Abstract
The aim of the study was to analyze the relationship between genotypic and phenotypic drug resistance profiles of human immunodeficiency virus type 1 (HIV-1) strains isolated from patients during double-analogue nucleoside therapy. A drug-resistant HIV strain was isolated from 20 out of 25 patients, with 16 (64%) subjects carrying a virus with multiple drug resistance mutations. The most frequent resistance mutations were M184V (18 isolates) and M41L (7 isolates). Discordance between the genotypic and phenotypic profile for at least one drug was detected in 16 out of 25 strains. Particularly, eight isolates had a discordant genotypic-phenotypic resistance pattern for two drugs and one isolate had such a pattern for three drugs. A genotypic resistance pattern with a phenotypic sensitivity profile was detected in six isolates (four resistant to zidovudine and two resistant to lamivudine). On the other hand for several strains a genotypic pattern of sensitivity pattern to abacavir (10 strains), didanosine (7 strains), stavudine (3 strains), zidovudine (2 strains), and lamivudine (1 strain) with a phenotypic resistance profile was detected. After a follow-up period of 8 months, an impairment of virological and immunological parameters was detected only in subjects with an HIV-1 isolate with a phenotypic resistance profile in despite of the genotypic results. Predicting resistance phenotype from genotypic data has important limitations. Despite the low number of patients and the short follow-up period, this study suggests that during failing therapy with analogue nucleosides, a phenotypic analysis could be performed in spite of an HIV genotypic sensitivity pattern.
Mutations in the human immunodeficiency virus (HIV) reverse transcriptase (RT) and protease genes are associated with reduced sensitivity to antiretroviral drugs (9, 15). Recently, two studies (3, 7) provided evidence that antiretroviral therapy adapted to genotypic resistance mutations gave more-effective results than therapy adapted to treatment history in patients who failed combination regimens.
Genotype- and phenotype-based assays are fundamentally different but yield complementary information. Phenotypic tests measure virus drug susceptibility, resulting from known or unknown resistance-related mutations and their interactions. Genotypic tests detect mutations in the viral genome that may be associated with decreased drug susceptibility.
In previous studies, during primary HIV infection, in antiretroviral-naïve patients, discordance between genotypic and phenotypic drug resistance analyses has been described (4, 13). However, the clinical relevance of a large number of mutations has not been established. Moreover, the level of phenotypic resistance predictive of therapy failure is not known and is probably dependent on the drug or antiviral combinations used. Both phenotypic and genotypic resistance assays should be interpreted with an understanding of all issues surrounding the efficacy of antiretroviral medications such as pharmacokinetics and adherence, both of which may confound the clinical interpretation of assay results. Although sequencing can detect all mutations present in the predominant virus population, the phenotypic effects of uncharacterized mutations and mutational interactions may be difficult to predict. Interpretation of genotypes is difficult, as there are large numbers of polymorphisms in both protease and RT that may or not may confer some degree of drug resistance.
The aim of the present study was to analyze the relationship between the genotypic and phenotypic drug resistance profiles of HIV type 1 (HIV-1) strains isolated from patients treated for an average period of 18 months with a double-analogue nucleoside therapy.
MATERIALS AND METHODS
Patients.
The 25 HIV-1-seropositive subjects enrolled in the study were selected from among 101 patients treated with two nucleoside RT inhibitors (NRTI) showing a progressive decline of HIV-1 RNA in plasma to <10,000 copies/ml and an increase of CD4+ cell count to>50 cells/ml from before treatment values. The selection criteria to identify the 25 patients were either the isolation of the HIV-1 strain from peripheral blood mononuclear cells (PBMC) and a titer of viral stock of the HIV-1 isolates of more than the prerequisite 4,000 50% tissue culture infective doses to perform the phenotypic assay. The majority of patients were treated with lamivudine (3TC) in combination with stavudine (d4T) (12 patients) or zidovudine (ZDV) (10 patients); further 3 patients had been treated with ZDV and didanosine (ddI). At enrollment after an average treatment period of 18 months (range, 6 to 74 months), median values of 2,000 HIV RNA copies/ml (range, <20 to 9,879 copies/ml) and 526 CD4+ cells/ml (range, 163 to 858 cells/ml) were detected. After enrollment, the 25 patients were monitored for a mean time of 7.7 (standard deviation, 1.5) months for clinical examination and evaluation of CD4+ cell count and plasma viral load. Informed consent was obtained from all subjects participating in this study.
Laboratory monitoring.
A blood sample was obtained from patients at enrollment for genotypic and phenotypic drug resistance analysis. Viral load and CD4 cell count were evaluated at base line and after a follow-up period.
HIV RNA was quantified with the Amplicor Monitor Assay (Roche Molecular System Branchburg, N.J.). When the level of HIV RNA in plasma dropped below 400 copies/ml, separate aliquots of plasma were assayed using the Roche Ultradirect Assay (limit of detection, 20 copies/ml).
HIV was isolated from CD8-depleted PBMC as previously described (2). Briefly, negative selection with magnetic beads (Miltenyi Biotec GmbH) was used to remove CD8+ T cells from PBMC, and the negative fraction, activated in the presence of human recombinant interleukin-2 (100 U/ml; Sigma) and phytohemagglutinin (5 μg/ml; Sigma), was cocultured with 107 CD8-depleted PBMC combined from two seronegative donors. Cultures, placed in a humidified chamber at 37°C with 5% CO2, were maintained for 60 days and monitored twice a week for p24 antigen production using a commercially available enzyme immunoassay (Abbott Laboratories, North Chicago, Ill.). A culture was considered positive if the concentrations of p24 exceeded 1,000 pg/ml in two consecutive determinations.
Positive supernatants were harvested by centrifugation and stored in liquid nitrogen.
Phenotypic assay.
HIV-1 strains isolated from PBMCs at enrollment were tested for sensitivity to NRTI according to AIDS Clinical Trials Group Consensus procedures (1). Briefly, phytohemagglutinin-stimulated donor PBMC (4 × 106 cells) were infected with 2 ml of medium containing viral stock adjusted to a multiplicity of infection of 2,000 50% tissue culture infective doses/ml. After a 2-h adsorption period, aliquots of the cells washed twice in phosphate-buffered saline were put into a 96-well plate containing five different concentrations of ZDV (0.001, 0.01, 0.1, 1, and 5 μM), 3TC (0.01, 0.1, 1, 5, and 25 μM), d4T (0.005, 0.05, 0.5, 5, and 25 μM), ddI (0.01, 0.1, 1, 5, and 25 μM), or abacavir (0.01, 0.1, 1, 5, and 25 μM). All culture assays were carried out in quadruplicate and monitored for p24 antigen production for 7 days after infection. Fifty percent inhibitory concentrations (IC50) of drug against virus were determined based on comparative growth of isolates in untreated control cultures. For phenotypic drug susceptibility testing, two strains, HIV-1MP27 and HIV-1GA61, isolated from antiretroviral-naïve patients in 1986, served as the susceptible control. An isolate, HIV-1ST543, with the complete Q151M multinucleoside resistance complex (A62V, V75I, F77L, F116Y, and Q151M) served as the resistant control.
Stocks of d4T and ddI were from Bristol Myers Squibb (Princeton, N.J.); ZDV, 3TC, and abacavir were from Glaxo Wellcome (Hertfordshire, United Kingdom). The ranges of concentrations tested were 0.005, 0.05, 0.5, 5, and 25 μM d4T; 0.01, 0.1, 1, 5, and 25 μM ddI; 0.001, 0.01, 0.1, 1, and 5 μM ZDV; 0.01, 0.1, 1, 5, and 25 μM 3TC; and 0.01, 0.1, 1, 5, and 25 μM abacavir.
An IC50 of more than 10-fold or less than 4-fold the average IC50 of the drug-susceptible reference strains indicated a resistance or susceptibility to drug, respectively. HIV isolates with a 4- to 10-fold IC50 were considered to have an indeterminate susceptibility to drug.
PCR and direct sequencing of RT gene.
HIV DNA was obtained from PBMC of HIV-seronegative blood donors infected with HIV strains isolated from the 25 enrolled patients. Total cellular DNA was prepared by resuspending 3 × 106 PBMC in 400 μl of lysis buffer containing 10 mM Tris-HCl (pH 8.3), 1 mM EDTA, 0.5% Triton X-100, 0.001% sodium dodecyl sulfate, and proteinase K (300 mg/ml). Lysed cells were digested with proteinase K overnight at 37°C, and at the end point the enzyme was inactivated for 15 min at 94°C. The lysates were stored at −20°C until they were used. In the first amplification for RT gene a fragment of 930 bp was obtained with the oligonucleotides JA99 (5"-GGG GGA ATT GGA GGT TTT ATC AAA G-3") and MM4 (5"-TTC TGT ATG TCA TTG ACA GTC CAG C-3") (0.2 mM), and 0.2 mM deoxynucleoside triphosphates. PCR was performed in 40 cycles, each consisting of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and elongation at 74°C for 30 s, plus a final extension at 74°C for 10 min. Five microliters was used as template for the second amplification performed under the same conditions by additional 30 cycles using the inner primers JA100 (5"-GAC CTA CAC CTG TCA ACA TAA TTG G-3") and MM3 (5"-GAT GGA GTT CAT AAC CCA TCC AAA G-3"). The final product consisted of a fragment of 750 bp (6).
For sequencing 200 ng of PCR product was used and a cycle sequencing kit (Prism Ready Reaction Big Dye Terminator; Applied Biosystems, Foster, City, Calif.) with DNA polymerase was used. The conditions for 25 cycles were 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. To obtain forward and reverse sequences of the PCR products, sequencing primers used in separate reactions were JA100 and MM3. Nucleotide sequencing was performed for codons 1 to 240 of the HIV-1 RT. Detection of sequencing products and generation of sequence data were done on an ABI 310 automated sequencer (Applied Biosystems).
Gene sequences were analyzed with DNAsis software and were related to the HIV-1LAI sequence (GenBank accession number K02013). Details of key mutations in the pol gene associated with reduced sensitivity to antiretroviral treatment were obtained from the literature (15).
Statistical analysis.
Patients were stratified in four groups according to the genotypic and phenotypic drug resistance profile. Statistical analysis of the variable related to the virological and imunological parameters of the four groups was performed by the analysis of variance for parametric data and by the Kruskal-Wallis test for nonparametric data. Student's t test was used for continuous measurements to test relationships in paired analysis.
RESULTS
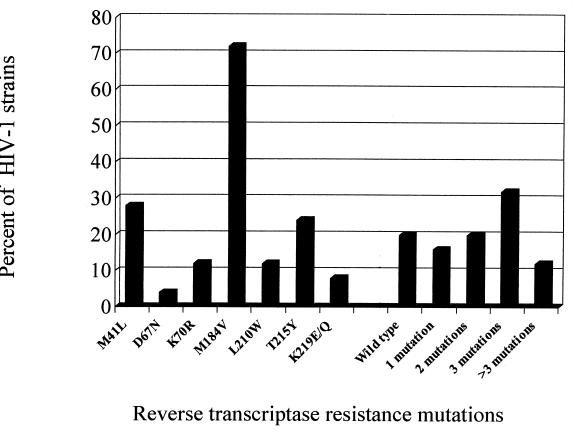
The genotypic mutations related to NRTI resistance of HIV-1 strains isolated from PBMC of the 25 patients are shown in Fig. 1. The most frequent resistance mutations were M184V (18 isolates) and M41L (7 isolates). No multidrug resistance mutations to NRTI (T69SSS and Q151M) were detected. Five patients (20%) harbored a wild-type strain, whereas 16 subjects (64%) carried an HIV-1 isolate with multiple mutations.
FIG. 1.
Prevalence of genotypic resistance mutations to NRTIs in 25 HIV-1 strains isolated from patients during antiretroviral therapy.
According to the resistance table for mutations in antiretroviral genes associated with drug resistance (15), 16 HIV-1 isolates harboring M41L, D67N, K70R, V118I, L210W, T215Y, K219Q, and M184V-R211K-L214F mutations alone or in combination were considered to be resistant to ZDV. Nineteen isolates harboring M184V or V118I in association with ZDV resistance mutations were defined as 3TC-resistant isolates, and three isolates with M184V combined with more than two ZDV resistance mutations were considered to be resistant to abacavir. No primary resistance mutations to ddI (K65R and L74V) and d4T (V75T) were detected.
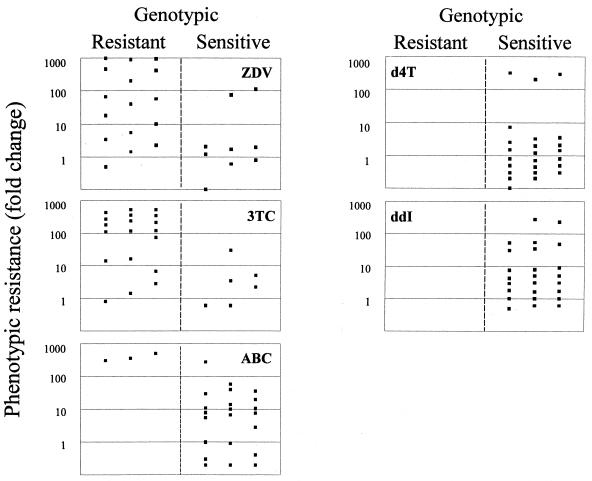
Sixteen out of 25 isolates showed discordance between the genotypic and phenotypic profiles for at least one drug. Particularly, eight isolates had a discordant genotypic-phenotypic resistance pattern for two drugs and one isolate had such a pattern for three drugs. The relationship between genotypic and phenotypic resistance of HIV-1 isolates to five NRTIs is shown in Fig. 2. A genotypic profile of resistance to ZDV with a sensitivity phenotype (<4-fold increase in IC50) was detected in four isolates (mean IC50, 0.012 ± 0.003 μM), and such a 3TC profile was detected in two isolates (mean IC50, 1.1±0.3 μM). On the other hand, an in vitro resistance to ZDV (>10-fold increase in IC50) with a sensitivity genotype was detected in two isolates (mean IC50, 0.47 ± 0.09 μM), and such profiles were detected for 3TC in one isolate (IC50, 38 μM), for abacavir in 10 isolates (mean IC50, 5.4 ± 8.0 μM), for ddI in 7 isolates (mean IC50, 10.4 ± 7.3 μM), and for d4T in 3 isolates (mean IC50, 22.2±2.6 μM). No phenotypic resistance to d4T was detected in HIV isolates with ZDV-like drug resistance mutations.
FIG. 2.
Comparison between genotypic and phenotypic resistance to analogue NRTIs of 25 HIV-1 strains. The following mutations were considered to confer resistance to the indicated drugs: M41L, D67N, K70R, L210W, T215(Y/F), and K219Q, ZDV; M184V, 3TC; M184V combined with three or more ZDV resistance mutations, abacavir; K65R and L74V, ddI; V75T, d4T.
A high phenotypic resistance (>50-fold increase in IC50) to the five NRTIs examined was detected in three isolates with M184V plus R211K alone (two isolates) or in combination with L214F (one isolate). Furthermore, the HIV-1 isolates with M184V mutation, alone or in combination with ZDV-like mutations (M41L, L210W, T215Y, or K219Q), showed a wide range of ddI or abacavir susceptibility. Nine isolates resistant to abacavir had M184V mutations alone (four isolates) or associated with one or two ZDV-like mutations. However, one isolate resistant to ddI (54-fold increase in IC50) showed only ZDV-like mutations (M41L-L210W-T215Y). Two isolates with V118I alone or in combination with M41L and T215Y mutations (one isolate) resulted in sensitivity to 3TC in the phenotypic drug assay. No E44D mutation was detected.
Among the three patients with a phenotypically susceptible strain, one subject with a plasma viremia level of 2,190 HIV RNA copies/ml had an HIV-1 isolate with resistance mutations (D67N, K70R, and K219E), whereas two subjects with undetectable viremia yielded a wild-type virus.
Table 1 shows the CD4+ cell count and HIV RNA viral load, at enrollment and after 8 months of follow-up, in patients according to genotypic and phenotypic drug resistance profile of HIV-1 isolates. No significant difference at base line and follow-up period was found among the four groups of patients. However, 14 patients with an HIV-1 isolate with a phenotypic and genotypic resistance profile had a significant increase in HIV RNA copy number (P = 0.004) and a significant CD4+ cell number decrease (P = 0.02) from the base line value. Five patients, with a phenotypic resistance and genotypic sensitivity profile of HIV isolates for at least one drug included in the patient treatment (three strains discrepant for d4T, one strain discrepant for ZDV, and one strain discrepant for 3TC), showed a significant increase of viral load (P = 0.04) and a decrease of CD4+ cell count. Conversely, four patients with phenotypic sensitivity and genotypic resistance profile of HIV isolates (two strains discrepant for ZDV and two strains discrepant for 3TC) showed a significant increase in CD4+ cell count (P = 0.02) with a persistent low HIV RNA copy number.
TABLE 1.
CD4+ cell count and HIV-RNA viral load, at enrollment and after 8 months of follow-up, for 25 patients according to genotypic and phenotypic drug resistance profile
| Resistance patterna
|
No. of patients | Median (range)/ml
|
|||||
|---|---|---|---|---|---|---|---|
| At enrollment
|
After 8 mo follow-up
|
||||||
| Genotype | Phenotype | CD4+ cell count | HIV RNA copies | CD4+ cell count | HIV RNA copies | ||
| R | R | 14 | 533 (163-730) | 2,200 (<20-6,500) | 445 (101-648) | 7,150 (610-26,700) | |
| S | R | 5 | 414 (213-858) | 4,666 (<20-9,879) | 368 (172-730) | 11,606 (500-27,144) | |
| R | S | 4 | 449 (295-602) | 550 (128-3,000) | 513 (336-704) | 470 (100-3,800) | |
| S | S | 2 | 324 (346-303) | 240 (<20-460) | 531 (691-372) | 470 (430-510) | |
Genotype and phenotype evidence of resistance (R) or sensitivity (S) to at least one drug in the patient treatment.
Virological and immunological data of patients with undetectable viral loads (<400 HIV RNA copies/ml) at enrollment are shown in Table 2. A relevant increase in viral load was detected in one patient (patient 18) with a resistant phenotypic profile for the two NRTIs (d4T and 3TC) included in the treatment.
TABLE 2.
Genotypic and phenotypic profiles of six patients with plasma HIV RNA levels of <400 copies/ml at enrollment
| Patient no. | Treatment | Enrollment profile
|
Follow-up profile
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype
|
Phenotype
|
CD4+ cells/ml | HIV-RNA copies/ml | Mo. | CD4+ cells/ml | HIV-RNA copies/ml | ||||||
| Mutation(s) | Interpretationa | Susceptibilitiesb | Interpretationc | |||||||||
| 9 | ZDV+ddI | D67N, K70R, K219E | ZDV resistant, ddI sensitive | ZDV, 2.3; ddI, 1 | ZDV sensitive, ddI sensitive | 697 | 98 | 9 | 579 | <20 | ||
| 11 | ZDV+3TC | None | ZDV sensitive, 3TC sensitive | ZDV, 2.1; 3TC, 9.3 | ZDV sensitive, 3TC indeterminate | 346 | <20 | 8 | 691 | 430 | ||
| 13 | d4T+3TC | M184V | d4T sensitive, 3TC resistant | d4T, 0.9; 3TC, 16.6 | d4T sensitive, 3TC resistant | 708 | <20 | 8 | 836 | 610 | ||
| 18 | d4T+3TC | M184V, R211K, L214F | d4T sensitive, 3TC resistant | d4T, 313; 3TC, 354 | d4T resistant, 3TC resistant | 858 | <20 | 10 | 675 | 7,400 | ||
| 27 | ZDV+3TC | M184V, L214F | ZDV sensitive, 3TC resistant | ZDV, 1.4; 3TC, 14 | ZDV sensitive, 3TC resistant | 602 | 300 | 7 | 704 | 440 | ||
| 43 | ZDV+3TC | M184V | ZDV sensitive, 3TC resistant | ZDV, 0.1; 3TC, 1.4 | ZDV sensitive, 3TC sensitive | 504 | 128 | 7 | 590 | 100 | ||
See reference 15.
Susceptibility for each indicated drug is expressed as the fold IC50 increase relative to drug susceptible reference strains.
Isolates with IC50 increases of 10-fold or more are interpreted as drug resistance. Isolates with IC50 increases of 4- to 10-fold are interpreted as drug intermediate. Isolates with IC50 increases of less than 4-fold are interpreted as drug sensitive.
DISCUSSION
In this study a discordance between the genotypic and phenotypic drug resistance profiles in 64% of HIV-1 strains isolated from 25 patients treated with two NRTIs was detected. Discordance between phenotypic and genotypic analyses has previously been demonstrated. Particularly, in two studies (4, 13) a high prevalence (26 and 15%, respectively) of reduced drug susceptibility to certain antiretroviral drugs was not associated with the presence of recognized drug resistance mutations. Differently from our study, these reports analyzed patients during primary HIV infection and who were naïve to antiretroviral therapy. Moreover, the phenotypic analyses were performed with recombinant virus assays.
The phenotypic susceptibility measured for a certain set of resistance mutations is probably the result of several factors, including the inherent sequence variability of patients strains, the reproducibility of the phenotypic test used, the presence of minority genotypes, and the sensitivity of the assay used to map the mutations under investigation in the presence of mixed genotypes (8, 16). Furthermore, genotypic mutational pattern does not necessarily correspond to a single viral clone, because the same pattern may result from the superimposition of the genotypes from different clones. It may be quite misleading to assume that all mutations belong to the same clone, as is often done in a more conservative therapeutic approach. On the other hand, given the low limit of detection for small viral populations and the known association of some primary and secondary mutations, it is unlikely that each mutation represents a separate viral clone. Intermediate interpretations could explain discordant clinical results in patients with similar genotypic resistance patterns.
A phenotypic profile of sensitivity to ZDV and 3TC was found in several isolates in spite of a demonstrated genotypic resistance pattern. Conversely, four wild-type strains showed a reduced sensitivity to ZDV (two isolates), 3TC (one isolate), and abacavir (one isolate). Moreover, a reduced susceptibility to abacavir has been detected in nine HIV-1 isolates with the M184V mutation alone or in combination with one or two ZDV-like resistant mutations. In a previous study, the virologic response to abacavir was inversely associated with the number of ZDV resistance mutations, and patients with isolates dually resistant to ZDV and 3TC had the worst response to abacavir, suggesting a cross-resistance among these drugs (10).
In isolates with a reduced phenotypic sensitivity to d4T (three strains) and to ddI (seven strains), no resistance mutation for these two drugs was found. The emergence of mutants resistant to d4T and ddI is a rare event, mainly described during monotherapy or prolonged virological therapy failure (5, 12, 14). Nonetheless, patients heavily pretreated with two NRTIs could have diminished virologic response to d4T-containing regimens (11). Particularly, in this study three patients failing therapy with d4T and 3TC yielded an HIV isolate with high-level phenotypic resistance to d4T in the absence of known mutations correlated with resistance to the drug.
A possible bias in this study is represented by the unsuccessful detection by genotypic analysis of minor subpopulations (minority viral quasispecies) of highly resistant virus subsequently selected in the presence of drug during the phenotype assay. On the other hand, a sensitive phenotypic profile of an HIV strain with mutations correlated to drug resistance could represent impaired fitness of a resistant mutant or the result of antagonistic interactions of some mutational patterns.
In our study, a sensitive phenotype, even in the presence of NRTI-resistant mutations, has a favorable predictive value of antiviral drug response. Conversely, some patients experiencing virologic failure during antiretroviral therapy harbored HIV-1 strains with a resistance of phenotype to antiretroviral drugs without any known drug resistance mutations. This study has been performed with patients treated with two NRTIs, whereas the current guidelines for the use of antiretroviral agents in HIV-infected adults and adolescents recommend the use of a triple therapy. Nevertheless, the backbone of antiretroviral regimens usually includes NRTIs, and the implication of a genotypic-phenotypic resistance to these drugs remains relevant for the clinical outcome. Thus, in adherent patients failing therapy with analogue nucleosides, a phenotypic analysis should be performed in spite of an HIV genotypic sensitivity pattern.
The current debate over the relative merits of phenotypic versus genotypic analysis suffers from a lack of large-scale, longitudinal studies aimed at correlating primary and secondary resistance mutations, phenotypic resistance patterns, and sustained virologic patient responses.
Acknowledgments
This work was supported by grants from the Italian Ministry of Health, Istituto Superiore di Sanità, AIDS Research Project 1997; from the Cassa di Risparmio of Verona, Vicenza, Belluno, Italy; and from the Ancona Foundation, Health Projects 1996-1997.
We thank Marco Montano for technical assistance.
REFERENCES
- 1.ACTG Virology Technical Advisory Committee and the Division of AIDS, National Institute of Allergy and Infectious Diseases. 1994. The ACTG virology manual for HIV laboratories. NIH publication 3828. National Institutes of Health, Bethesda, Md.
- 2.Andreoni, M., S. G. Parisi, L. Sarmati, F. Nicastri, L. Ercoli, G. Mancino, G. Sotgiu, M. Mannazzu, M. Trevenzoli, G. Tridente, E. Concia, and A. Aceti. 2000. Cellular proviral HIV-DNA decline and viral isolation in naive subjects with <5000 copies/ml of HIV-RNA and >500 × 10(6)/1 CD4 cells treated with highly active antiretroviral therapy. AIDS 14:23-29. [DOI] [PubMed] [Google Scholar]
- 3.Baxter, J. D., D. L. Mayers, D. N. Wentworth, J. D. Neaton, M. L. Hoover, M. A. Winters, S. B. Mannheimer, M. A. Thompson, D. T. Abrams, B. J. Brizz, J. P. Ioannidis, and T. C. Merigan. 2000. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. CPCRA 046 Study Team for the Terry Beirn Community Programs for Clinical Research on AIDS. AIDS 14:F83-F93. [DOI] [PubMed] [Google Scholar]
- 4.Boden, D., A. Hurley, L. Zhang, C. M. D. Yunzhen, G. Yong, E. Jones, J. Tsay, J. Ip, C. Farthing, K. Limoli, N. Parkin, and M. Markowitz. 1999. HIV-1 drug resistance in newly infected individuals. JAMA 282:1135-1141. [DOI] [PubMed] [Google Scholar]
- 5.Coakley, E. P., J. M. Gillis, and S. M. Hammer. 2000. Phenotypic and genotypic resistance patterns of HIV isolates derived from individuals treated with didanosine and stavudine. AIDS 14:F9-F15. [DOI] [PubMed] [Google Scholar]
- 6.Di Stefano, M., F. Sabri, T. Leitner, B. Svennerholm, L. Hagberg, G. Norkrans, and F. Chiodi. 1995. Reverse transcriptase sequence of paired isolates of cerebrospinal fluid and blood from patients infected with human immunodeficiency virus type 1 during zidovudine treatment. J. Clin. Microbiol. 33:352-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duran, J., P. Clevenbergh, P. Halfon, P. Delgiudice, S. Porsin, P. Simonet, N. Montagne, C. A. B. Boucher, J. M. Schapiro, and P. Dellamonica. 1999. Drug-resistance genotyping in HIV-1 therapy: the VIRADAPT randomised controlled trial. Lancet 353:2195-2199. [DOI] [PubMed] [Google Scholar]
- 8.Hertogs, K., M. P. de Bethune, V. Miller, T. Ivens, P. Schel, A. Van Cauwenberge, C. Van Den Eynde, V. Van Gerwen, H. Azijn, M. Van Houtte, F. Peeters, S. Staszewski, M. Conant, S. Bloor, S. Kemp, B. Larder, and R. Pauwels. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42:269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirsch, M. S., F. Brun-Vezinet, R. T. D'Aquila, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, C. Loveday, J. W. Mellors, B. Clotet, B. Conway, L. M. Demeter, S. Vella, D. M. Jacobsen, and D. D. Richman. 2000. Antiretroviral drug resistance testing in adults with HIV infection. Recommendations of an international AIDS Society USA panel. JAMA 283:2417-2426. [DOI] [PubMed] [Google Scholar]
- 10.Jorgensen, L. B., T. L. Katzenstein, J. Gerstoft, L. R. Mathiesen, C. Pedersen, and C. Nielsen. 2000. Genotypic and phenotypic nevirapine resistance correlates with virological failure during salvage therapy including abacavir and nevirapine. Antivir. Ther. 5(3):187-194. [PubMed]
- 11.Katlama, C., M. A. Valantin, S. Matheron, A. Coutellier, V. Calvez, D. Descamps, C. Longuet, M. Bonmarchand, R. Tubiana, M. De Sa, R. Lancar, H. Agut, F. Brun-Vezinet, and D. Costagliela. 1998. Efficacy and tolerability of stavudine plus lamivudine in treatment-naive and treatment-experienced patients with HIV-1 infection. Ann. Intern. Med. 129:525-531. [DOI] [PubMed] [Google Scholar]
- 12.Lin, P. F., H. Samantha, R. E. Rose, A. K. Patick, J. Trimble, C. M. Bechtold, D. R. Revie, N. C. Khan, M. E. Federici, H. Li, A. Lee, R. E. Anderson, and R. J. Colonno. 1994. Genotypic and phenotypic analysis of human immunodeficiency virus type 1 isolates from patients on prolonged stavudine therapy. J. Infect. Dis. 170:1157-1164. [DOI] [PubMed] [Google Scholar]
- 13.Little, S. J., E. S. Daar, R. T. D'Aquila, H. P. Keiser, E. Connick, J. M. Whitcomb, N. S. Hellmann, C. J. Petropoulos, L. Sutton, J. A. P. Eric, S. Rosenberg, R. A. Koup, B. D. Walker, and D. D. Richman. 1999. Reduced Antiretroviral drug susceptibility among patients with primary HIV infection. JAMA 282:1142-1149. [DOI] [PubMed] [Google Scholar]
- 14.Milazzo, L., S. Rusconi, L. Testa, S. La Seta-Catamancio, M. Galazzi, S. Kurtagic, P. Citterio, M. Gianotto, A. Grassini, F. Adorni, A. D'Arminio- Monforte, M. Galli, and M. Moroni. 1999. Evidence of stavudine-related phenotypic resistance among zidovudine-pre-treated HIV-1-infected subjects receiving a therapeutic regiment of stavudine plus lamivudine. J. Acquir. Immune Defic. Syndr. 22:101-103. [DOI] [PubMed] [Google Scholar]
- 15.Schinazi, R. F., B. A. Larder, and J. Mellors. 2000. Mutations in retroviral genes associated with drug resistance: 2000-2001 update. Int. Antivir. News 8:65-91. [Google Scholar]
- 16.Van Laethem, K., K. Van Vaerenbergh, J.-C. Schmit, S. Sprecher, P. Hermans, V. De Vroey, R. Schuurman, T. Harrer, M. Witvrouw, E. Van Wijngaerden, L. Stuyver, M. Van Ranst, J. Desmyter, E. De Clercq, and A. M. Vandamme. 1999. Phenotypic assays and sequencing are less sensitive than point mutation assays for the detection of resistance in mixed HIV-1 genotypic populations. J. Acquir. Immune Defic. Syndr. 22:107-118. [DOI] [PubMed] [Google Scholar]