Abstract
An assay that uses heminested PCR-restriction fragment length polymorphism analysis for the detection and genotyping of Giardia duodenalis on the basis of polymorphism in the triose phosphate isomerase (tpi) gene was developed. This assay was evaluated with DNA extracted from purified parasite material, bacterial cultures, whole human feces containing G. duodenalis and other parasites, and their corresponding immunofluorescence-stained fecal smears on glass microscope slides. The assay was specific and discriminated between G. duodenalis assemblages A and B. RFLP analysis further distinguished two groups (designated groups I and II) within assemblage A. Among 35 DNA samples extracted from whole feces from patients with confirmed sporadic giardiasis, the tpi gene was amplified from 33 (94%). Of these, nine (27%) samples contained assemblage A group II, 21 (64%) contained assemblage B, and 3 (9%) contained a mixture of assemblage A group II and assemblage B. The tpi gene of G. duodenalis assemblage B was amplified from 21 of 24 (88%) DNA samples extracted from whole feces from patients with confirmed cases of infection in a nursery outbreak. No amplification was detected from the remaining three DNA samples. Overall, analysis of DNA extracted from material recovered from stained microscope slides identified identical G. duodenalis genotypes in 35 (65%) of the 54 samples for which a genotype was established with DNA from whole feces. The heminested PCR method developed is sensitive, simple, and rapid to perform and is applicable for the analysis of other intestinal pathogens.
Giardia duodenalis (synonym of G. intestinalis and G. lamblia [25, 27, 29]) is an intestinal protozoan found in a wide range of mammalian hosts (7, 28). In humans, giardiasis is a common cause of parasitic gastroenteritis and is a major health concern worldwide (30). The disease is principally acquired by oral ingestion of G. duodenalis cysts, and the clinical manifestations vary from asymptomatic infection to acute diarrheal illness (4, 21). In immunocompetent individuals, giardiasis is usually self-limited but can develop into persistent and life-threatening diarrhea for both immunodeficient individuals (7, 8) and malnourished children in developing countries (16).
Isoenzyme and DNA analyses indicate that G. duodenalis is heterogeneous (6, 20, 22). Isolates identified as infectious for humans are classified into two major groups designated assemblages A and B. G. duodenalis assemblage A has been further separated into two subgroups I and II (22, 27). Assemblage A has also been detected in the feces of livestock, cats, dogs, beavers, guinea pigs, and slow lorises, with assemblage A group I having a broad host range and assemblage A group II being confined to humans (28). G. duodenalis assemblage B has a broad host range and has been recovered from dogs, beavers, rats, slow lorises, chinchillas, and siamangs (28). Other genotypes of G. duodenalis occur; and these are specific to their named hosts and are designated the Dog, Cat, Hoofed Animal, Rat, and Muskrat genotypes (11, 28). It has been suggested that for humans, other humans are the main reservoir of infection, with zoonotic sources constituting a minor reservoir (26). However, application of genetic analysis to G. duodenalis parasites from humans has been limited, and there is considerable uncertainty as to the relative role of animals as reservoirs of human infection as well as whether the anthroponotic or zoonotic origins of this parasite are reflected genetically (11).
Current methods for the detection of Giardia in the stool are usually based on visual recognition by light microscopy of stained or unstained Giardia cysts or trophozoites (1, 13). However, these methods are time-consuming, require experienced microscopists, are of low sensitivity, and are unable to distinguish between genetically distinct G. duodenalis isolates. Molecular biology provides powerful analytical tools that can be used to develop new and nonsubjective tests that have not yet had widespread application to the study of the molecular epidemiology of human giardiasis.
We previously described a method for the successful extraction of cryptosporidial DNA from whole feces (19) and from stained fecal smears on microscope slides (3). Initial evaluations of some of the published PCR-based protocols for amplification of giardia-specific gene sequences found that they were unsuitable for amplification of giardial DNA recovered from feces and stained smears by the extraction procedures described above. The purpose of the study described here was to develop a highly sensitive PCR technique that can be used to detect and distinguish G. duodenalis assemblages A and B as well as the two subgroups within assemblage A by using DNA extracted by the procedures described above.
MATERIALS AND METHODS
DNA samples.
DNA (concentration, 1 ng/μl) extracted from cultured G. duodenalis trophozoites of reference strains was kindly provided by W. Homan (Laboratory for Parasitology and Mycology, Bilthoven, The Netherlands). These included isolates 265KA1184 (Hoofed Animal genotype), Dog1 (Dog genotype), AMC13 (assemblage A), VNB3 (assemblage A), and AMC9 (assemblage B). DNA recovered from purified oocysts of Cryptosporidium parvum Iowa strain (strain 1372; AIDS Research Reference Reagent Program, National Institutes of Health), Cryptosporidium baileyi, Cryptosporidium muris, and Eimeria tenella (23), tachyzoites of Toxoplasma gondii (23), and bacterial suspensions of Escherichia coli (strain N211; PHLS Food External Quality Assessment Scheme, London, United Kingdom) and Clostridium perfringens type A (NCTC 8237) was also included.
Fecal samples.
Fecal samples from patients with diarrhea in which Giardia cysts had been detected by conventional techniques by clinical microbiology laboratories were collected at the Food Safety Microbiology Laboratory, Public Health Laboratory Service, London, United Kingdom. The samples originated from patients in England and Wales with sporadic cases of giardiasis diagnosed between September 1995 and March 2000. Details about the age, sex, and recent foreign travel of the patients were obtained from the original request forms. Samples were also collected from individuals involved in a nursery outbreak of giardiasis that occurred in North Wales during April 2000 (17), where children, child care workers, and parents had confirmed giardiasis. A retrospective case-control study identified sitting in paddling pools without nappies (diapers) as a risk factor for illness.
Fecal samples from patients with diarrhea were also collected. C. parvum genotype 1 (five samples), C. parvum genotype 2 (five samples), or Cyclospora (three samples) had been detected in these samples by conventional techniques.
Prior to DNA extraction, all samples were stored as whole feces at 4°C without preservatives for up to 4 years.
DNA extraction and polyvinylpyrrolidone (PVP) treatment.
Cyst disruption and DNA purification from whole feces and from immunofluorescence (IF)-stained smears on glass microscope slides were performed as described before (3, 19).
For samples for which the PCR amplification (described below) was unsuccessful, a further DNA purification was achieved by PVP treatment performed as described by Lawson and colleagues (15).
Microscopy.
Smears were produced from all fecal samples (3) and were reexamined by IF microscopy with an anti-Giardia cyst monoclonal antibody (clone 22A6; Novocastra Laboratories Ltd., Newcastle upon Tyne, United Kingdom) and fluorescein isothiocyanate-conjugated antimouse antibody (Biosource International, Camarillo, Calif.). The numbers of cysts detected were estimated by calculation of the mean for 20 microscope fields by use of a ×40 objective (Zeiss, Welwyn Garden City, United Kingdom).
PCR amplification and restriction fragment length polymorphism (RFLP) analysis. (i) Oligonucleotide primers.
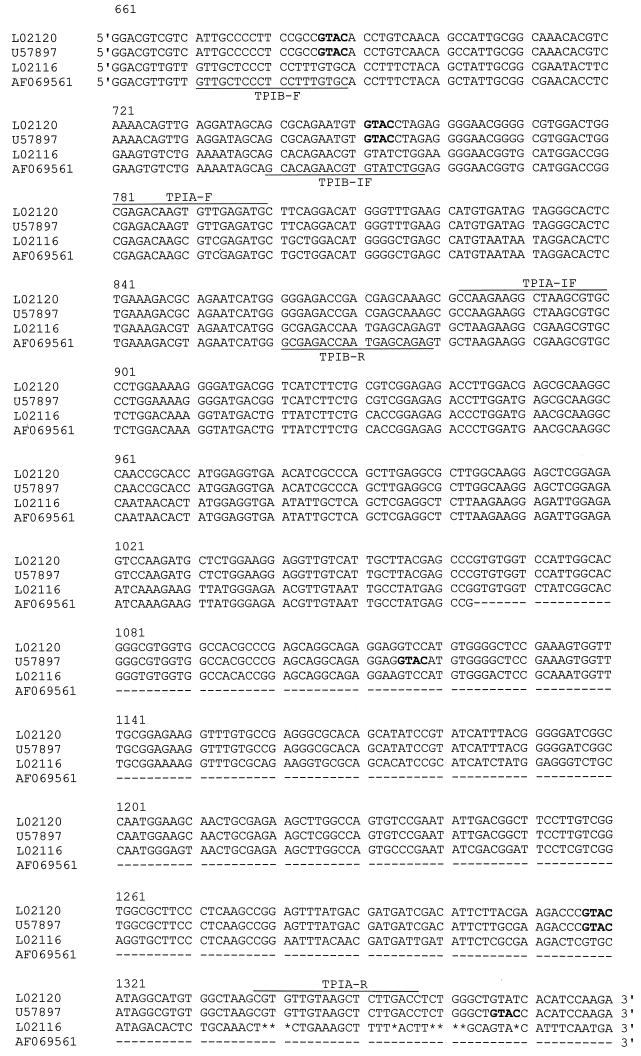
Two triplets of oligonucleotide primers (forward [F], inner forward [IF], and reverse [R]) were designed with the Hemi-Nested Oligo Selection Program (P. H. Dear, unpublished data) on the basis of the DNA sequences of the triose phosphate isomerase (tpi) gene of G. duodenalis. The tpi gene of assemblage A was designated TPIA (GenBank accession numbers L02120 and U57897), and that of assemblage B was designated TPIB (GenBank accession numbers L02116 and AF069561) (Fig. 1). Primers were obtained from GIBCO/Life Technologies (Paisley, United Kingdom); and their designations, corresponding nucleotide sequences, and positions are shown in Fig. 1.
FIG. 1.
Alignments, determined with the BioEdit program, of DNA sequences of the tpi gene used in the present study and retrieved from the GenBank database (G. duodenalis assemblage A group I, GenBank accession number L02120; G. duodenalis assemblage A group II, GenBank accession number U57897; G. duodenalis assemblage B, GenBank accession numbers L02116 and AF069561). The numbers designate the base pair positions of the longest sequence, that with GenBank accession number L02116. The underscores show the positions of the primers in the sequences. Boldface letters indicate the RsaI restriction sites (GTAC) in the G. duodenalis assemblage A group I and group II sequences. The asterisks indicate base deletions, and the hyphens indicate unknown bases.
(ii) PCR amplification.
Amplification of the tpi gene was performed as a two-step PCR, with phase I comprising a single duplex reaction and phase II comprising two individual reactions. In the phase I duplex, a 576-bp fragment of TPIA and a 208-bp fragment of TPIB were amplified simultaneously with forward and reverse primer sets TPIAF-TPIAR and TPIBF-TPIBR, respectively (Fig. 1). PCR amplification was performed in 10-μl volume with 5 μl of DNA in 1× PCR buffer, 2 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 0.25 mM, each primer at a concentration of 0.3 μM, and 0.5 U of Taq DNA polymerase (all reagents were from GIBCO/Life Technologies). Samples were subjected to an initial denaturation of 94°C for 1 min, followed by 25 cycles of 94°C for 20 s, 52°C (whole feces DNA) or 50°C (fecal smear DNA) for 30 s, and 72°C for 1 min, with a final extension at 72°C for 5 min.
Phase II comprised two separate heminested PCRs designated the TPIA-PCR and the TPIB-PCR, respectively. In the TPIA-PCR, a 476-bp fragment of the TPIA gene was amplified by use of inner forward primer and reverse primer set TPIAIF-TPIAR (Fig. 1). PCR amplification was performed in a 20-μl volume with 10 μl of the duplex amplicon diluted 10 times in 1× PCR buffer, 1 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 0.25 mM, each primer at a concentration of 1 μM, and 1 U of Taq DNA polymerase. Samples were subjected to an initial denaturation of 94°C for 1 min, followed by 33 cycles of 94°C for 20 s, 56°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 5 min.
In the separate TPIB-PCR, a 140-bp fragment of the TPIB gene was amplified with primers TPIBIF and TPIBR (Fig. 1). Amplification was performed under the same conditions used for the TPIA-PCR described above, except that the MgCl2 concentration in the PCR mixture was 1.5 mM. Controls were included in each batch of tests. Ten picograms of DNA recovered from a purified G. duodenalis trophozoite culture of assemblage A (strain AMC13) and 10 pg of DNA recovered from a purified G. duodenalis trophozoite culture of assemblage B (strain AMC9) were used as the templates for the positive controls, and distilled water was used as the template for the negative controls throughout.
RFLP analysis was performed by digesting 5 μl of the TPIA-PCR product with 5 U of RsaI in 1× enzyme buffer (GIBCO/Life Technologies) in a final volume of 30 μl for 3 h at 37°C.
(iii) PCR product and restriction fragment detection.
PCR products and restriction fragments were separated by horizontal electrophoresis in 1 and 3.2% agarose gels, respectively, with ethidium bromide staining and were recorded by UV transillumination with type 667 film (Polaroid Ltd., St. Albans, United Kingdom).
DNA sequencing.
The PCR products obtained from strains AMC13 and VNB3 by TPIA-PCR and the PCR product obtained from DNA extracts of the AMC9 reference strain by TPIB-PCR were cloned by use of the TOPO-TA cloning kit (Invitrogen, Leek, The Netherlands). Plasmid DNA was purified by using the Promega Wizard SV miniprep kit (Promega UK Ltd., Southampton, United Kingdom), and cloned DNA was sequenced on an ABI 377 automated sequencer by use of BigDye terminator chemistry with M13 primers at the Single Reaction DNA Sequencing Service (Cambridge Bioscience).
Multiple alignment and restriction map analysis were performed with the BioEdit Sequence Alignment Editor (9).
RESULTS
Initial studies were performed with DNA extracted from purified trophozoite preparations. By using 10 pg of DNA obtained from reference strain AMC13 (assemblage A), the predicted 476-bp product was obtained by TPIA-PCR, but no product was amplified by TPIB-PCR. Conversely, by using 10 pg of DNA from reference strain AMC9 (assemblage B), the predicted 140-bp product was obtained by TPIB-PCR, but no product was amplified by TPIA-PCR. No product was amplified by either TPIA-PCR or TPIB-PCR with DNA extracted from purified C. baileyi, C. muris, C. parvum, T. gondii, E. tenella, E. coli, C. perfringens, G. duodenalis Dog genotype (strain Dog1), and G. duodenalis Hoofed Animal genotype (strain 265KA1184). Results of the sequencing analysis of the products obtained from G. duodenalis AMC13 and VNB3 DNAs by TPIA-PCR showed 100% matches with the sequences with GenBank accession numbers U57897 (G. duodenalis assemblage A group II) and L02120 (G. duodenalis assemblage A group I), respectively. Results of the sequencing analysis of the product obtained from G. duodenalis AMC9 DNA by TPIB-PCR showed a 100% match with the sequence with GenBank accession number AF069561 (G. duodenalis assemblage B).
To estimate the sensitivities of the assays, decimal dilutions of the AMC13 and AMC9 reference strain DNA extracts at 1 ng/μl were prepared. The tpi gene fragment from assemblages A and B could be amplified by using 0.5 and 0.05 pg of DNA per reaction mixture, respectively, equivalent to 50 and 5 copies of the tpi gene, respectively, based on a genome size of 1.2 × 107 bp (2).
Restriction maps were constructed from an alignment of the G. duodenalis tpi gene sequences of assemblage A group I (GenBank accession number L02120) and assemblage A group II (GenBank accession number U57897) (Fig. 1). RsaI restriction sites were identified to distinguish the two groups that gave the predicted restriction digestion products of 437 and 39 bp for assemblage A group I and 235, 202, and 39 bp for assemblage A group II. Digestion of the products from VNB3 and AMC13 extracts obtained by TPIA-PCR with the RsaI enzyme showed the predicted restriction fragment pattern but without the 39-bp fragment, which was not resolved in this system. This allowed the identification of G. duodenalis VNB3 as assemblage A group I and AMC13 as assemblage A group II (Fig. 2).
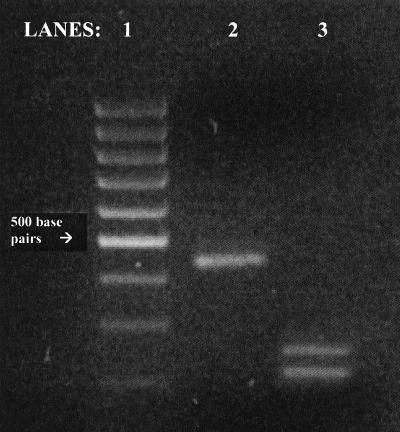
FIG. 2.
RsaI digestion of TPIA-PCR products on an ethidium bromide-stained 3.2% agarose gel. Lane 1, 100-bp marker (GIBCO/Life Technologies) (the 500-bp fragment is indicated); lane 2, G. duodenalis assemblage A group I (strain VNB3); lane 3, G. duodenalis assemblage A group II (strain AMC13).
The heminested PCR protocol was applied to fecal samples and their corresponding IF-stained smears. Among 68 fecal samples previously identified as containing Giardia cysts (37 from patients with sporadic cases and 31 from individuals involved in the nursery outbreak), cysts were reconfirmed by IF microscopy in 59 (87%) of the samples (35 from the patients with sporadic cases and 24 from individuals involved in the nursery outbreak). The numbers of cysts per microscope field ranged between 0.1 and 50 in samples from the patients with sporadic cases and between 0.1 and 20 from those involved in the outbreak. No Giardia cysts were detected in fecal samples containing C. parvum or Cyclospora.
Among the 35 samples from patients with sporadic cases of giardiasis in which the presence of Giardia cysts was reconfirmed, the tpi gene was amplified from 33 (94%) when DNA was extracted from whole feces. Assemblage A was detected in 9 samples, assemblage B was detected in 21 samples, and a mixture of assemblages A and B was detected in 3 samples (Table 1). All G. duodenalis assemblage A strains were identified as assemblage A group II on the basis of their fragment patterns obtained by RFLP analysis. Among the 24 samples from the nursery outbreak in which the presence of Giardia cysts was reconfirmed, the tpi gene was amplified from 21 (88%) of the DNA samples extracted from whole feces. G duodenalis assemblage B alone was detected in the DNA from all 21 samples (Table 1).
TABLE 1.
Results of PCR-RFLP analysis of the G. duodenalis tpi gene amplified from DNA extracted from whole feces and IF-stained fecal smears
| Sample and type of DNA extracted from whole feces (no. of samples) | No. of samples from which DNA was extracted from corresponding stained smears
|
|||
|---|---|---|---|---|
| Assemblage A group II | Assemblage B | Assemblage A group II and assemblage B | None | |
| Samples from patients with sporadic cases (n = 35) | ||||
| Assemblage A group II (9) | 5 | 0 | 0 | 4 |
| Assemblage B (21) | 0 | 17 | 0 | 4 |
| Assemblage A group II and assemblage B (3) | 0 | 0 | 2 | 1 |
| None (2) | 0 | 0 | 0 | 2 |
| Samples from individuals involved in nursery outbreakb (n = 24) | ||||
| Assemblage A (0) | 0 | 0 | 0 | NAc |
| Assemblage B (21) | 0 | 10 | 0 | 11 |
| None (3) | 0 | 1 | 0 | 2 |
DNA was amplified from 33 (94%) of the 35 whole fecal samples and 24 (69%) of the 35 corresponding stained smears.
DNA was amplified from 21 (88%) of the 24 whole fecal samples and 11 (46%) of the 24 corresponding stained smears.
NA, not applicable.
None, not amplified.
DNA was not amplified from any of the 9 samples from the patients with sporadic cases and from the individuals involved in the nursery outbreak in which the presence of the cysts could not be reconfirmed or from the 13 samples in which C. parvum or Cyclospora was detected.
Among all the DNA samples extracted from whole feces, amplification of the tpi gene was obtained only after PVP treatment for 18% of the DNA samples. The proportions of DNA extracts in which G. duodenalis assemblage A group II and assemblage B could be identified only after PVP treatment were 44 and 14%, respectively; this association was not statistically significant (P = 0.06 by Fisher's exact two-tailed test).
DNA was extracted from stained smears prepared from the same fecal samples mentioned above. The tpi gene fragments were amplified from 34 (59%) of the 59 samples (Table 1); for 8% of these 34 samples amplification was achieved after PVP treatment. The TPIB fragment was amplified from one stained smear prepared from a sample collected during the outbreak but from which the tpi gene was not amplified by use of DNA extracted from whole feces. Identical genotyping results were obtained with DNA extracted from whole feces and from stained smears, including two samples in which a mixture of G. duodenalis assemblage A group II and assemblage B was detected (Table 1).
Successful tpi gene amplification was achieved by seeding giardial DNA into the five DNA samples from whole feces in which the presence of Giardia was reconfirmed but from which the tpi gene fragment could not be amplified.
The effect of the numbers of Giardia cysts detected and the percentage of samples from which the tpi gene fragments that were amplified were investigated. Among all samples in which Giardia cysts were detected, 28 had a mean of less than five cysts per microscope field and 31 had a mean of five or more cysts per microscope field. Among the samples with an average of less than five cysts per field, amplification was achieved for 82 and 50% of the DNA samples extracted from whole feces and smears, respectively. However, when the average numbers of cysts per field was five or more, the amplification frequencies were 100 and 68% with DNA recovered from whole feces and smears, respectively.
To investigate the reproducibility of tpi gene fragment amplification, triplicate tests were performed with DNA extracted from reference strains AMC13, AMC9, Dog1, and 265KA1184 (2 pg/μl); 20 whole fecal samples (9 containing assemblage A group II, 9 containing assemblage B, and 2 containing a mixture of assemblage A group II and assemblage B); 16 smears (5 containing assemblage A group II, 9 containing assemblage B, and 2 containing a mixture of assemblage A group II and assemblage B); and feces containing C. parvum or Cyclospora oocysts. The results were 100% reproducible with DNA extracted from reference strains AMC13 and AMC9. The amplification was 77% reproducible when the PCR was performed with DNA derived from whole feces or smears. The results of replicate genotyping tests were always consistent. The amplifications achieved in triplicate, duplicate, and single tests were independent of the number of cysts present in the stool or the use of PVP treatment (data not shown). Successful amplification of the tpi gene was achieved by seeding giardial DNA into extracts for which this gene could not be amplified by PCR (data not shown). No amplification was achieved in triplicate tests with DNA from G. duodenalis Dog1 and 265KA1184 or DNA from whole feces containing C. parvum or Cyclospora.
No significant differences in the distributions of patients infected with the different G. duodenalis assemblages were detected (data not shown).
DISCUSSION
The 18% DNA sequence divergence within a fragment of the tpi gene of G. duodenalis assemblages A and B (2) available from GenBank allowed the development of discriminatory primers for PCR amplification, as described here. The resulting heminested PCR was evaluated with DNA extracted from cultured G. duodenalis trophozoites, purified eukaryotic and prokaryotic intestinal pathogens of different genera, and whole feces and stained smears containing Giardia or other intestinal parasites. The present study demonstrates that the PCR-RFLP procedure described here reliably identifies both G. duodenalis strains of assemblage A and assemblage B, with differentiation of groups I and II within assemblage A. This heminested PCR procedure had a high degree of sensitivity and is capable of amplifying giardial DNA from 0.05 pg of DNA derived from cultured trophozoites, which is equivalent to 5 copies of the tpi gene. The tpi gene fragment was amplified from 91 and 59% of the DNA samples extracted from whole feces and stained smears, respectively, in which the presence of Giardia cysts had been reconfirmed. The proportion of samples from which the tpi gene could be amplified was reduced by a quarter when less than five cysts per microscopic field were detected.
Fecal samples initially identified in hospital laboratories as containing Giardia were reexamined in the present study, and the presence of the parasite could not be reconfirmed for 13% of the specimens. Possible reasons for this may include initial misidentification, the presence of the parasite at very low levels, or degradation of parasite material during storage. We previously reported that DNA from Cryptosporidium oocysts is stable for >4 years in whole feces stored at 4°C (19). Preliminary data suggest that Giardia cysts are less robust (C. F. L. Amar, unpublished data), and degradation of this parasite during storage may contribute to the inability to reconfirm the presence of the parasite by both microscopy and PCR after initial recognition.
The primers used in the PCR assay described here were specific for G. duodenalis assemblages A and B since no amplification was observed when either DNA from the G. duodenalis Dog and Hoofed Animal genotypes or DNA from other eukaryotes and prokaryotes was used. In addition, no tpi gene amplification was successful when DNA recovered from feces in which no giardial cysts had been detected or in which C. parvum and Cyclospora oocysts had been detected was used.
The PCR results were reproducible both when DNA from cultured trophozoites was used and in the assemblages identified from all samples. However, the reproducibility of the results by replicate testing of DNA derived from feces and stained smears was found to be 77%. The reproducibility was independent of the number of cysts present in the samples tested and was also independent of the use of DNA that had been further purified by PVP treatment. This reduced fidelity may be due to the presence in feces of inhibitors of the PCR which are copurified in the DNA extraction or to the presence of DNA from the fecal microflora (5). However, successful amplification of the tpi gene was achieved in experiments by seeding giardial DNA into extracts from which this gene could not be amplified. Furthermore, similar analyses of feces containing C. parvum that used DNA extraction techniques identical to those described here showed very high sensitivities with respect to the number of samples from which cryptosporidial genes could be amplified (3, 24). Hence, the poorer reproducibility reported here may be due to a very small template number even in the presence of morphologically entire cysts and/or degradation of giardial nucleic acid during storage (as suggested above). A modification of the extraction procedure is being explored to improve the reproducibility of this assay.
The authors have previously reported a method for the extraction of cryptosporidial DNA from stained fecal smears on glass microscope slides (3; C. Amar, R. M. Chalmers, K. Elwin, P. Tynan, and J. McLauchlin, submitted for publication). By that technique, the correct genotype of C. parvum was recognized in 85% of 105 smears. Similar techniques are described here for the amplification of G. duodenalis DNA from stained smears.
The present study provides, for the first time, information on the distribution of the genotypes of G. duodenalis from humans with both sporadic and outbreak-associated giardiasis in the United Kingdom. Among 35 fecal samples from patients with confirmed sporadic cases of giardiasis, assemblage B, assemblage A group II, and both assemblage B and assemblage A group II were detected in 60, 26, and 8% of the samples, respectively; the tpi gene was not detected in the remaining 6% of the samples. Among the 24 samples from individuals involved in the nursery outbreak, G. duodenalis assemblage B was detected in 88% of the samples and the tpi gene fragment was not amplified from the remaining 12%. G. duodenalis assemblage A group II has been isolated only from humans, while assemblage A group I and assemblage B have much broader host ranges including humans (28). The data from the present study, even though they are based on the results for a relatively small group of patients, together with the known host ranges of the different G. duodenalis assemblages, suggest that the origins of infection are either anthroponotic (assemblage A group II and assemblage B) or zoonotic from dogs or rats (assemblage B). The study does not support the role of livestock animals as a reservoir for human giardiasis in the United Kingdom. Further studies with a larger series of humans as well as potential host reservoirs are required, and these are planned.
It is of note that in the present study, G. duodenalis assemblage A group II and assemblage B together were detected in three samples, and a similar mixture has been reported previously (12, 18). These multiple infections may reflect ingestion of sources contaminated by heterogeneous mixtures of parasites, such as water contaminated by sewage or slurry. In one of these samples in which a heterogeneous giardial infection was suggested, Cryptosporidium was detected by microscopy; and the isolate was confirmed to be C. parvum genotype 1 (Human type) by PCR (data not shown), further supporting the hypothesis that the patient had been exposed to a sewage-contaminated source.
Further analysis of G. duodenalis from humans with molecular characterization systems with greater discriminatory powers than the system described here suggests that some strains appear to be host specific and others produce asymptomatic or mild infections (4, 10, 14, 28). However, since these studies have been conducted with relatively small numbers of samples from humans with giardiasis, more wide-scale analyses are required to verify these observations. We are extending the results of the present study by testing larger numbers of specimens from humans with giardiasis and applying methods with increased discriminatory powers.
In summary, a sensitive method for the identification of G. duodenalis assemblage A groups I and II and assemblage B that uses DNA extracted from human fecal samples is reported. The PCR method is specific, robust, and reproducible. The protocol is applicable to the testing of DNA recovered from whole feces and stained fecal smears, and previous experience would suggest that the method is therefore applicable to the analysis of a relatively large series of samples. The authors believe that this approach, together with the development of more discriminatory typing methods, will vastly increase the understanding of the epidemiology of giardiasis. Methods with improved sensitivities, such as the one described here, will also be invaluable in the detection and characterization of Giardia in nonhuman hosts and in the environment.
Acknowledgments
We thank colleagues in clinical microbiology laboratories for the donation of specimens, W. Homan (Laboratory for Parasitology and Mycology, Bilthoven, The Netherlands) for purified giardial DNA, and the Department of Public Health Medicine, North Wales Health Authority, where the outbreak was investigated, for support.
C.F.L.A. is funded by a PHLS Ph.D. studentship.
REFERENCES
- 1.Adam, R. D. 1991. The biology of Giardia spp. Microbiol. Rev. 55:706-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam, R. D. 2000. The Giardia lamblia genome. Int. J. Parasitol. 30:475-484. [DOI] [PubMed] [Google Scholar]
- 3.Amar, C., S. Pedraza-Díaz, and J. McLauchlin. 2001. Extraction and genotyping of Cryptosporidium parvum DNA from faecal smears on glass slides stained conventionally for direct microscope examination. J. Clin. Microbiol. 39:401-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Astiazarán-García, H., M. Espinosa-Cantellano, G. Castañón, B. Chávez-Munguia, and A. Martinez-Palomo. 2000. Giardia lamblia: effect of infection with symptomatic and asymptomatic isolates on the growth of gerbils (Meriones unguiculatus). Exp. Parasitol. 95:128-135. [DOI] [PubMed] [Google Scholar]
- 5.Da Silva, A. J., F. J. Bornay-Llinares, I. N. Moura, S. B. Slemenda, J. L. Tuttle, and N. J. Pieniazek. 1999. Fast and reliable extraction of protozoan parasite DNA from faecal specimens. Mol. Diagn. 4:57-64. [DOI] [PubMed] [Google Scholar]
- 6.Ey, P. L., M. Mansouri, J. Kulda, E. Nohýnková, P. T. Monis, R. H. Andrews, and G. Mayrhofer. 1997. Genetic analysis of Giardia from hoofed farm animals reveals artiodactyl-specific and potentially zoonotic genotypes. J. Eukaryot. Microbiol. 44:626-635. [DOI] [PubMed] [Google Scholar]
- 7.Farthing, M. J. G. 1995. Giardia lamblia, p. 1081-1106. In M. J. Blaser, P. D. Smith, J. I. Ravdin, H. B. Greenberg, and R. L. Guerrant (ed.), Infections of the gastrointestinal tract. Raven Press, New York, N.Y.
- 8.Fontanet, A. L., T. Sahlu, T. Rinke de Wit, T. Messele, W. Masho, T. Woldemichael, H. Yeneneh, and R. A. Coutinho. 2000. Epidemiology of infections with intestinal parasites and human immunodeficiency virus (HIV) among sugar-estate residents in Ethiopia. Ann. Trop. Med. Parasitol. 94:269-278. [DOI] [PubMed] [Google Scholar]
- 9.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Window 95/98NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 10.Homan, W. L., and T. G. Mank. 2001. Human giardiasis: genotype linked differences in clinical symptomatology. Int. J. Parasitol. 31:822-826. [DOI] [PubMed] [Google Scholar]
- 11.Hopkins, R. M., B. P. Meloni, D. M. Groth, J. D. Wetherall, J. A. Reynoldson, and R. C. Thompson. 1997. Ribosomal RNA sequencing reveals differences between the genotypes of Giardia isolates recovered from humans and dogs living in the same locality. J. Parasitol. 83:44-51. [PubMed] [Google Scholar]
- 12.Hopkins, R. M., C. C. Constantine, D. A. Groth, J. D. Wetherall, J. A. Reynoldson, and R. C. A. Thompson. 1999. PCR-based DNA fingerprinting of Giardia duodenalis isolates using the intergenic rDNA spacer. Parasitology 118:531-539. [DOI] [PubMed] [Google Scholar]
- 13.Isaac-Renton, J. L. 1991. Laboratory diagnosis of giardiasis. Clin. Lab. Med. 11:811-827. [PubMed] [Google Scholar]
- 14.Karanis, P., and P. L. Ey. 1998. Characterisation of axenic isolates of Giardia intestinalis established from humans and animals in Germany. Parasitol. Res. 84:442-449. [DOI] [PubMed] [Google Scholar]
- 15.Lawson, A. J., D. Linton, J. Stanley, and R. J. Owen. 1997. Polymerase chain reaction detection and speciation of Campylobacter upsaliensis and C. helveticus in human faeces and comparison with culture techniques. J. Appl. Microbiol. 83:375-380. [DOI] [PubMed] [Google Scholar]
- 16.Lima, A. A., S. R. Moore, M. S. Barboza, Jr., A. M. Soares, M. A. Schleupner, R. D. Newman, C. L. Sears, J. P. Nataro, D. P. Fedorko, T. Wuhib, J. B. Schorling, and R. L. Guerrant. 2000. Persistent diarrhea signals a critical period of increased diarrhea burdens and nutritional shortfalls: a prospective cohort study among children in northeastern Brazil. J. Infect. Dis. 181:1643-1651. [DOI] [PubMed] [Google Scholar]
- 17.Linnane, E., R. Roberts, and N. Looker. 2001. Nappies and transmission of Giardia lamblia between children. Lancet 358:507.. [DOI] [PubMed] [Google Scholar]
- 18.Lu, S. Q., A. C. Baruch, and R. D. Adam. 1998. Molecular comparison of Giardia lamblia isolates. Int. J. Parasitol. 28:1341-1345. [DOI] [PubMed] [Google Scholar]
- 19.McLauchlin, J., S. Pedraza-Díaz, C. Amar-Hoetzeneder, and G. L. Nichols. 1999. Genetic characterisation of Cryptosporidium strains from 218 patients with diarrhea diagnosed as having sporadic cryptosporidiosis. J. Clin. Microbiol. 37:3153-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meloni, B. P., A. J. Lymbery, and R. C. A. Thompson. 1995. Genetic characterisation of isolates of Giardia duodenalis by enzyme electrophoresis: implications for reproductive biology, population structure, taxonomy, and epidemiology. J. Parasitol. 81:368-383. [PubMed] [Google Scholar]
- 21.Meyer, E. A., and E. L. Jarroll. 1980. Giardiasis. Am. J. Epidemiol. 111:1-12. [DOI] [PubMed] [Google Scholar]
- 22.Monis, P. T., G. Mayrhofer, R. H. Andrews, W. L. Homan, L. Limper, and P. L. Ey. 1996. Molecular genetic analysis of Giardia intestinalis isolates at the glutamate dehydrogenase locus. Parasitology 112:1-12. [DOI] [PubMed] [Google Scholar]
- 23.Patel, S., S. Pedraza-Díaz, and J. McLauchlin. 1999. The identification of Cryptosporidium species and Cryptosporidium parvum directly from whole faeces by analysis of a multiplex PCR of the 18S rRNA gene and by PCR/RFLP of the Cryptosporidium outer wall protein (COWP) gene. Int. J. Parasitol. 29:1241-1247. [DOI] [PubMed] [Google Scholar]
- 24.Pedraza-Díaz, S., C. Amar, G. L. Nichols, and J. McLauchlin. 2001. The development of a nested PCR procedure for amplification of the Cryptosporidium oocyst wall protein (COWP) gene, and analysis of 2128 cryptosporidiosis cases. Emerg. Infect. Dis. 7:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith, H. V., L. J. Robertson, A. T. Campbell, and R. W. A. Girdwood. 1995. Giardia and giardiasis: what's in a name? Microbiol. Europe 3:22-29. [Google Scholar]
- 26.Thompson, R. C. A. 1998. Giardia infections, p. 545-561. In S. R. Palmer, E. J. L. Soulsby, and D. I. H. Simpson, Zoonoses, biology, clinical practice and public health control. Oxford University Press, Oxford, United Kingdom.
- 27.Thompson, R. C. A. 2000. Giardiasis as a re-emerging infectious disease and its zoonotic potential. Int. J. Parasitol. 30:1259-1267. [DOI] [PubMed] [Google Scholar]
- 28.Thompson, R. C. A., R. M. Hopkins, and W. L. Homan. 2000. Nomenclature and genetic groupings of Giardia infecting mammals. Parasitol. Today 16:210-213. [DOI] [PubMed] [Google Scholar]
- 29.Upcroft, P. 1991. DNA fingerprinting of the human intestinal parasite Giardia intestinalis with hypervariable minisatellite sequences, p. 70-84. In T. Burke, G. Dolf, A. J. Jeffreys, and R. Wolff (ed.), DNA fingerprinting: approaches and applications. Birkhäuser Verlag, Basel, Switzerland. [DOI] [PubMed]
- 30.Wolfe, M. S. 1992. Giardiasis. Clin. Microbiol. Rev. 5:93-100. [DOI] [PMC free article] [PubMed] [Google Scholar]