk111h5, and yk35b11 were obtained from Y. Kohara (National Institute of Genetics, Mishima, Japan). Phagemid DNA and sense and antisense strands of RNA were prepared as described previously (Strome et al., 2001). dsRNA from the three clones gave identical phenotypes. Clone yk111h5, which BLAST comparisons indicate will produce gene-specific transcripts, was used for all klp-19 RNAi experiments. dsRNA was introduced into worms by injection or feeding. For injection, 0.5–1 mg/ml of dsRNA was injected into young adult hermaphrodites. RNAi embryos were obtained from injected mothers 22–28 h after injection. For feeding, a 756-bp fragment of yk111h5 was inserted into the L4440 feeding vector and transformed into the RNase III-deficient E. coli strain HT115, and feeding plates were prepared as in protocol I of Kamath et al. (2001). L1 stage larvae were placed on feeding plates and grown to adulthood at 22°C. For klp-12 RNAi, the primer pair 5′-CCACGTGCAATCCAACATAC-3′ and 5′-TTTTCCGTTCGAAGGATGTC-3′ was used to amplify a gene-specific template that was used to generate dsRNA for injection (Kamath and Ahringer, 2003). For hcp-3 RNAi, the primer pair 5′-ATGGCCGATGACACCCC-3′ and 5′-TCAGAGATGTCGAAGGC-3′ was used to amplify full-length hcp-3 from N2 genomic DNA. The ∼1.0-kb product was inserted into the L4440 feeding vector. HCP-3 RNAi was done by feeding.
Antibodies and immunofluorescence microscopy
To generate antisera against KLP-19, a COOH-terminal peptide (amino acids 1061–1080) was synthesized, conjugated to keyhole limpet hemocyanin (Research Genetics), and injected into rabbits (Cocalico Biologicals). Antibodies were affinity purified by passing serum over a column of the peptide coupled to epoxy-activated agarose (Pierce Chemical Co.), eluting with 0.2 M glycine and 150 mM NaCl, pH 2.0, dialysis in PBS, and concentration. Western blot analysis of embryonic protein extracts showed specificity for a single band of ∼120 kD, which is consistent with the predicted size of KLP-19. Staining of the band was reduced substantially by preincubation of the anti-KLP-19 antibody with a molar excess of the peptide. These results and elimination of KLP-19 immunofluorescence by RNAi (Fig. 2 G) demonstrated that the affinity-purified antibodies were specific for KLP-19.
For immunofluorescence staining of gonads and embryos, gravid adult hermaphrodites were cut just behind the pharynx, fixed, and stained as described previously (Strome et al., 2001). Primary antibodies used were affinity-purified rabbit anti–KLP-19 at 1:4,000, mouse 4A1 anti–α-tubulin at 1:45 (Piperno and Fuller, 1985), mouse E7 anti–β-tubulin at 1:100 (Developmental Studies Hybridoma Bank), mouse PA3 anti-nucleosome at 1:100 (Monestier et al., 1994), and rat anti–KLP-7 (CeMCAK) at 1:50. Secondary antibodies used were Alexa Fluor 594–conjugated goat anti–mouse and goat anti–rabbit IgG at 1:250, and Alexa Fluor 488–conjugated goat anti–rat and goat anti–rabbit IgG at 1:500 (Molecular Probes).
Images of fixed specimens were collected on one of three microscopes. For two-wavelength images, a scanning confocal microscope (model MRC600; Bio-Rad Laboratories) was used with a 60× (1.4 NA) objective. Stacks of 0.5-μm optical sections were collected and displayed as projections. For three-wavelength images, either a microscope (model Eclipse E800; Nikon) equipped with a CCD camera (model Orca-ER; Hamamatsu) and Metamorph Imaging software (Universal Imaging Corp.) or a widefield deconvolution system (Deltavision; Applied Precision) was used. On the Deltavision system, stacks of 0.2-μm optical sections were collected, deconvolved using the Softworx (Applied Precision) software, and displayed as projections. Images for figures were processed using Photoshop (Adobe Systems).
Imaging live embryos
Embryos were mounted in M9 buffer on 2% agarose pads and covered with a coverslip. Observation by Nomarski optics was done on a microscope (model Axioplan; Carl Zeiss MicroImaging, Inc.). Images were captured using a camera (model C2400-00; Hamamatsu) and video controller with an Argus-10 image processor (Hamamatsu) and NIH Image (version 1.62f). Time-lapse imaging of GFP::γ-tubulin and GFP::histone fluorescence was done using a scanning confocal microscope. To minimize photodamage, a single optical plane was imaged once every 3 s, except as noted. Images were captured at the maximum pinhole aperture to enhance depth of field. Stacks of images were manipulated in NIH Image (version 1.62f, available at http://rsb.info.nih.gov/nih-image/). Images for figures were processed using Adobe Photoshop.
Online supplemental material
Fig. S1 shows comparisons of structural and sequence features of chicken chromokinesin, mouse Kif4, Drosophila KLP3A, C. elegans KLP-19, C. elegans KLP-12, and human Kid. Table S1 displays timing of mitotic events and measurements of pole-to-chromosome distances from fluorescence movies of GFP::histone and GFP::γ-tubulin in C. elegans embryos. All videos show time-lapse confocal microscopy of mitosis in C. elegans embryos. Videos 1 and 2 show GFP::histone in untreated and KLP-19–depleted two-cell embryos. Video 3 shows mitosis of paternal chromosomes after depletion of KLP-19. Videos 4 and 5 show GFP::histone and GFP::γ-tubulin in untreated and KLP-19–depleted embryos. Video 6 shows chromosome behavior after partial depolymerization of microtubules by Nocodazole. Video 7 shows mitosis in a P0 cell after inhibition of kinetochore assembly. Videos 8 and 9 show two pairs of movies comparing monopolar AB mitosis in control and KLP-19–depleted zyg-1 mutant embryos. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200403036/DC1.
Acknowledgments
We thank Kevin O'Connell, Paul Mains, Arshad Desai, Yuji Kohara, Margaret Fuller, Marc Monestier, and the Caenorhabditis Genetics Center for reagents and helpful advice.
This work was supported by National Institutes of Health grant GM58811 to W.M. Saxton and S. Strome, GM46295 to W.M. Saxton, and GM34059 to S. Strome.
Abbreviations used in this paper: AP, antipoleward; P, poleward.
References
- Afshar, K., J. Scholey, and R.S. Hawley. 1995. Identification of the chromosome localization domain of the Drosophila nod kinesin-like protein. J. Cell Biol. 131:833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson, D.G., and J.N. Thomson. 1982. The kinetochores of Caenorhabditis elegans. Chromosoma. 86:409–428. [DOI] [PubMed] [Google Scholar]
- Antonio, C., I. Ferby, H. Wilhelm, M. Jones, E. Karsenti, A.R. Nebreda, and I. Vernos. 2000. Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell. 102:425–435. [DOI] [PubMed] [Google Scholar]
- Bringmann, H., G. Skiniotis, A. Spilker, S. Kandels-Lewis, I. Vernos, and T. Surrey. 2004. A kinesin-like motor inhibits microtubule dynamic instability. Science. 303:1519–1522. [DOI] [PubMed] [Google Scholar]
- Carpenter, A.T. 1991. Distributive segregation: motors in the polar wind? Cell. 64:885–890. [DOI] [PubMed] [Google Scholar]
- Chang, C.J., S. Goulding, W.C. Earnshaw, and M. Carmena. 2003. RNAi analysis reveals an unexpected role for topoisomerase II in chromosome arm congression to a metaphase plate. J. Cell Sci. 116:4715–4726. [DOI] [PubMed] [Google Scholar]
- Cimini, D., B. Howell, P. Maddox, A. Khodjakov, F. Degrassi, and E.D. Salmon. 2001. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J. Cell Biol. 153:517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini, D., D. Fioravanti, E.D. Salmon, and F. Degrassi. 2002. Merotelic kinetochore orientation versus chromosome mono-orientation in the origin of lagging chromosomes in human primary cells. J. Cell Sci. 115:507–515. [DOI] [PubMed] [Google Scholar]
- Cimini, D., B. Moree, J.C. Canman, and E.D. Salmon. 2003. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J Cell Sci. 116:4213–4225. [DOI] [PubMed] [Google Scholar]
- Dagenbach, E.M., and S.A. Endow. 2004. A new kinesin tree. J. Cell Sci. 117:3–7. [DOI] [PubMed] [Google Scholar]
- Dernburg, A.F. 2001. Here, there, and everywhere: kinetochore function on holocentric chromosomes. J. Cell Biol. 153:F33–F38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., S. Rybina, T. Muller-Reichert, A. Shevchenko, A. Hyman, and K. Oegema. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki, H., and A.W. Murray. 2000. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 102:411–424. [DOI] [PubMed] [Google Scholar]
- Goshima, G., and R.D. Vale. 2003. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162:1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom, K.A., V.F. Holmes, N.R. Cozzarelli, and B.J. Meyer. 2002. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 16:729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald, R. 2000. Motor function in the mitotic spindle. Cell. 102:399–402. [DOI] [PubMed] [Google Scholar]
- Kamath, R.S., and J. Ahringer. 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 30:313–321. [DOI] [PubMed] [Google Scholar]
- Kamath, R.S., M. Martinez-Campos, P. Zipperlen, A.G. Fraser, and J. Ahringer. 2001. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans Genome Biol. 2:RESEARCH0002.1–0002.10. [DOI] [PMC free article] [PubMed]
- Kline-Smith, S.L., A. Khodjakov, P. Hergert, and C.E. Walczak. 2004. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol. Cell. 15:1146–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, M., S. Morales-Mulia, I. Brust-Mascher, G.C. Rogers, D.J. Sharp, and J.M. Scholey. 2004. The chromokinesin, KLP3A, drives mitotic spindle pole separation during prometaphase and anaphase and facilitates chromatid motility. Mol. Biol. Cell. 15:219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C.J., R.L. Malmberg, M.G. Muszynski, and R.K. Dawe. 2002. Maximum likelihood methods reveal conservation of function among closely related kinesin families. J. Mol. Evol. 54:42–53. [DOI] [PubMed] [Google Scholar]
- Lawrence, C.J., R.K. Dawe, K.R. Christie, D.W. Cleveland, S.C. Dawson, S.A. Endow, L.S.B. Goldstein, H.V. Goodson, N. Hirokawa, J. Howard, et al. 2004. A standardized kinesin nomenclature. J. Cell Biol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque, A.A., and D.A. Compton. 2001. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J. Cell Biol. 154:1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., and R.B. Nicklas. 1995. Mitotic forces control a cell-cycle checkpoint. Nature. 373:630–632. [DOI] [PubMed] [Google Scholar]
- Lima-de-Faria, A. 1949. Genetics, origin and evolution of kinetochores. Hereditas. 35:422–444. [Google Scholar]
- Mains, P.E., K.J. Kemphues, S.A. Sprunger, I.A. Sulston, and W.B. Wood. 1990. Mutations affecting the meiotic and mitotic divisions of the early Caenorhabditis elegans embryo. Genetics. 126:593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, W.F. 2002. Polar wind left flapping in the breeze? Trends Cell Biol. 12:9. [DOI] [PubMed] [Google Scholar]
- McIntosh, J.R., E.L. Grishchuk, and R.R. West. 2002. Chromosome-microtubule interactions during mitosis. Annu. Rev. Cell Dev. Biol. 18:193–219. [DOI] [PubMed] [Google Scholar]
- Monestier, M., K.E. Novick, and M.J. Losman. 1994. D-penicillamine- and quinidine-induced antinuclear antibodies in A.SW (H-2s) mice: similarities with autoantibodies in spontaneous and heavy metal-induced autoimmunity. Eur. J. Immunol. 24:723–730. [DOI] [PubMed] [Google Scholar]
- Moore, L.L., and M.B. Roth. 2001. HCP-4, a CENP-C–like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J. Cell Biol. 153:1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas, R.B. 1997. How cells get the right chromosomes. Science. 275:632–637. [DOI] [PubMed] [Google Scholar]
- O'Connell, K.F., C. Caron, K.R. Kopish, D.D. Hurd, K.J. Kemphues, Y. Li, and J.G. White. 2001. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 105:547–558. [DOI] [PubMed] [Google Scholar]
- O'Toole, E.T., K.L. McDonald, J. Mantler, J.R. McIntosh, A.A. Hyman, and T. Muller-Reichert. 2003. Morphologically distinct microtubule ends in the mitotic centrosome of Caenorhabditis elegans. J. Cell Biol. 163:451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema, K., A. Desai, S. Rybina, M. Kirkham, and A.A. Hyman. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergren, G., J. Mole-Bajer, and A. Bajer. 1960. An interpretation of transport phenomena at mitosis. Ann. NY Acad. Sci. 90:381–408. [DOI] [PubMed] [Google Scholar]
- Peretti, D., L. Peris, S. Rosso, S. Quiroga, and A. Caceres. 2000. Evidence for the involvement of KIF4 in the anterograde transport of L1-containing vesicles. J. Cell Biol. 149:141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, G., and M.T. Fuller. 1985. Monoclonal antibodies specific for an acetylated form of α-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J. Cell Biol. 101:2085–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, J., O. Bossinger, D. Rose, S. Strome, and W. Saxton. 1998. A nematode kinesin required for cleavage furrow advancement. Curr. Biol. 8:1133–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis, V., E. Casey, D. Collar, and J. Austin. 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 157:1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, C.L., and E.D. Salmon. 1998. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 8:310–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, C.L., E.A. Davison, L.C. Jensen, L. Cassimeris, and E.D. Salmon. 1986. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J. Cell Biol. 103:581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina, D., P. Enarson, J.B. Rattner, and B. Burke. 2003. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol. 162:991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segbert, C., R. Barkus, J. Powers, S. Strome, W.M. Saxton, and O. Bossinger. 2003. KLP-18, a Klp2 kinesin, is required for assembly of acentrosomal meiotic spindles in Caenorhabditis elegans. Mol. Biol. Cell. 14:4458–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine, Y., Y. Okada, Y. Noda, S. Kondo, H. Aizawa, R. Takemura, and N. Hirokawa. 1994. A novel microtubule-based motor protein (KIF4) for organelle transports, whose expression is regulated developmentally. J. Cell Biol. 127:187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D.J., H.M. Brown, M. Kwon, G.C. Rogers, G. Holland, and J.M. Scholey. 2000. Functional coordination of three mitotic motors in Drosophila embryos. Mol. Biol. Cell. 11:241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stear, J.H., and M.B. Roth. 2002. Characterization of HCP-6, a C. elegans protein required to prevent chromosome twisting and merotelic attachment. Genes Dev. 16:1498–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome, S., and W. B. Wood 1983. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell. 35:15–25. [DOI] [PubMed] [Google Scholar]
- Strome, S., J. Powers, M. Dunn, K. Reese, C.J. Malone, J. White, G. Seydoux, and W. Saxton. 2001. Spindle dynamics and the role of γ-tubulin in early Caenorhabditis elegans embryos. Mol. Biol. Cell. 12:1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf, W.E., and R.S. Hawley. 1992. Meiotic spindle assembly in Drosophila females: behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J. Cell Biol. 116:1167–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokai, N., A. Fujimoto-Nishiyama, Y. Toyoshima, S. Yonemura, S. Tsukita, J. Inoue, and T. Yamamota. 1996. Kid, a novel kinesin-like DNA binding protein, is localized to chromosomes and the mitotic spindle. EMBO J. 15:457–467. [PMC free article] [PubMed] [Google Scholar]
- Vale, R.D. 2003. The molecular motor toolbox for intracellular transport. Cell. 112:467–480. [DOI] [PubMed] [Google Scholar]
- Vale, R.D., and R.J. Fletterick. 1997. The design plan of kinesin motors. Annu. Rev. Cell Dev. Biol. 13:745–777. [DOI] [PubMed] [Google Scholar]
- Vernos, I., and E. Karsenti. 1995. Chromosomes take the lead in spindle assembly. Trends Cell Biol. 5:297–301. [DOI] [PubMed] [Google Scholar]
- Vernos, I., J. Raats, T. Hirano, J. Heasman, E. Karsenti, and C. Wylie. 1995. Xklp1, a chromosomal Xenopus kinesin-like protein essential for spindle organization and chromosome positioning. Cell. 81:117–127. [DOI] [PubMed] [Google Scholar]
- Wang, S.Z., and R. Adler. 1995. Chromokinesin: a DNA-binding, kinesin-like nuclear protein. J. Cell Biol. 128:761–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasser, M., and W. Chia. 2003. The Drosophila EAST protein associates with a nuclear remnant during mitosis and constrains chromosome mobility. J. Cell Sci. 116:1733–1743. [DOI] [PubMed] [Google Scholar]
- Waters, J.C., R.H. Chen, A.W. Murray, and E.D. Salmon. 1998. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 141:1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B.C., M.F. Riedy, E.V. Williams, M. Gatti, and M.L. Goldberg. 1995. The Drosophila kinesin-like protein KLP3A is a midbody component required for central spindle assembly and initiation of cytokinesis. J. Cell Biol. 129:709–723. [DOI] [PMC free article] [PubMed] [Google Scholar]




