Short abstract
A relational database, called the General Repository for Interaction Datasets (The GRID) has been developed to archive and display physical, genetic and functional interactions.
Abstract
We have developed a relational database, called the General Repository for Interaction Datasets (The GRID) to archive and display physical, genetic and functional interactions. The GRID displays data-rich interaction tables for any protein of interest, combines literature-derived and high-throughput interaction datasets, and is readily accessible via the web. Interactions parsed in The GRID can be viewed in graphical form with a versatile visualization tool called Osprey.
Rationale
Physical, genetic and functional interactions between biological molecules are being discovered at an ever-increasing rate through proteomic and functional genomic approaches [1]. As a result, large-scale datasets containing many thousands of interactions have been deposited in publicly available databases. However, none of the extant datasets is systematically linked, and most data are presented only in a rudimentary format. Thus, data analysis is often tedious and incomplete. To alleviate this bottleneck, we developed a generic interaction database called The GRID [2], which can be used to collate and display interactions from any data source.
Software platform
The GRID uses MySQL version 3.23 as its underlying database [3], which is freely available from the MySQL homepage. The web-based user interface is implemented with Java Servlet technology, and the Java SDK version 1.4.0_02 [4]. These tools provide a facile interface for parsing interactions. Graphical representation of user defined interaction networks is achieved with a new visualization tool called Osprey [5], which can be used to construct elaborate interaction networks from any set of interactions in the database.
Data structure
The GRID is built on a master look-up table of all primary and secondary Saccharomyces cerevisiae gene names and corresponding open reading frame (ORF) names. Valid name lists are compiled via the open file transfer protocol (FTP) provided by the Saccharomyces Genome Database (SGD) [6]. Currently, The GRID recognizes 6,355 unique ORFs. Each gene entry in the GRID is presented in a data-rich tabular format that includes a description of gene function, Gene Ontology (GO) annotation [7], experimental system(s) on which associated interactions are based, the source of interaction data and publication links. Each row in the table represents a unique interaction, which is further divided into additional subsections corresponding to each experimental system in which the interaction is reported. Defined physical and genetic experimental systems currently include: affinity precipitation, affinity chromatography, two-hybrid, purified complex, reconstituted complex, biochemical assay, synthetic lethality, synthetic rescue, dosage lethality, dosage suppression, chemical lethality and chemical rescue. Additional systems may be added as needed.
Datasets
The GRID is periodically updated to contain all published large-scale interaction datasets, as well as available curated interactions from the primary literature. At present The GRID contains a total of 13,830 unique interactions and 21,839 total interactions, including most interactions deposited in BIND [8] and MIPS [9], as well as large-scale protein interaction datasets generated by Uetz et al. [10], Ito et al. [11,12], Gavin et al. [13] and Ho et al. [14] and a synthetic lethal interaction dataset produced by Tong et al. [15]. An upload interactions option allows new interactions to be added from a tab-delimited text file that contains the interaction pair, the experimental system and the data source. The GRID only accepts new interactions, so redundant interactions are excluded during the upload process. Details for upload format are provided at The GRID website.
Searches
Any valid gene or ORF name can be searched for to yield a comprehensive list of known interactions and associated annotations in tabular format (Figure 1). The search result table provides direct links for recursive searches, PubMed citations and data-rich graphical visualization with Osprey. In addition to standard keyword searches, an advanced search option allows keywords to be combined with Boolean operators to expand or reduce the number of recovered interactions. Results from advanced searches can be displayed using Osprey or saved as a tab-delimited text file.
Figure 1.
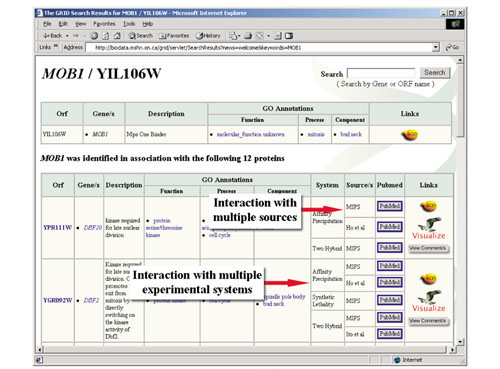
Search result page from The GRID. Multiple experimental systems and sources are indicated.
Access and software requirements
All relevant information on The GRID can be retrieved from The GRID website [2]. The GRID runs with the most recent versions of popular web browsers on all major platforms. An online version of the Osprey network visualization system is available as an add-on to The GRID that is automatically launched from a link on The GRID search result page. The Osprey add-on requires version 1.4.0_02 of the Java plug-in [4]. A full-featured application version of Osprey is available for non-profit use at [5] (see accompanying software article).
Private versions
Individual laboratories at not-for-profit institutions may request a private version of The GRID that can be easily customized to allow storage and manipulation of unpublished datasets, including integration and comparison with all publicly available interactions. The GRID is designed to work with any set of interactions, including those derived from other model organisms, combinations of organism systems and even social or commercial networks for which interaction data are available. For more information about hosting a private version of The GRID, please contact the authors.
Acknowledgments
Acknowledgements
We thank Lorrie Boucher, Ashton Breitkreutz and Paul Jorgensen for suggestions on GRID features. Development of The GRID was supported by the Canadian Institutes of Health Research. M.T. is a Canada Research Chair in Biochemistry.
A previous version of this manuscript was made available before peer review at http://genomebiology.com/2002/3/12/preprint/0013/
References
- Vidal M. A biological atlas of functional maps. Cell. 2001;104:333–339. doi: 10.1016/s0092-8674(01)00221-5. [DOI] [PubMed] [Google Scholar]
- The GRID http://biodata.mshri.on.ca/grid
- MySQL http://www.mysql.com/
- Sun Microsystems Java Standard Development Kit 1.4.0_02 http://java.sun.com
- Osprey http://biodata.mshri.on.ca/osprey
- Cherry JM, Ball C, Dolinski K, Dwight S, Harris M, Matese JC, Sherlock G, Binkley G, Jin H, Weng S, Botstein D. Saccharomyces Genome Database June 2002 ftp://genome-ftp.stanford.edu/pub/yeast/SacchDB/ [DOI] [PMC free article] [PubMed]
- The Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. BIND - a data specification for storing and describing biomolecular interactions, molecular complexes and pathways. Bioinformatics. 2000;16:465–477. doi: 10.1093/bioinformatics/16.5.465. [DOI] [PubMed] [Google Scholar]
- Munich Information Center for Protein Sequences http://mips.gsf.de/
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Tashiro K, Muta S, Ozawa R, Chiba T, Nishizawa M, Yamamoto K, Kuhara S, Sakaki Y. Toward a protein-protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc Natl Acad Sci USA. 2000;97:1143–1147. doi: 10.1073/pnas.97.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]


