Abstract
A regulatory system for the in-depth study of gene functions in higher eukaryotic cells has been developed. It is based on the tetracycline-controlled transactivators and reverse tTA, which were remodeled to discriminate efficiently between two different promoters. The system permits one to control reversibly the activity of two genes, or two alleles of a gene, in a mutually exclusive way, and also allows one to abrogate the activities of both. This dual regulatory circuit, which can be operated by a single effector substance such as doxycycline, overcomes limitations of conventional genetic approaches. The conditional mutants that can now be generated will be useful for the study of gene function in vitro and in vivo. In addition, the system may be of value for a variety of practical applications, including gene therapy.
The potential to control the activity of individual genes in higher eukaryotes quantitatively and reversibly has opened up new perspectives for the in-depth study of gene function. A decisive feature of this approach is that phenotypically the state of a null-mutation can be induced without altering the target gene itself, while the possibility to subsequently return to the wild-type state makes the mutant phenotype truly conditional. Equally important is the quantitative nature of this approach: Gene activities can be varied by small increments, a feature that is of particular interest for the study of gene products that are involved in a variety of intracellular equilibria. Whereas in general they will participate in such interactions with different affinities, a gradual change of their intracellular concentration may reveal phenotypes and targets of interaction that remain obscure when the respective gene is simply inactivated.
Here we describe a regulatory system that permits one to control reversibly the activity of two genes in a mutually exclusive way. It thus allows one to switch between the expression, e. g., of two alleles of a gene or of two independent genes of interest and allows one to analyze the accompanying phenotypic changes. Moreover, both genes under study can be kept inactive. All three states of gene activity can be brought about simply by varying the concentration of a single nontoxic effector compound.
This development is based on the two tetracycline (Tc)-controlled transcription activation systems (1, 2) that have found widespread application in the study of gene function and have been used successfully at the level of cultured cells of mammalian, plant (for review, see ref. 3), or amphibian (4) origin, as well as in whole organisms such as Saccharomyces cerevisiae (5), Drosophila melanogaster (6), plants (7, 8), and mammals such as mice (9–13) and rats (14). Exploiting the differential susceptibilities of the previously described Tc-controlled transactivator (tTA) toward Tcs, a dual regulatory system has been developed that allows transactivators to discriminate between two transcription units and thus to control two gene activities in a mutually exclusive way. The system described herein will enable an analysis of in vivo gene function at an unprecedented level of resolution.
MATERIALS AND METHODS
Generation of tTAs with Altered DNA-Binding Specificity.
DNA fragments encoding the mutated DNA-binding domain of TetR conferring specificity for 4C tetO or 6C tetO sequences were obtained from plasmids pWH520[EA37PQ39YM42] and pWH520[ES37WR43HN44], respectively, upon cleavage of the plasmid DNA with XbaI and EcoNI. The electrophoretically purified fragments were inserted into pUHD15–1 (1), which had been digested with XbaI and EcoNI resulting in pUHD15–14 and pUHD15–16 that encode tTA4C and tTA6C, respectively. In an analogous way, the fragments encoding the mutated HTH motifs were inserted into plasmid pUHD20–1 carrying the tTA2 gene (15) yielding the plasmids pUHT2–2 and pUHT2–3. These plasmids encode tTA24C and tTA26C, respectively, which differ from the above transactivators by containing three minimal transcriptional activation domains instead of the C-terminal portion of VP16 (15). The reverse tTA (rtTA)26C was generated by transferring the mutations of the rtTA gene that confer DNA binding in the presence of doxycycline (Dox) (2) into the coding sequence of tTA26C. Thus the moiety conferring the reverse phenotype was isolated from pUHD17–1 by EcoNI/Eco47III cleavage and introduced into plasmid pUHT2–3 digested previously with EcoNI/Eco47III. The resulting plasmid pUHrT2–1 carries the gene for rtTA26C.
Construction of the Coding Sequence for tTA2E4C.
The sequence encoding the HTH motif specific for 4C tet operators was retrieved from pUHT2–2 by PCR (upstream primer: 5′-GACACCGGGACCGATCCAGGC-3′; downstream primer: 5′-ATTTTTCACGTGCCACATCAATGTCTGC-3′). The amplified material that encodes amino acids 1 to 44 of TetRB was recovered after cleavage with XbaI and PmlI. The DNA fragment coding for amino acid 45 to 211 of TetRE was recovered from pWH610 also by PCR (upstream primer: 5′-GTATTGGCACGTGCGCAACAAGC-3′; downstream primer: 5′-GGCGTCGGCCGGCTTATTACCATCCTCAATGGG-3′) and digested with PmlI and NaeI. The two fragments were simultaneously inserted into pUHD20–1 (15) previously cleaved with XbaI and NaeI. The resulting DNA sequence encodes a chimeric Tet repressor (TetRB/E) that is fused to three minimal activation domains and that specifically recognizes 4C tetO sequences, i.e. tTA2E4C. The corresponding plasmid is designated pUHT2–1. The backbone of all transactivator plasmids described herein is identical to pUHD15–1 (1).
Generation of Ptet4 and Ptet6.
Promoter sequences responsive for transactivators binding to 4C and 6C tetO sequences were obtained by oligomerizing synthetic 42-bp double stranded oligonucleotides containing the 4C (5′-TCCCTGTCAGTGACAGAGA-3′, upper strand) and the 6C (5′-TCCGTATCAGTGATACAGA-3′, upper strand) version of the 19-bp tet operator as described previously (1). Heptamerized oligonucleotides were inserted into the XhoI cleavage site of pUHD13–2 (1), yielding plasmids pUHC13–8 and pUHC13–9 that contain Ptet4 and Ptet6, respectively, directing the transcription of the luc gene. Ptet6 was also transferred to pUHG16–3 (16) by means of XhoI/SacII cleavage yielding pUHG16–9, where it controls the expression of the lacZ gene.
Quantitation of Luciferase Activity and DNA Retardation Analysis.
The determination of luciferase activity in cell extracts is described in detail in ref. 1. Analysis of protein DNA complexes by retardation experiments were performed as described in ref. 15. The 4C and 6C operator sequences were isolated from pUHC13–8 and pUHC13–9 as 42 bp TaqI fragments, respectively. The double-stranded asymmetric 4C/6C tetO variant was synthesized. Protruding ends were filled in by T4 DNA polymerase in the presence of [α-32P]dCTP.
Generation of HeLa Cell Line HT/rT-1.
A cell line constitutively synthesizing both transactivators, tTA2E4C and rtTA2B6C, was generated by first cotransfecting HeLa cells by means of the calcium phosphate method with plasmids pUHT2–1 and pSV2neo (17). Twenty clones resistant to G418 (500 μg/ml) were isolated and assayed for constitutive production of tTA2E4C by transient transfection with luciferase reporter plasmid pUHC13–8, as described previously (1). Of these clones, six showed high luciferase activity that was susceptible to Dox. Clone HA-6, which exhibited a high transactivation potential (2,700-fold in transient assays) was transfected with plasmids pUHrT2–1 and pHMR272 (15) conferring resistance to hygromycin B. Twelve clones resistant to hygromycin (300 μg/ml) were isolated and assayed for constitutive production of rtTA2B6C by transient transfection of the cells with pUHC13–9. Of three clones that exhibited efficient and selective activation of both Ptet4 and Ptet6 in a Dox-dependent manner, HeLa cell line HT/rT-1 was derived by subcloning.
Indirect Immunofluorescence.
HT/rT-1 cells were seeded into two-well chamber slides (Nunc) and grown to 30–40% confluency. Cultures were transfected with a DNA mixture containing 0.4 μg of luciferase reporter construct pUHC13–8, 0.5 μg of β-galactosidase expression vector pUHG16–9, and 0.6 μg of pUC18 as carrier DNA per well and kept in the absence of Dox or in its presence at 30 ng/ml or at 3 μg/ml. After 36 h, the cells were analyzed by indirect immunofluorescence according to ref. 18, with the following modifications: Cells were fixed in 3.7% formaldehyde/PBS for 10 min and were permeabilized by treatment with PBS containing 0.2% Triton X-100 for 5 min. On blocking of unspecific sites with PBS/10% BSA (fraction V, Sigma), the cells were incubated with a mixture of polyclonal anti-luciferase antibodies from rabbit serum and a monoclonal mouse-anti-β-galactosidase antibody (Promega), both diluted 1:750 in PBS/2% BSA. After additional blocking for 15 min, antigen–antibody complexes were detected by a mixture of donkey-anti-rabbit IgG conjugated with dichlorotriazinyl aminofluorescein (Sigma) and donkey-anti-mouse IgG conjugated with Cy3 (Sigma), both diluted 1:1,000 in PBS/2% BSA. Immunocomplexes were visualized by fluorescence microscopy (Leica DM-RXE) by using a charge-coupled device camera system (Photometrics CH250, Tucson, AZ).
RESULTS
Experimental Strategy.
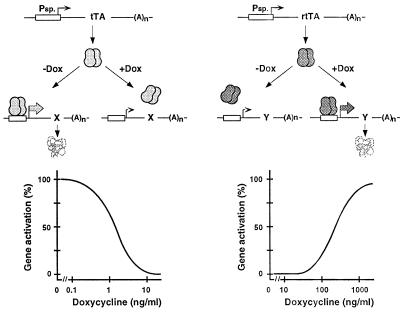
Two complementary versions of the Tc-controlled transcription activation system have been developed (1, 2). As detailed in Fig. 1, tTA activates transcription from a minimal RNA polymerase II promoter fused to tet operator (tetO) sequences in absence of Tc, whereas in presence of the antibiotic, binding of the transactivator to the promoter and thus its activation are prevented. By contrast, the rtTA requires the presence of certain Tc derivatives for the binding to tetO and thus activates transcription only in presence of , for example, Dox or anhydrotetracycline (ATc). Interestingly, tTA and rtTA differ in their susceptibility to Dox and ATc. As indicated in Fig. 1, the tTA system responds to Dox at concentrations between 0.1 and 10 ng/ml, whereas the dose response of rtTA shows an effective range for Dox of between 100 and 3,000 ng/ml. These findings suggest that when placed under the control of tTA or rtTA, respectively, the activity of two genes may be controlled in a mutually exclusive way just by varying the Dox concentration. Moreover, using the proper concentration of Dox, the expression of both genes would be prevented.
Figure 1.
Schematic outline of the Tet regulatory systems. (Upper Left) The mode of action of the tTA. tTA, a fusion protein between the Tet repressor of the Tn10 Tc resistance operon from Escherichia coli and the C-terminal portion of VP16 from Herpes simplex virus, binds in the absence of the effector molecule Dox to multiple tet operator sequences (tetO) placed upstream of a minimal human cytomegalovirus promoter and activates transcription of gene x. Addition of Dox prevents tTA from binding and thus the initiation of transcription. (Lower Left) The dose response of Dox on the tTA-dependent gene expression. Gene activity is maximal in the absence of the antibiotic, whereas increasing effector concentrations gradually decrease expression to background levels at concentrations ≥10 ng/ml. (Upper Right) The mechanism of action of the rtTA. rtTA is identical to tTA with the exception of four amino acid substitutions in the TetR moiety that convey a reverse phenotype. rtTA requires Dox for binding to tetO sequences to activate transcription of gene y. (Lower Right) The dose response of Dox on the rtTA-dependent transcription activation. There is no gene expression in the absence of the antibiotic. By increasing the effector concentration beyond 100 ng/ml Dox, rtTA-dependent gene expression is gradually stimulated.
To convert the two Tet regulatory systems into a “double switch” as outlined, two conditions have to be fulfilled: (i) The two transactivators tTA and rtTA have to be altered to discriminate efficiently between two new cognate operator sequences. These new operator sequences would then specify a tTA- and a rtTA-responsive promoter, respectively. (ii) Both transactivators function as dimeric proteins. Accordingly, the coproduction of the tTA and rtTA proteins would also result in tTA/rtTA heterodimers that would reduce the effective intracellular concentration of the two transactivators. Moreover, the heterodimers may reduce the apparent discrimination between the two novel operator sequences. Therefore, dimerization properties of the TetR moiety have to be altered to warrant the exclusive formation of tTA and rtTA homodimers.
Below we describe the stepwise conversion of tTA and rtTA into transactivators with the required properties and demonstrate their potential by controlling indicator genes whose expression products can be monitored with high sensitivity.
Altering the DNA-Binding Specificity of tTA.
Thorough structure–function analyses of the interaction between TetR and tetO have revealed (19, 20) that some mutations introduced into the helix-turn-helix motif of TetR that abolish the binding to the tetO can be compensated by alterations within the operator sequence. Two mutant repressor/operator combinations exhibited particularly remarkable properties. By exchanging the three amino acids, E, W, and H, at positions 37, 43, and 44 of TetR for S, R, and N, respectively, a mutant protein was obtained that specifically recognizes an operator sequence where the G at position 6 of the 19-bp-long sequence of dyad symmetry has been replaced by a C (21). Similarly, the exchange of amino acids E, P, and Y at positions 37, 39, and 42 for A, Q, and M, respectively, yielded a TetR mutant that binds with high specificity to a tetO sequence that contains a C instead of a T at position 4 (22). The two TetR mutants were converted into transcriptional activators for RNA polymerase II promoters by C-terminal fusions with a portion of Herpes simplex virus protein 16 (VP16), as described previously (1), resulting in tTA4C and tTA6C.
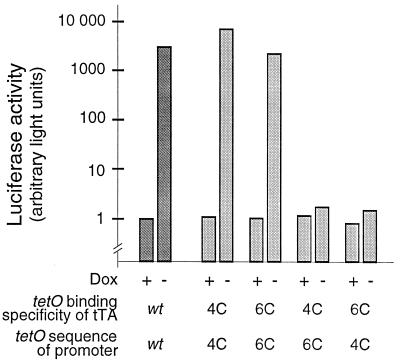
Promoters specifically responsive to these new transactivators were obtained by combining a minimal promoter sequence derived from the human cytomegalovirus promoter IE with heptamerized 4C and 6C operator sequences, respectively, as reported previously (1). Placed upstream of the luciferase reporter gene (luc), the two promoters, designated Ptet4 and Ptet6, were used to examine the specificity of the new transactivator promoter combinations in transient expression experiments. The results in Fig. 2 show that the new redesigned control elements have the same regulatory potential as the originally described tTA system, i. e., under the experimental conditions used they achieve regulation factors between 2,000 and 8,000. Most importantly, when Ptet6 was exposed to tTA4C and Ptet4 to tTA6C, the background activities were elevated only marginally (less than 2-fold), demonstrating that both transactivators discriminate between the two promoters better than 1,000-fold.
Figure 2.
Specificity of transcription activation by transactivators with new DNA-binding properties. HeLa cells kept in the absence or presence of Dox (3 μg/ml) were transiently transfected with a combination of plasmids encoding transactivators and plasmids containing the luc gene under control of tTA/rtTA responsive promoters, as indicated. The operator binding specificities of the transactivators were wt, 4C, or 6C, respectively. Correspondingly, the promoters driving the luc gene expression contained wt, 4C, or 6C tet operator sequences (PCMV*-1, ref. 1, Ptet4, Ptet6). For normalization of transfection efficiencies, all DNA mixtures contained also pUHD16–1 (23) constitutively expressing the lacZ gene. After 30 h, luciferase activity in all extracts was measured and normalized to β-galactosidase activity. The figure shows transactivation of PhCMV*-1, Ptet4, and Ptet6 by the differently specified tTAs.
To reduce side effects known to occur when VP16 fusion proteins or other transcription factors are produced above certain intracellular threshold concentrations, the TetR mutants were fused to three 12 amino acid-long minimal activation domains. Fusions between TetR and such minimal domains are tolerated at higher intracellular concentrations when compared with the original TetR VP16 fusions (15). The two resulting transactivators, designated tTA24C and tTA26C, exhibited the same specificity for their promoters and showed the same regulatory factors as their tTA parent constructs (data not shown). They were used in all further experiments described henceforth.
Altering the Dimerization Properties of tTA24C and tTA26C.
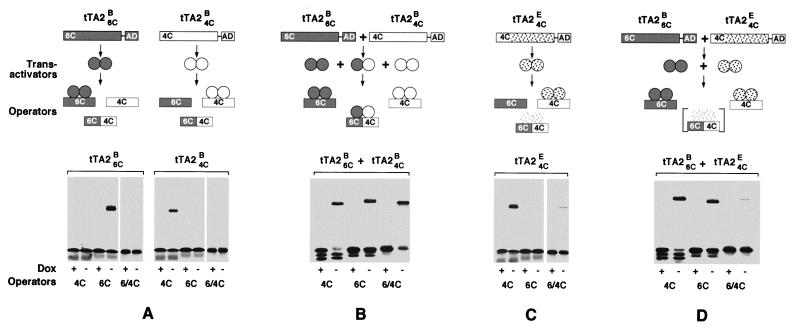
When tTA24C and tTA26C are produced simultaneously in HeLa cells, homodimeric as well as heterodimeric transactivators will form, as is revealed by DNA retardation experiments where the transactivators are exposed to 4C and 6C tet operator sequences as well as to an operator composed of a 4C and a 6C halfside (6C/4C). Whereas the two homodimeric transactivators produced individually in HeLa cells discriminate well between the operator sequences offered (Fig. 3A), coexpression of the tTA24C and the tTA26C gene leads to heterodimeric transactivators that bind the 6C/4C operator (Fig. 3B). These heterodimers may not only reduce the specificity of the system; they also lower the concentration of effective homodimers that would necessitate an overproduction of transactivators above the threshold required. Therefore, to avoid heterodimer formation, the dimerization specificities of the transactivators were altered.
Figure 3.
DNA-binding and dimerization specificities of various Tc-controlled transactivators. (A) (Upper) Schematic outline of the genes encoding tTA2B6C and tTA2B4C. The 4C and 6C DNA-binding domains are located N-, the transcriptional activation domain (AD) C-terminally. When exposed to operator DNA, the two transactivators discriminate between 6C (dark), 4C (light), and 6C/4C (dark/light) operator DNA. (Lower) DNA retardation experiments (for experimental detail, see ref. 15) in which the three radio-labeled tet operator DNAs were exposed to tTA2B6C and tTA2B4C, respectively, in presence (1 μg/ml) and absence of Dox. Complex formation was detected exclusively between transactivators and their cognate operator sequences in absence of Dox. Neither tTA2B6C nor tTA2B4C binds the hybrid 6C/4C operator DNA. (B) Demonstration of heterodimer formation between tTA2B6C and tTA2B4C. (Upper) Outline of the experiment in which the two transactivators were produced simultaneously in HeLa cells. The expected heterodimers bind to the hybrid 6C/4C operator DNA, as verified in the DNA retardation experiment depicted in the lower part where 6C/4C operator DNA is shifted in absence of Dox. All designations and symbols are as in A. (C) Interaction of tTA2E4C with its cognate operator DNA. (Upper) Schematic outline of the experiment as in A and B; the E-class Tet repressor is indicated by stipples. The retardation experiment (Lower) shows that tTA2E4C discriminates well between 4C and 6C operator DNA. A faint band is, however, visible also with the composite 6C/4C operator, indicating some affinity between tTA2E4C and the 6C/4C operator DNA. (D) The E- and B-type transactivators do not heterodimerize (outlined, Upper). The two transactivators tTA2E4C and tTA2B6C were produced simultaneously as in B and were exposed to the various operator DNAs. Whereas both selectively bind their cognate operators, there is no sign of heterodimerization. The faint band indicating a 6C/4C operator DNA transactivator complex is caused by the low affinity of tTA2E4C to this DNA, as seen in C.
The Tet repressors used so far for the generation of Tc-controlled transactivators belong to the B class, one of several classes of naturally occurring Tet repressors that resulted from comparisons of primary sequences (24). Detailed structural and functional analyses of Tet repressors (20, 25) indicated sequence elements involved in the dimerization and suggested that monomers of repressors of class E and D may be least capable of forming dimers with repressors of class B. It was, therefore, attempted to confer the dimerization specificities of the E and D repressors to tTA24C and tTA26C. Here we describe the transfer of the dimerization specificity of the Tet repressor of class E (TetRE) to tTA24C. In a first step, the N-terminal portion of TetRE conferring DNA-binding specificity was replaced by the corresponding domain of tTA2B4C. The fusion point of this chimeric repressor lies between amino acids 44 and 45, a region that connects the DNA-binding domain with the protein core (25). Such fusions have previously been shown to transfer operator-binding specificities to TetR core proteins (26). To convert TetRE4C into a transcriptional activator, it was combined with three minimal activation domains (FFF) as described above for TetRB, resulting in tTA2E4C. When the specificity of this transactivator was examined by DNA retardation experiments, it showed excellent discrimination between the 4C and the 6C operator sequences (Fig. 3C). As with tTA2B4C, no interaction with the wt tetO was detected (data not shown). However, a faint band indicating a low-affinity interaction between tTA2E4C and the 4C/6C hybrid operator is revealed in Fig. 3C.
Whether monomers of tTA2B6C and tTA2E4C would form heterodimers was examined by producing both transactivators simultaneously in HeLa cells and subjecting the respective cell extracts to DNA retardation experiments. As shown in Fig. 3D, when exposed to 4C or 6C operator DNA in absence of Dox, the expected complexes are formed between the transactivators tTA2B6C and tTA2E4C and their respective operators. However, when the extracts were exposed to 6C/4C operator DNA, no tTA2B6C/tTA2E4C heterodimers were detected (Fig. 3D). The faint band visible in the 6C/4C lane reflects the residual binding of tTA2E4C to this sequence, as observed in Fig. 3C. These findings show that no heterodimers are detectable when the two transactivators tTA2B6C and tTA2E4C are synthesized simultaneously in a cell.
Introduction of the Reverse Phenotype into tTA2B6C.
The transactivators and their respective DNA target sequences described so far exhibit two essential properties required for a dual regulatory system, as initially outlined: they discriminate efficiently between their respective operators and do not heterodimerize. To introduce the reverse phenotype for Dox-dependent operator binding into one of them, the mutations conferring the reverse phenotype were transferred from rtTA (2) to tTA2B6C. The resulting sequence codes for rtTA2B6C. Interestingly, while both the 4C and the 6C binding specificities could be combined with tTA2B to yield well-functioning transactivators, only rtTA2B6C but not rtTA2B4C exhibited the discrimination and induction properties required (data not shown).
Mutually Exclusive Regulation of Two Genes by tTA2E4C and rtTA2B6C.
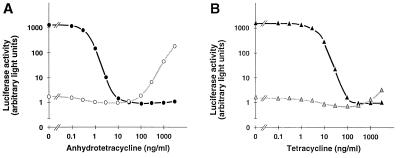
To examine the characteristics of the two new control systems in vivo, two types of experiments were conceived. In the first, both transactivators tTA2E4C and rtTA2B6C were transiently produced in HeLa cells in the presence of the luc gene controlled by either Ptet4 or Ptet6. These cultures were incubated at different concentrations of either ATc or Tc, and luciferase activity was monitored. The results of this experiment are summarized in Fig. 4. They show that luciferase activity controlled by Ptet4 is reduced to background levels at ATc concentrations ≥10 ng/ml because of the inactivation of tTA2E4C (Fig. 4A). By contrast, when the luc gene is controlled by Ptet6, luciferase is detected only at ATc concentrations ≥100 ng/ml, which enable rtTA2B6C to bind to Ptet6. These dose-response studies clearly show that the two regulatory systems can be operated separately and quantitatively. They reveal also a sizable window of ATc concentration where both transactivators are prevented from binding to their respective promoters and thus both the Ptet4- and the Ptet6-controlled genes are inactive. The different susceptibility of the two transactivators for Tcs can be exploited even further. Repeating the above experiment with Tc instead of ATc (Fig. 4B) shows that the tTA2E4C–Ptet4 circuit can be regulated over its full range at Tc concentrations ≤1 μg/ml. By contrast, the rtTA2B6C responsive promoter Ptet6 remains inactive at these concentrations of the antibiotic. Thus, an even more stringent differentiation between the two regulatory circuits may be achieved by using different Tcs.
Figure 4.
Transcription activation by tTA2E4C and rtTA2B6C at different concentrations of ATc and Tc. (A) The two plasmids encoding tTA2E4C and rtTA2B6C, respectively, were transferred into HeLa cells together with pUHC13–8 containing the luc gene controlled by Ptet4 and, for standardization of transfection efficiency, plasmid pUHD16–1 constitutively expressing lacZ (23). The transfected cultures were incubated for 36 h at the ATc concentrations indicated before cells were harvested and luciferase activity was monitored (filled circles). The result of the analogous experiment in which Ptet6 controlled the luc gene (pUHC13–9) is depicted by open circles. (B) Same experiments as described in A, except that ATc is replaced by Tc.
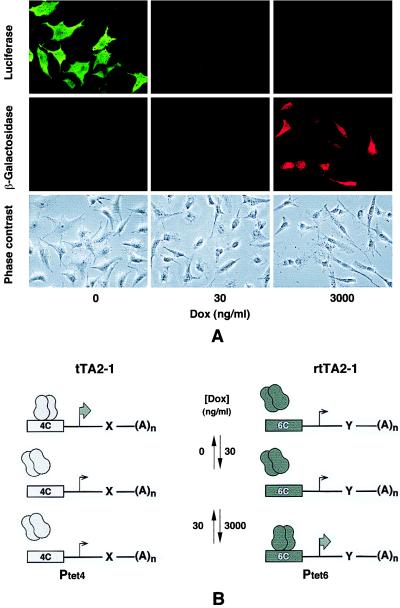
In the second experiment, HeLa cells were stably transfected in a two-step procedure with DNA encoding tTA2E4C and rtTA2B6C, respectively, to obtain cell lines that would constitutively produce both transactivators under the control of the human cytomegalovirus promoter IE. Indeed, the HT/rT-1 cell line that was characterized in more detail exhibited the properties of a novel cell type for Tet regulation. When transfected with DNA encoding a Ptet4 and a Ptet6 controlled expression unit, a differential regulation of the respective genes can be demonstrated. As shown in Fig. 5A, Ptet4-directed synthesis of luciferase occurs only in absence of Dox, whereas the Ptet6-controlled lacZ gene is active only at elevated Dox concentrations. At 30 ng/ml of Dox, both genes are inactive. In transient expression experiments using the Ptet4- or Ptet6-controlled luc gene, regulation factors of up to 1,000-fold were monitored (data not shown). Thus, cell lines can be established that constitutively produce both transactivators in sufficient amounts to warrant the efficient regulation of two expression units as schematically outlined in Fig. 5B.
Figure 5.
Mutually exclusive regulation of two genes by Dox. (A) Mutually exclusive expression of the luc and lacZ genes at different Dox concentrations in HT/rT-1 cells constitutively producing tTA2–1 and rtTA2–1. The plasmids carrying the luc gene under control of Ptet4 (pUHC13–8) and the lacZ gene controlled by Ptet6 (pUHG16–9) were transferred to the HT/rT-1 cells in a transient expression experiment. After incubation of the cultures for 36 h at the Dox concentrations indicated, luciferase (Top) and β-galactosidase (Middle) synthesis was monitored by indirect immunofluorescence. (Bottom) Cultures by means of phase-contrast microscopy. (B) Principle of action of the dual regulatory system. The transactivator tTA2E4C (simplified nomenclature: tTA2–1) activates Ptet4 in absence of Dox, whereas rtTA2B6C (simplified nomenclature: rtTA2–1) stimulates Ptet6 in the presence of Dox (3,000 ng/ml). At intermediate Dox levels (e. g. 30 ng/ml or at Tc concentrations between 100 and 1,000 ng/ml), none of the promoters is active. The different states of activation of the two genes x and y are indicated by small and large arrows, respectively.
DISCUSSION
The remodeling of the transactivators, tTA and rtTA, described here has led not only to a regulatory system with unusual properties, but it is also an interesting paradigm for respecifying regulatory proteins. Considering the complex molecular dynamics that constitute the allosteric properties of a protein like TetR, it is actually surprising that both new DNA-binding and dimerization specificities could be introduced that are of the same quality as those of the original transactivators tTA and rtTA. It should be noted, however, that the two transactivator/promoter combinations tTA2–1/Ptet4 and rtTA2–1/Ptet6 are the best of a number of combinations examined (and not reported here), a finding that clearly indicates the interdependence between parameters involved in dimerization and those determining the positioning of the DNA-recognition helices in the induced and uninduced state (27). Moreover, superimposing the mutations conferring the reverse phenotype affected differentially the properties of the various constructs, again underlining the intricate dynamics of this allosteric regulatory protein. Nevertheless, the two transactivator/promoter combinations selected are capable of controlling the expression of genes tightly and over a wide range, depending only on the concentration of a single effector molecule such as Dox or ATc. Actually, the results obtained in transient expression experiments as depicted in Figs. 2 and 4 leave little doubt that on integration into proper chromosomal loci, Ptet4- and Ptet6-controlled transcription units can be regulated as efficiently by tTA2–1 and rtTA2–1 as has been shown for the original tTA/rtTA responsive promoters.
By exploiting the different susceptibilities for Tcs, the combination of the two new regulatory circuits constitutes a unique system for controlling in a mutually exclusive way the expression of two genes quantitatively and reversibly. The level of resolution at which gene functions may be analyzed in vivo thus will be raised, since it will be possible to repeatedly switch between different states of gene activity and to follow the phenotypic consequences. Numerous experimental scenarios can be envisioned among which the generation of novel conditional mutants at the level of transgenic organisms appears particularly attractive. For example, to switch from a wild-type to a mutant allele at a defined developmental state of the organism and possibly to revert to wild-type again or to a null phenotype (both alleles turned off) promises new insights. Equally important is the quantitative nature of the regulatory principle that allows the ridgeless tuning of tTA/rtTA-responsive promoters by Dox, as demonstrated by fluorescence-activated cell sorter analysis at the single-cell level (28). It thus will be feasible to perturb intracellular equilibria by small increments and to follow the impact on the phenotype. In such studies, it will be of great advantage that the same effector substances, such as ATc or Dox, can be used to operate both regulatory systems. The well known pharmacological properties of these chemicals, such as tissue distribution, biological and chemical half-life, etc., will ensure a high degree of predictability and reliability in the application of these regulatory systems. Moreover, it is of particular advantage for in vivo applications that the tTA circuit can be operated by Tc alone without affecting the rtTA circuit. The latter may then be activated at will by Dox.
Although establishing this double regulatory system at the cellular or organismal level may be considered as experimentally demanding, it should be pointed out that the genes encoding the two transactivators can be transferred simultaneously, e. g. by means of a single plasmid or viral vector. Thus, cell lines, embryonic stem cells, and transgenic mice constitutively expressing the two transactivator genes will be available in future. The HT/rT-1 HeLa cell line is a first example, demonstrating that both transactivators can be stably produced at intracellular concentrations sufficient for the control of the two respective expression units. Such cell lines provide an excellent base for the transient (e. g., episomal) or permanent regulation of two expression units of interest. Moreover, for controlling just a single expression unit, these cell lines will allow control of a target gene by either tTA or rtTA. If proper precautions are taken (29), it will also be feasible to transfer both the gene for a transactivator such as tTA2–1 and its cognate promoter Ptet4 controlling the gene of interest simultaneously in a single DNA construct.
The dissection of gene functions in vivo by systems capable of quantitatively controlling gene expression will profit greatly from recent progress in targeted gene inactivation. For example, a gene under investigation can be targeted such that it is placed under control of one of the transactivators described. A different allele of this gene under control of the complementary transactivator can then be introduced by standard transgenic techniques, allowing the mutually exclusive expression of the two alleles. Many other applications also in combination with site-specific recombinases appear obvious. For example, a dual control system as specified here provides another degree of freedom for gene therapeutic strategies, while one regulatory circuit may be used to control a therapeutic gene, the other may be exploited for the termination of the regimen.
Finally, the results presented demonstrate the potential of redesigning specificities of regulatory proteins by genetic means. In this context, the central moiety, the Tet repressor, appears to be a particularly promising example since many well characterized effector molecules are known that, together with increasing insights into structural and functional parameters of this allosteric protein, may lead to further useful modifications of the system.
Acknowledgments
We are grateful to S. Freundlieb for stimulating discussions. We thank W. Just for the gift of antiluciferase antibodies and A. Pujol for advice in immunofluorescence experiments. The patient help of S. Reinig in preparing the manuscript is gratefully acknowledged. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 229), the Volkswagenstiftung, and the Fonds der Chemischen Industrie Deutschlands.
ABBREVIATIONS
- tTA
tetracycline-controlled transactivator
- rtTA
reverse tTA
- Tc
tetracycline
- Dox
doxycycline
- ATc
anhydrotetracycline
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
A Commentary on this article begins on page 797.
References
- 1.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, Bujard H. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 3.Freundlieb S, Baron U, Bonin A, Gossen M, Bujard H. Methods Enzymol. 1997;283:159–172. doi: 10.1016/s0076-6879(97)83014-5. [DOI] [PubMed] [Google Scholar]
- 4.Camacho-Vanegas O, Mannucci L, Amaldi F. In Vitro Cell Dev Biol Anim. 1998;34:14–15. doi: 10.1007/s11626-998-0043-8. [DOI] [PubMed] [Google Scholar]
- 5.Garí E, Piedrafita L, Aldea M, Herrero E. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 6.Girard F, Bello B, Laemmli U, Gehring W J. EMBO J. 1998;17:2079–2085. doi: 10.1093/emboj/17.7.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinmann P, Gossen M, Hillen W, Bujard H, Gatz C. Plant J. 1994;5:559–569. doi: 10.1046/j.1365-313x.1994.5040559.x. [DOI] [PubMed] [Google Scholar]
- 8.Zeidler M, Gatz C, Hartmann E, Hughes J. Plant Mol Biol. 1996;30:199–205. doi: 10.1007/BF00017815. [DOI] [PubMed] [Google Scholar]
- 9.Kistner A, Gossen M, Zimmermann F, Jerecic J, Ullmer C, Lübbert H, Bujard H. Proc Natl Acad Sci USA. 1996;93:10933–10938. doi: 10.1073/pnas.93.20.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efrat S, Fusco-DeMane D, Lemberg H, Emran O A, Wang X. Proc Natl Acad Sci USA. 1995;92:3576–3580. doi: 10.1073/pnas.92.8.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewald D, Li M, Efrat S, Auer G, Wall R J, Furth P A, Hennighausen L. Science. 1996;273:1384–1386. doi: 10.1126/science.273.5280.1384. [DOI] [PubMed] [Google Scholar]
- 12.Mayford M, Bach M E, Huang Y, Wang L, Hawkins R, Kandel E. Science. 1996;274:1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- 13.Kolster, K., Kistner, A., Baron, U., Freundlieb, S., Gossen, M. & Bujard, H. (1998) Transgenic Techniques, in press.
- 14.Fishman G I, Kaplan M L, Buttrick P M. J Clin Invest. 1994;93:1864–1868. doi: 10.1172/JCI117174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baron U, Gossen M, Bujard H. Nucleic Acids Res. 1997;25:2723–2729. doi: 10.1093/nar/25.14.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Resnitzky D, Gossen M, Bujard H, Reed S I. Mol Cell Biol. 1994;14:1669–1679. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Southern P J, Berg P. J Mol Appl Genet. 1982;1:327–341. [PubMed] [Google Scholar]
- 18.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 19.Baumeister R, Helbl V, Hillen W. J Mol Biol. 1992;226:1257–1270. doi: 10.1016/0022-2836(92)91065-w. [DOI] [PubMed] [Google Scholar]
- 20.Helbl V, Berens C, Hillen W. J Mol Biol. 1995;245:538–548. doi: 10.1006/jmbi.1994.0044. [DOI] [PubMed] [Google Scholar]
- 21.Helbl V, Tiebel B, Hillen W. J Mol Biol. 1998;276:319–324. doi: 10.1006/jmbi.1997.1539. [DOI] [PubMed] [Google Scholar]
- 22.Helbl V, Hillen W. J Mol Biol. 1998;276:313–318. doi: 10.1006/jmbi.1997.1540. [DOI] [PubMed] [Google Scholar]
- 23.Bonin A, Gossen M, Bujard H. Gene. 1994;141:75–77. doi: 10.1016/0378-1119(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 24.Hillen W, Berens C. Annu Rev Microbiol. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- 25.Hinrichs W, Kisker C, Duvel M, Muller A, Tovar K, Hillen W, Saenger W. Science. 1994;264:418–420. doi: 10.1126/science.8153629. [DOI] [PubMed] [Google Scholar]
- 26.Altschmied L, Baumeister R, Pfleiderer K, Hillen W. EMBO J. 1988;7:4011–4017. doi: 10.1002/j.1460-2075.1988.tb03290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baron U. Ph.D. thesis. Univ. Heidelberg; 1998. [Google Scholar]
- 28.Kringsten A M, Rossi F M V, Hofmann A, Blau H N. Proc Natl Acad Sci USA. 1998;95:13670–13675. doi: 10.1073/pnas.95.23.13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freundlieb, S., Schirra, Ch. & Bujard, H. (1999) J. Gene Medicine, in press. [DOI] [PubMed]