Abstract
FIP-3 (14.7K interacting protein) was discovered during a search for cell proteins that could interact with an adenovirus protein (Ad E3–14.7K) that had been shown to prevent tumor necrosis factor (TNF)-α-induced cytolysis. FIP-3, which contains leucine zippers and a zinc finger domain, inhibits both basal and induced transcriptional activity of NF-κB and causes a late-appearing apoptosis with unique morphologic manifestations. Ad E3–14.7K can partially reverse apoptotic death induced by FIP-3. FIP-3 also was shown to bind to other cell proteins, RIP and NIK, which previously had been described as essential components of TNF-α-induced NF-κB activation. In addition, FIP-3 inhibited activation of NF-κB induced by TNF-α, the TNFR-1 receptor, RIP, NIK, and IKKβ, as well as basal levels of endogenous NF-κB in 293 cells. Because the activation of NF-κB has been shown to inhibit apoptosis, FIP-3 appears both to activate a cell-death pathway and to inhibit an NF-κB-dependent survival mechanism.
Adenoviruses (Ads) contain a variety of immunoregulatory genes, many of which are clustered in early-region three (E3) and are not required for viral replication in tissue culture (1, 2). These genes include the Ad E3–14.7K and the complex of the Ad E3–14.5K/10.4K proteins, which inhibit tumor necrosis factor (TNF)-α cytolysis in a variety of human and mouse cells (3, 4). The 14.5K/10.4K complex also inhibits Fas-mediated cell killing and promotes the internalization and degradation of both Fas and the epidermal growth factor receptors (5–7)
The control of TNF-α during the host response to viral infection must be very important, as evidenced by the number and diversity of viruses that affect these processes (8–15). Baculovirus and poxviruses code for proteins p35 and crmA, respectively, which directly affect the caspases and prevent proteolysis that results in apoptosis (9, 15). The mechanism of inhibition of TNF-α cytolysis by the Ad E3 proteins is not well understood; however, Ad E3–14.7K has been shown to bind to caspase 8 (ref. 16; M.S.H., unpublished observations). Although another Ad protein (E1B-19K) is a homologue of Bcl-2 and presumably acts in a similar manner to its cellular homologue to inhibit apoptosis, there appear to be differences in the ways that Bcl-2 or Ad E1B 19K and the Ad E3–14.7K inhibit cytolysis (17–21).
There has been a large amount of information published recently about cell proteins whose overexpression can cause cell death (22–29). Some of these studies initially have used the intracellular domains of one of the two TNF-α receptors (TNFR-1/TR55) or of the Fas molecule in the yeast two-hybrid system to find interacting cell proteins. Such studies have discovered molecules such as TRADD and MORT1/FADD, whose overexpression during transient transfection causes cell death (23, 25–27). The use of MORT1/FADD proteins as “baits” in subsequent yeast two-hybrid searches of interacting proteins has led to discovery of molecules such as MACH/FLICE (caspase 8), which has protease activity (28, 29).
In contrast to the pathways that lead to cell death, it has been shown recently that the induction of NF-κB inhibits apoptosis (30, 31). TRAF2, which interacts with the TNF receptor, activates both NF-κB and JNK pathways (24). TRAF2 associates with NIK, a MAP3K homologue, which increases NF-κB levels when overexpressed (32). NIK activates the IκB kinase, which phosphorylates IκBα on serines 32 and 36, causing degradation of IκBα and subsequent activation of NF-κB (33). Two kinases, IKKα and IKKβ, have been identified in the IkB kinase complex. Some of the death-inducing molecules, such as TNF-α or the transfected TNFR-1, also can induce NF-κB activity, even though the net effect of each of these overexpressed molecules appears to be cytolysis (24). Thus, it seems that TNF-α initiates a cascade in which cell death or survival rests on a delicate balance between opposing pathways. RIP, originally isolated by its interaction with the intracellular domain of Fas, also has been shown to interact with TRADD and FADD (23). These interactions potentially placed RIP as a mediator of both TNF-α- and Fas-ligand-induced signal transduction. Overexpressed RIP has been shown to cause apoptosis as well as to activate NF-κB (23, 34); however, recent gene-deletion studies have shown that RIP is essential for TNF-α-induced activation of NF-κB but not for cell death (35).
Our studies were initiated to determine the mechanism of action of the Ad E3–14.7K inhibitor of TNF-α-induced cytolysis by isolating cell proteins that bound to the viral molecule. Such studies have recognized a series of proteins called FIPs (14.7K-interacting proteins) (18, 36). FIP-3 is a protein containing leucine-zippers and a zinc finger. When FIP-3 is expressed by transient transfection, it causes cell death and inhibits the transcriptional activity of NF-κB. FIP-3 binds to RIP and to NIK and inhibits NF-κB activation by both of these molecules. FIP-3 also inhibits NF-κB activity induced by the addition of TNF-α to 293 cells and by the cotransfection of the TNFR-1 receptor (TR55) or the IKKβ.
EXPERIMENTAL PROCEDURES
Cell Lines.
The human 293 cell line was maintained in DMEM supplemented with 10% fetal bovine serum, 50 units of penicillin, and 50 μg/ml streptomycin. The mouse fibroblast C3HA cell lines with or without constitutively expressed Ad-E3–14.7K (obtained from Linda Gooding of Emory University) were maintained in DMEM with the same supplements as above (37).
Plasmid Constructs.
The bait vector expressing the Ad E3–14.7K protein in yeast, the expression plasmids pcDNA-FLAG-14.7 in mammalian cells, and pGST-14.7K (glutathione S-transferase fusion protein) in bacteria as well as the negative control plasmid pcDNA-T7 were constructed as described (18). pcDNA-T7-FIP-3Δ179 was made by inserting the FIP-3Δ179 cDNA sequence between the BamHI and XhoI sites 3′ to the T7 tag. For in vitro transcription and translation of FIP-3Δ179, the FIP-3 cDNA released from the “target” vector by BamHI/XhoI was cloned into pCITE-4b (Novagen) at the corresponding sites. The full length FIP-3 was inserted into the BamHI/XhoI site of the pcDNA-T7 vector for expression in mammalian cells and into the corresponding sites of pCITE-4b for expression of 35S-labeled FIP-3 in vitro. The fidelity of all of the constructs was confirmed by sequencing. The full length FIP-3 cDNA was obtained by 5′-rapid amplification of cDNA ends and was cloned into pcDNA-T7 as described below. The GST-NIK expression plasmid was constructed by cloning NIK into the BamHI/XhoI sites at the C terminus of the GST vector pGEX-5x-1 (Amersham Pharmacia).
The NF-κB-dependent luciferase reporter plasmid pIgκ-Luc was kindly provided by Bruce Horwitz of the Whitehead Institute at the Massachusetts Institute of Technology (Cambridge, MA). The pFLAG-CMV-IKKβ plasmid was generously provided by Jun Li (Boehringer Ingelheim), and the pcDNA-FLAG-RIP domain constructs were obtained from David Goeddel (Tularik, South San Francisco, CA). The FIP-1-expressing plasmid and the N-terminal deletion mutant of FIP-2 (FIP-2Δ134) used in coimmunoprecipitation reactions have been reported (18, 36). The plasmid (pGreen Lantern-1) expressing the green fluorescent protein (GFP) was purchased from GIBCO.
Immunofluorescent Studies for Colocalization Between Ad E3–14.7K and FIP-3.
Immunohistochemical colocalization studies in C3HA cells expressing the Ad E3–14.7K were done by following previously reported procedures (18). C3HA cells grown on chamber slides (Nunc) were transfected with pcDNA-T7-FIP-3Δ179 DNA by using the LipofectAmine technique (GIBCO) and were analyzed on a confocal microscope. The antibody to the E3–14.7K was a generous gift from William Wold, Saint Louis University.
Preparation and Use of GST Fusion Protein.
The expression and absorption of the Ad E3–14.7-GST fusion protein to GST-beads, in vitro labeling of the FIP-3Δ179 with 35S-methionine, and the in vitro protein–protein interaction assay followed protocols previously described (18). A similar protocol was applied to the interaction between full length 35S-labeled FIP-3 from a pCITE vector and GST-NIK expressed in bacteria.
Apoptosis Studies of the FIP-3 Protein.
The cell morphology studies were facilitated by using coexpressed GFP as a transfection marker (38). Forty hours after transfection, the cells were observed and photographed with a fluorescein isothiocyanate filter by using a Zeiss Axiophot 1 fluorescent microscope. The 293 cells for DNA fragmentation assays were harvested 48 hours after transfection by scraping into the medium. After centrifugation, pelleted cells were lysed by using 1 ml of ice-cold lysis buffer (50 mM Tris⋅HCl/20 mM EDTA/0.5% Triton X-100, pH8). Four hours after incubation on ice, cell debris was removed by brief centrifugation (12,000 × g), and the supernatant was subjected to two round of phenol extraction. After ethanol precipitation, DNA was treated with RNase and was analyzed on 2% agarose gels.
Apoptosis also was quantified by the Cell Death Detection ELISA Plus as described by the manufacturer (Boehringer) by using mAbs against DNA and histones to detect mono- and oligonucleosomes. The amount of substrate bound to lysates of the experimental divided by the controls was calculated as the apoptotic index, a measure of the number of nucleosomes released during apoptosis.
RESULTS
Identification of FIP-3 as a Specific Target of E3–14.7K in the Yeast Two-Hybrid System.
By using the E3–14.7K as bait in the yeast two-hybrid system as described (18), FIP-3 was identified in 9 of the original 107 colonies as a strongly interacting protein. The specificity of the interaction between E3–14.7K and FIP-3 in the yeast two-hybrid system was demonstrated by testing the FIP-3 against a number of other baits (lamin, TAD, bHLH, MaxI, Bcl2, E1B-19K, and BIK-1). FIP-3 did not interact with any of the other baits, including Bcl2 or Ad E1B-19K, two known inhibitors of TNF-α cytolysis. These data suggest that E3–14.7K and E1B-19K/Bcl2 are acting on different cellular targets.
FIP-3 Is a Cellular Protein Containing Multiple Leucine-Zipper Domains and a Zinc Finger.
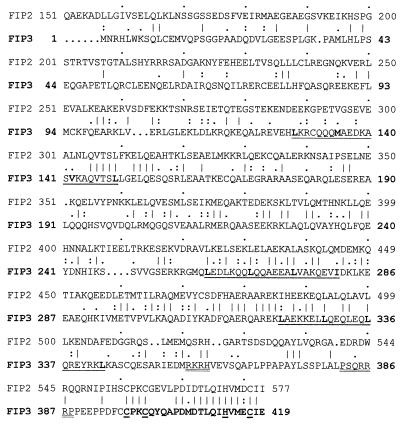
Full length FIP-3 is a protein containing 419 amino acids (Fig. 1), and the cDNA clone we isolated was 2,009 nucleotides in length with the start and stop codons at positions 110 and 1,368 respectively (GenBank accession no. AF062089). By comparing the full length sequence and the sequence obtained from the yeast two-hybrid screening, we found that the latter lacked the N-terminal 179 amino acids of the protein (FIP-3Δ179). By computer-aided sequence analysis, it was found that FIP-3 had some domains that were similar to a protein, FIP-2, that also had been isolated by binding to AdE3–14.7K (36). FIP-3 has one perfect leucine-zipper domain and two similar domains containing hydrophobic amino acids, as indicated. Both FIP-3 and FIP-2 also contained a putative zinc finger domain at their C termini; however, amino acid 404 in FIP-3 is not a hydrophobic or aromatic amino acid as is generally found in other zinc-finger proteins, including FIP-2. Putative nuclear localization signals are indicated by double underline (Fig. 1) and were detected by using the program psort ii (http://psort.nib.ac.jp/psort/helpwww2.html).
Figure 1.
Polypeptide sequence of FIP-3 derived from cDNA sequence. The figure shows the polypeptide sequence of FIP-3, which was deduced from the sequence of the yeast two-hybrid clone starting at amino acid 180 plus the 5′ rapid amplification of cDNA ends/PCR-derived sequences. Gene-specific primers (5′-CTCCTCGGCCTGCTGGAGCTGCTG 3′) and adapter primers (5′-CCATCCTAATACGACTCACTATAGGGC 3′) present in rapid amplification of cDNA ends–ready heart cDNA templates (CLONTECH) were used to identify the missing 5′ sequence (18). Three continuously underlined regions are three putative leucine-zipper regions. A zinc finger domain is shown in bold at the C terminus of the protein with the canonical C and H residues underlined, and putative nuclear localization signals are double-underlined. The sequence of FIP-2 is juxtaposed with the homologous amino acids, indicated by conventional symbols to indicate identity (27%) or degrees of similarities (38%).
FIP-3 mRNA was present in the eight human tissues (heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas) studied by Northern blot analysis using the 32P-labeled FIP-3 cDNA (data not shown). The major transcript of FIP-3 mRNA was a 2.4-kilobase band in all of the human tissues tested.
Interaction of FIP-3 with E3–14.7K.
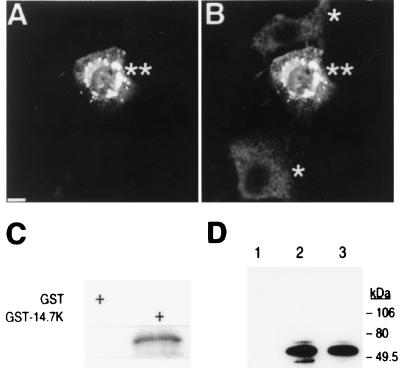
In addition to the yeast two-hybrid assay, the interaction between FIP-3 and E3–14.7K was shown in multiple protein–protein interaction analyses. First, FIP-3 (FIP-3Δ179 form used here) after transient transfection was shown to colocalize with E3–14.7K intracellularly (Fig. 2). Because E3–14.7K was expressed constitutively in all C3HA cells but FIP-3 was expressed only in a subpopulation of these cells after transient transfection, the redistribution of some of the E3–14.7K in the presence of FIP-3 could be detected. The appearance of the perinuclear bead-like structures having both E3–14.7K and FIP-3 staining suggests that FIP-3 and E3–14.7K interact intracellularly.
Figure 2.
Interaction of FIP-3 and Ad E3–14.7K proteins. Immunohistochemical studies of mouse C3HA cells, which constitutively express Ad E3–14.7K protein, were performed after transient transfection with a T7-tagged FIP-3 (FIP-3Δ179, 1 μg)-expressing plasmid. The relationship of these two proteins in the cells was shown by the use of anti-T7 mAbs (A) or anti-14.7K polyclonal antibodies (B). The immunoreactivities were visualized by confocal microscopy after staining with fluorescein-conjugated anti-mouse antibodies (A) or Texas red-conjugated anti-rabbit sera (B). The single asterisks in B denote cells that were expressing E3–14.7K in a diffuse distribution pattern in the cytoplasm in the absence of FIP-3. The double asterisks refer to cells coexpressing FIP-3 and E3–14.7K. The dense perinuclear bead-like structures were stained heavily with antibodies directed against the FIP-3 and E3–14.7K proteins after transient transfection of the cells with a T7-FIP-3 expressing plasmid. (Bar = 10 μm.) Direct in vitro and in vivo interactions between FIP-3 and E3–14.7K also were demonstrated as described (36). 35S-met-labeled FIP-3 from an in vitro translation reaction was incubated with either bacterial-expressed GST or GST-14.7K protein immobilized on glutathione beads. After extensive washes, labeled FIP-3 retained on the beads was analyzed by SDS/PAGE and autoradiography (C). Human 293 cells were seeded on 100-mm dishes and were transfected by using the LipofectAmine technique with 2 μg each of the following plasmids (D). Lanes: 1, FLAG-FIP-2Δ134 plus T7-FIP3; 2, FLAG-14.7 plus T7-FIP3; 3, FLAG-14.7 plus T7-FIP-3, treated with 20 ng/ml TNF-α for 20 min before harvesting the cells. Twenty-four hours after transfection, cell lysates were prepared and treated with 2 μl of anti-FLAG antibody followed by incubation with protein A beads. After several washes, the protein complex bound to the beads was analyzed by SDS/PAGE. Immunoprecipitated T7-tagged FIP-3 protein was identified by Western blotting with horseradish peroxidase-conjugated anti-T7 antibody (Novagen).
The direct interaction between FIP-3 and E3–14.7K was demonstrated further by a GST protein binding assay. As shown in Fig. 2C, FIP-3Δ179 directly interacted with E3–14.7K and was retained by the beads containing GST-E3–14.7K (Fig. 2C, right lane) but not by the beads containing GST protein alone (Fig. 2C, left lane). In the coimmunoprecipitation studies using the cell lysate of transfected cells, T7-FIP-3 was coimmunoprecipitated with FLAG-14.7K (Fig. 2D, lane 2). In contrast, T7-FIP-3 was not coprecipitated with FLAG-FIP-2Δ134, an N-terminal deletion mutant of FIP-2 (36) (Fig. 2D, lane 1), when using the same anti-FLAG antibody. TNF-α treatment did not significantly affect the interaction between the FIP-3 and 14.7K proteins under the conditions used for these assays (Fig. 2D, lane 3). The interaction between E3–14.7K and endogenous FIP-3 in C3HA cells was confirmed by coimmunoprecipitation and Western blotting using antibodies against a FIP-3 peptide (data not shown).
FIP-3 Is a Death Protein That Causes Unique Cell Morphology, and Ad E3–14.7K Can Reverse the FIP-3 Phenotype.
Because E3–14.7K inhibits TNF-α cytolysis, FIP-3 was tested as an inducer of cell death in transient transfection experiments. GFP, a product of the Green Lantern plasmid, was used as a marker of the transfected cells (38). Examination of the FIP-3 expressing cells revealed a unique morphology. Although the cells remained normal for ≈24 hours after FIP-3 transfection, they subsequently developed unique spherical structures that stained green with the fluorescent GFP (Fig. 3A, arrows). Other cells at this time showed typical signs of apoptotic disintegration with blebbing of the cytoplasm and nuclear fragmentation (Fig. 3A, arrowhead). The controls transfected with the pcDNA-T7 plasmid showed flat cells with normal pseudopod-like projections (Fig. 3B) and diffuse GFP fluorescent staining. To examine whether the spherical structures were in the nucleus or in cytoplasm, the transfected GFP-positive cells were counterstained with Hochest, a fluorescent nuclear stain. The unique round structures were present primarily in the cytoplasm (data not shown).
Figure 3.
Overexpression of FIP3 induces a unique morphology and intracytoplasmic inclusion bodies. Studies of FIP-3 morphology by using a GFP marker are shown in A. Human 293 cells on 6-well plates were transfected with 2 μg of pcDNA-T7-FIP3 and 1 μg of pGFP plasmid DNA. In B, cells transfected with 2 μg of pcDNA-T7 and 1 μg of pGFP plasmid as normal controls are shown. Forty hours after transfection, cells were examined and photographed under a fluorescent microscope with a fluorescein isothiocyanate filter. The cells with the intracellular round structures, stained green in the original and typical of the FIP-3 phenotype, are indicated by arrows (A). Another cell in an advanced stage of disintegration is designated by an arrowhead. (Bar = 10 μm.)
Evidence that FIP-3 killed cells by apoptosis and that Ad E3–14.7K could partially reverse this effect was provided by using the DNA fragmentation assay and the ELISA-based determination of mono- and oligonucleosomes and also by observing the morphologic effects of E3–14.7K on the reversal of FIP-3-induced cell death. As shown in Fig. 4A, the overexpression of FIP-3 could induce DNA fragmentation (Fig. 4A, lane 2). In Fig. 4B, the ELISA-based determination of mono- and oligonucleosomes in cytoplasmic extracts was used to quantify the reversal of FIP-3-induced apoptosis by E3–14.7K. The results of inducing cell death with FIP-3 are shown (Fig. 4B, lane 2) in relationship to killing with the TR55 (Fig. 4B, lane 5) as a positive control and to the pcDNA-T7 plasmid alone (Fig. 4B, lane 1) as a negative control. It can be seen that addition of Ad E3–14.7 inhibits cell death by ≈70% (Fig. 4A, lane 3), and there was no induction of apoptosis by an N-terminal 179-aa deletion of FIP-3 (Fig. 4B, lane 4). These results were concordant with morphologic determination of cell death after transient transfection with a plasmid expressing FIP-3 and its reversal by the addition of a plasmid expressing the Ad E3–14.7K protein (data not shown).
Figure 4.
Induction of apoptosis by FIP-3 and prevention of cell death by Ad E3–14.7K. Human 293 cells were seeded onto 100-mm dishes and were transfected by using the LipofectAmine protocol with plasmids as indicated below. In addition, each plate contained 0.3 μg of the GFP plasmid for monitoring transfection efficiency. Forty hours after transfection, the transfected cells were lysed (A). Low molecular weight nuclear DNAs were isolated and analyzed on 2% agarose gel as described in Materials and Methods. Lanes: 1, 4.7 μg of pcDNA-T7; 2, 1.5 μg of pcDNA-T7-FIP3 plus 3.2 μg pcDNA T7. In B, 293 cells were transfected similarly, but apoptosis was assayed and quantified by an ELISA technique as described in Materials and Methods. Lanes: 1, 4.5 μg of pcDNA T7; 2, 1.5 μg of pcDNA FIP-3; 3, 1.5 μg of pcDNA FIP-3 and 3 μg of pcDNA E3–14.7K; 4, 1.5 μg of pcDNA FIP-3Δ1–179; 5, 2.0 μg of pcDNA TR55. The total amount of transfected DNA in each lane was adjusted to 4.5 μg by the addition of pcDNA T7.
Intracellular Localization of FIP-3 Affected by Ad E3–14.7K.
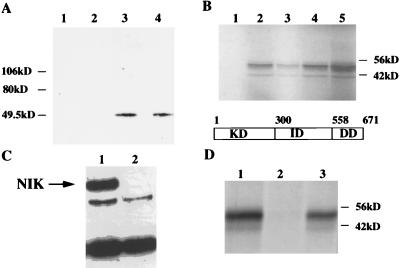
Although FIP-3 and Ad E3–14.7K proteins colocalized in the cytoplasm in perinuclear bead-like structures (Fig. 2 A and B), FIP-3 alone was demonstrated in the nucleus as well as in the cytoplasm (Fig. 5, lane 3) by Western blotting. On the addition of increasing amounts of Ad E3–14.7K (Fig. 5, lanes 4–7), the amount of FIP-3 in the nucleus decreased dramatically.
Figure 5.
Ad E3 14.7K affects the intracellular localization of FIP-3. 293 cells were transfected with plasmids expressing various combinations of pcDNA-T7-FIP-3 and pcDNA-FLAG-14.7K. The nuclear and cytoplasmic fractions were prepared by Dounce homogenization in hypotonic buffer, and each was analyzed for FIP-3 protein 24 hours later by Western blots using antibody to the T7 tag conjugated with horseradish peroxidase (Novagen) at a dilution of 1:2,000 for 1.5 hours at room temperature. The blots were washed 3–4 times in PBS with 0.1% Tween 20 and were developed with enhanced chemiluminescence reagents (Boehringer Mannheim). Lanes: 1 and 2, negative controls that contain 5 μg of pcDNA-T7 and 3 μg of pcDNA-14.7K, respectively; 3–7, 1 μg of pcDNA-FIP-3; 4–7, increasing amounts of pcDNA-14.7K; 4, 1 μg; 5, 2 μg; 6, 3 μg; 7, 4 μg. All transfections were normalized to contain 5 μg of DNA by the addition of pcDNA-T7.
FIP-3 Interacts with RIP and NIK.
In a series of studies of interactions between FIP-3 and other cell signal transduction proteins, it was found that FIP-3 could interact with RIP in the yeast two-hybrid system (data not shown). The RIP/FIP-3 interaction was confirmed by coimmunoprecipitation from 293 cell lysates transiently transfected with both of these proteins. As shown in Fig. 6A, FIP-3 could be coimmunoprecipitated by FLAG-RIP (Fig. 6A, lanes 3 and 4) but not by FLAG-FIP-2Δ134 (Fig. 6A, lane 2) or by FLAG-FIP-1 (Fig. 6A, lane 1). The addition of TNF-α had little or no effect on the interaction between the FIP-3 and RIP (Fig. 6A, lane 4). The RIP/FIP-3 interaction also was studied between 35S-FIP-3 synthesized in the rabbit reticulocyte system and the various domains of FLAG-RIP, which were made in transfected 293 cells and were bound to a Sepharose column by FLAG antibody (Fig. 6B). FIP-3 bound strongly to intact RIP (Fig. 6B, lane 4) and its intermediate domain (Fig. 6B, lane 2) but not to the kinase domain (Fig. 6B, lane 1). There was only a weak interaction to the death domain (Fig. 6B, lane 3). A strong interaction between FIP-3 and NIK after transient transfection of 293 cells followed by coimmunoprecipitation and Western blotting also could be demonstrated (Fig. 6C). FIP-3 also was shown to bind efficiently to NIK expressed as a GST-NIK fusion protein in bacteria (Fig. 6D, lane 3) but not to the GST protein alone (Fig. 6D, lane 2).
Figure 6.
Association of FIP-3 with RIP and NIK in 293 cells. In A, 293 cells were transfected with the following plasmids for coimmunoprecipitation and Western blot analysis. Lanes: 1, 2 μg of pcDNA-FLAG-RIP plus 2 μg of pcDNA-T7-FIP1; 2, 2 μg of pcDNA-FLAG-FIP-2Δ134 plus 2 μg of pcDNA-T7-FIP3; 3, 2 μg of pcDNA-FLAG-RIP plus 2 μg of pcDNA-T7-FIP3; 4, same as lane 3 plus TNF-α (20 ng/ml for 20 min before harvest). Cell lysate preparation, immunoprecipitation with anti-FLAG antibody, and Western blots with anti T7 antibody were done as described in Fig. 2D. In B, cytoplasmic extracts of 293 cells transiently transfected in 100 mm dishes with 8–10 μg of plasmids containing various RIP domains were added to anti-FLAG M2 affinity gels for 2 hours at 4°C. After washing the gels three times, 35S-FIP-3 made in reticulocyte lysates was added for an additional 2 hours, and the washing was repeated. The labeled proteins were eluted from the gel with SDS sample buffer, were separated by PAGE, and were detected by autoradiography. Lanes: 1, the RIP kinase domain (KD); 2, the intermediate domain (ID); 3, the death domain (DD); 4, intact RIP; lane 5, the labeled FIP-3 added directly to the SDS/PAGE. In C, 293 cell lysate preparation, immunoprecipitation with anti-HIS antibody, and Western blots with anti-FLAG antibody were done as described in A and Fig. 2D. Lanes: 1, the extracts of cells cotransfected with FLAG-NIK and HIS-FIP-3; 2 , the FLAG-NIK alone. The position of the NIK protein is shown. The lower two bands are nonspecific. In D, 35S-FIP-3 was added for 4 hours to extracts derived from Escherichia coli transfected with GST-NIK or GST in pGEX plasmids. The GST proteins were immobilized on glutathione Sepharose 4B before the addition of the radioactive extract, were washed five times, and were eluted for SDS/PAGE. Lanes: 1, 35S-FIP-3 added directly to SDS/PAGE; 2, the GST protein; 3, the GST-NIK protein.
FIP-3 Blocks NF-κB Activation by TNF-α, the TNF-α Receptor, RIP, NIK, and IKKβ.
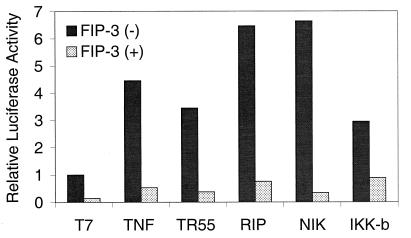
It has been reported previously that the addition of TNF-α or over-expression of either the TNFR-1 (TR55), RIP, NIK, or IKKβ by transient transfection could activate NF-κB (23, 32, 34, 39). To test whether FIP-3 could block the activation of NF-κB by a variety of signal transduction molecules, we studied the effect of FIP-3 on NF-κB activation by TNF-α, TNFR-1, RIP, NIK, or IKKβ. The experiments were done in the presence of the baculovirus p35 protein to prevent apoptosis by TR55 or RIP (40). NF-κB activity was assessed at 18 hours (or 24 hours for experiments with TNF-α) after plasmid transfection, significantly before the cells demonstrated the FIP-3-induced cell death. As shown in Fig. 7, TNF-α (15 ng/ml) as well as overexpression of the TR55, RIP, NIK, or IKKβ by transient transfection could activate NF-κB and increased NF-κB-dependent luciferase activity 3- to 6.5-fold over the basal levels achieved with the negative control plasmid. However, the addition of 1 μg of the FIP-3 plasmid to any of these transfected or TNF-α-treated cells completely blocked the activation of NF-κB to levels below basal values. As the amount of FIP-3 plasmid DNA was reduced below 10 ng, the inhibitory effects of FIP-3 were no longer observed.
Figure 7.
Inhibition of TNF-α, TNFR-1, RIP, NIK, or IKK-β-induced transcriptional activation of NF-κB by FIP-3. 293 cells on 6-well plates were transfected with one of the following plasmids: 1.0 μg pcDNA-T7; 0.3 μg of pcDNA-TR55; 0.1 μg of pcDNA-FLAG-RIP; 0.5 μg of pcDNA-NIK; or 0.4 μg of pcDNA-IKKβ. One microgram of pcDNA-T7-FIP3 was added to one of each paired samples as designated. The amount of DNA in each transfection was normalized to 2.5 μg with control plasmid pcDNA-T7. In addition, each well contained the following plasmids: 0.2 μg pGreen Lantern and 0.1 μg pAd-CMV-LacZ to monitor transfection efficiency, 1 μg of baculovirus p35, and 0.2 μg of the NF-κB-dependent luciferase reporter construct pIgκ-Luc. Eighteen hours after transfection, cells were harvested, and luciferase activity was assayed by using the Luciferase Reporter Assay Kit (Boehringer Mannheim) following the manufacturer’s protocol. In addition, TNF-α (15 ng/ml) was added at 18 hours to paired samples previously transfected with pcDNA-T7 or FIP-3 plus the additional indicator plasmids. These cells were harvested at 24 hours and were processed for luciferase activity. The experiment was repeated at least three times, and similar results were obtained. Data shown here represented the luciferase activities from one of these experiments.
DISCUSSION
Apoptotic cell death is used as one of the host defense mechanisms to control infection by viruses. To counter this protective measure, viruses encode genes whose products can inhibit apoptosis in the host cells. Ad E3–14.7K is one of several adenovirus proteins that inhibit TNF-α-induced apoptosis. In an effort to elucidate the mechanism by which viral proteins block apoptosis, we used the yeast two-hybrid system to isolate and characterize FIP-3, which interacts with Ad E3–14.7K. The specificity of this interaction was further confirmed by a number of assays, including the lack of reactivity to a series of other baits in the yeast two-hybrid system, in vitro GST protein–protein interaction, intracellular colocalization, and in vivo coimmunoprecipitation. In addition, the intracellular redistribution of E3–14.7K or FIP-3 on the addition of both of these proteins together suggested an interaction between FIP-3 and E3–14.7K.
FIP-3 has 27% identity with FIP-2, another Ad E3–14.7K binding protein that has been described (36). FIP-2 recently had been shown to bind to the huntingtin protein (41) but does not directly cause cell death or affect NF-κB (36). Computer-aided sequence analysis showed that FIP-3 contains a zinc finger, a leucine-zipper domain, and two similar domains with isoleucine, valine, and methionine substituting for some of the leucines. We have found that all three leucine-zipper domains were not required for its interaction with E3–14.7K, because the clones identified in the yeast two-hybrid system lacked the first leucine-zipper domain.
FIP-3 does not contain the so-called “death domain,” which is present in a number of cell death proteins, such as RIP, TNFR-1, Fas, TRADD, and FADD/MORT1. However, FIP-3 could cause apoptosis, inducing a unique morphology before fragmentation and eventual cell death. Most of the cells with intracytoplasmic GFP-staining bodies died between 48 and 72 hours after transfection. The mechanism of inducing apoptotic death by FIP-3 is not known, but it does not appear to be inhibitable by the p35 baculovirus protein, YVAD-CHO, DEVD-CHO, or Z-VAD-FMK caspase inhibitors. Because there are reports of caspase-independent effectors of apoptosis, some of which are blocked by Bcl-2, FIP-3 may belong to such a group of proteins (42). However, it has been found that Ad E3–14.7K also can bind to caspase 8 (ref. 16; M.S.H., unpublished observations). Thus, it is possible that Ad E3–14.7K may interact and inhibit both caspase-dependent as well as caspase-independent pathways that induce apoptosis. Because NF-κB appears to activate unknown genes whose products can prevent apoptosis, the inhibition of NF-κB activation by FIP-3 may augment signals required for cells to undergo apoptosis. Thus, FIP-3 might provide important clues for further understanding of the control of apoptosis.
It is not known which of the interactions between FIP-3 and the various signal transduction molecules on the NF-κB activation pathway are most important for the modulation of NFκB. We have shown interactions between FIP-3 and RIP or NIK as well as effects of FIP-3 on NF-κB activity induced by TNFα, RIP, NIK, and IKKβ. All of these interactions presumably occur in the cytoplasm. FIP-3 also appears in the nucleus, where its leucine zipper or zinc-finger domains might promote interactions with the NF-κB transcriptional apparatus (43–45). Recently, while this manuscript was in preparation, the mouse homologue of FIP-3 (named NEMO) was isolated by its ability to complement cells deficient in NF-κB activity (46). NEMO was essential for the HTLV-Tax-induction of NF-κB in cells deficient in this activity; however, the corresponding effects of NEMO on cytokine-induced activation of NF-κB in wild-type cells containing endogenous NEMO activity were not reported (46). In addition, NEMO was shown to bind to IKKβ. We also had isolated the FIP-3 homologue (mFIP-3) from a mouse cDNA library and found that its activity was identical to human FIP-3 in the inhibition of basal and induced NF-κB activities. The reasons that mFIP3/NEMO is needed to complement cells deficient in NF-κB activity, but both human and mouse FIP-3 inhibit NF-κB when added to cells expressing endogenous levels of FIP-3, are being explored. Because we have shown that FIP-3 can assemble into multimeric complexes (data not shown) and NEMO also has been shown to dimerize (46), such a model would place FIP-3 at an essential checkpoint to orchestrate either the inhibition or augmentation of NF-κB activity. One possible explanation is that the composition or steric configuration of a multiprotein complex of signal transduction molecules containing FIP-3 could be altered as the amount of FIP-3 is increased.
Acknowledgments
We acknowledge with gratitude the HeLa cDNA library for yeast two-hybrid screening as a gift from Greg Hannon and David Beach of the Cold Spring Harbor Laboratory; nonspecific baits, lamin, TAD, bHLH, and MaxI provided by Ron DePinho and described previously (47); and Bcl-2, E1B-19K, and BIK-1 made available to us by R. Chinnaduri, Saint Louis University (48, 49). This work was supported by National Institutes of Health Grant RO1 CA72963 (to M.S.H., L.T., and J.Y), National Institutes of Health Cancer Center Core Grant CA13330 (to M.S.H.), the Oncology Research Faculty Development Program (L.T.), the Forchheimer Foundation (M.S.H.), and by National Institutes of Health Training Grants 5T32 CA 09060 (to Y.L.) and 1T32 AI 07506 (to J.F.). The data in this paper was submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Y.L.) in the Sue Golding Graduate Division of Medical Sciences, Albert Einstein College of Medicine, Yeshiva University.
ABBREVIATIONS
- TNF
tumor necrosis factor
- GST
glutathione S-transferase
- GFP
green fluorescent protein
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF062089).
References
- 1.Wold W S M, Gooding L R. Virology. 1991;184:1–8. doi: 10.1016/0042-6822(91)90815-s. [DOI] [PubMed] [Google Scholar]
- 2.Wold W S M, Tollefson A E, Hermiston T W. In: The E3 Transcription Unit of Adenovirus. Doerfler W, editor. 199/1. Berlin: Springer; 1995. pp. 237–274. [DOI] [PubMed] [Google Scholar]
- 3.Gooding L R, Sofola I O, Tollefson A E, Duerksen-Hughes P J, Wold W S M. J Immunol. 1990;145:3080–3086. [PubMed] [Google Scholar]
- 4.Gooding L R, Ranheim T S, Tollefson A E, Aquino L, Duerksen-Hughes P J, Horton T M, Wold W S M. J Virol. 1991;65:4114–4123. doi: 10.1128/jvi.65.8.4114-4123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlin C R, Tollefson A E, Brady H A, Hoffman B L, Wold W S M. Cell. 1989;57:135–144. doi: 10.1016/0092-8674(89)90179-7. [DOI] [PubMed] [Google Scholar]
- 6.Tollefson A E, Hermiston T W, Lichtenstein D L, Colle C F, Tripp R A, Dimitrov T, Toth K, Wells C E, Doherty P C, Wold W S. Nature (London) 1998;392:726–730. doi: 10.1038/33712. [DOI] [PubMed] [Google Scholar]
- 7.Shisler J, Yang C, Walter B, Ware C F, Gooding L R. J Virol. 1997;71:8299–8306. doi: 10.1128/jvi.71.11.8299-8306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. Nature (London) 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 9.Smith G L. Curr Opin Immunol. 1996;8:467–471. doi: 10.1016/s0952-7915(96)80032-x. [DOI] [PubMed] [Google Scholar]
- 10.Kettle S, Alcami A, Khanna A, Ehret R, Jassoy C, Smith G L. J Gen Virol. 1997;78:677–685. doi: 10.1099/0022-1317-78-3-677. [DOI] [PubMed] [Google Scholar]
- 11.Sedger L, McFadden G. Immunol Cell Biol. 1996;74:538–545. doi: 10.1038/icb.1996.87. [DOI] [PubMed] [Google Scholar]
- 12.McFadden G, Schreiber M, Sedger L. J Neuroimmunol. 1997;72:119–126. doi: 10.1016/s0165-5728(96)00177-4. [DOI] [PubMed] [Google Scholar]
- 13.Wiertz E, Hill A, Tortorella D, Ploegh H. Immunol Lett. 1997;57:213–216. doi: 10.1016/s0165-2478(97)00073-4. [DOI] [PubMed] [Google Scholar]
- 14.Smith C A, Davis T, Wignall J M, Din W S, Farrah T, Upton C, McFadden G, Goodwin R G. Biochem Biophys Res Commun. 1991;176:335–342. doi: 10.1016/0006-291x(91)90929-2. [DOI] [PubMed] [Google Scholar]
- 15.Du X, Thiem S M. J Virol. 1997;71:7866–7872. doi: 10.1128/jvi.71.10.7866-7872.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen P, Tian J, Kovesdi I, Bruder J T. J Biol Chem. 1998;273:5815–5820. doi: 10.1074/jbc.273.10.5815. [DOI] [PubMed] [Google Scholar]
- 17.Perez D, White E. J Cell Biol. 1998;141:1255–1266. doi: 10.1083/jcb.141.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Kang J, Horwitz M S. J Virol. 1997;71:1576–1582. doi: 10.1128/jvi.71.2.1576-1582.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gooding L R, Aquino L, Duerksen-Hughes P J, Day D, Horton T M, Yei S, Wold W S M. J Virol. 1991;65:3083–3094. doi: 10.1128/jvi.65.6.3083-3094.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farrow S N, White J H, Martinou I, Raven T, Pun K T, Grinham C J, Martinou J C, Brown R. Nature (London) 1995;374:731–733. doi: 10.1038/374731a0. [DOI] [PubMed] [Google Scholar]
- 21.Han J, Wallen H D, Nunez G, White E. Mol Cell Biol. 1998;18:6052–6062. doi: 10.1128/mcb.18.10.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cleveland J L, Ihle J N. Cell. 1995;81:479–482. doi: 10.1016/0092-8674(95)90068-3. [DOI] [PubMed] [Google Scholar]
- 23.Stanger B Z, Leder P, Lee T H, Kim E, Seed B. Cell. 1995;81:513–523. doi: 10.1016/0092-8674(95)90072-1. [DOI] [PubMed] [Google Scholar]
- 24.Liu Z, Hsu H, Goeddel D V, Karin M. Cell. 1996;87:565–575. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 25.Hsu H, Xiong J, Goeddel D V. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 26.Chinnaiyan A M, O’Rourke K, Tewari M, Dixit V M. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 27.Boldin M P, Varfolomeev E E, Pancer Z, Mett I L, Camonis J H, Wallach D. J Biol Chem. 1995;270:7795–7798. doi: 10.1074/jbc.270.14.7795. [DOI] [PubMed] [Google Scholar]
- 28.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 29.Muzio M, Chinnaiyan A M, Kischkel F C, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, Mann M, Krammer P H, et al. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 30.Beg A A, Baltimore D. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 31.Van Antwerp D J, Martin S J, Kafri T, Green D R, Verma I M. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 32.Malinin N L, Boldin M P, Kovalenko A V, Wallach D. Nature (London) 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 33.Regnier C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 34.Hsu H, Huang J, Shu H-B, Baichwal V, Goeddel D V. Immunity. 1996;4:387–396. doi: 10.1016/s1074-7613(00)80252-6. [DOI] [PubMed] [Google Scholar]
- 35.Kelliher M A, Grimm S, Ishida Y, Kuo F, Stanger B Z, Leder P. Immunity. 1998;3:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Kang J, Horwitz M S. Mol Cell Biol. 1998;18:1601–1610. doi: 10.1128/mcb.18.3.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zilli D, Voelkel-Johnson C, Skinner T, Laster S M. Biochem Biophys Res Commun. 1992;188:177–183. doi: 10.1016/0006-291x(92)92366-6. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Horwitz M S. BioTechniques. 1997;23:1026–1029. doi: 10.2144/97236bm12. [DOI] [PubMed] [Google Scholar]
- 39.Zandi E, Rothwarf D M, Delhase M, Hayakawa M, Karin M. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 40.Xue D, Horvitz H R. Nature (London) 1995;377:248–251. doi: 10.1038/377248a0. [DOI] [PubMed] [Google Scholar]
- 41.Faber P W, Barnes G T, Srinidhi J, Chen J, Gusella J F, MacDonald M E. Hum Mol Genet. 1998;7:1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- 42.Monney L, Otter I, Olivier R, Ozer H L, Haas A L, Omura S, Borner C. J Biol Chem. 1998;273:6121–6131. doi: 10.1074/jbc.273.11.6121. [DOI] [PubMed] [Google Scholar]
- 43.Schule R, Evans R M. Trends Genet. 1991;7:377–381. doi: 10.1016/0168-9525(91)90259-s. [DOI] [PubMed] [Google Scholar]
- 44.Barnikol-Watanabe S, Gross N A, Gotz H, Henkel T, Karabinos A, Kratzin H, Barnikol H U, Hilschmann N. Biol Chem Hoppe-Seyler. 1994;375:497–512. doi: 10.1515/bchm3.1994.375.8.497. [DOI] [PubMed] [Google Scholar]
- 45.Alber T. Curr Opin Genet Dev. 1992;2:205–210. doi: 10.1016/s0959-437x(05)80275-8. [DOI] [PubMed] [Google Scholar]
- 46.Yamaoka S, Courtois G, Bessia C, Whiteside S T, Weil R, Agou F, Kirk H E, Kay R J, Israel A. Cell. 1998;93:1231–1240. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- 47.Schreiber-Agus N, Chin L, Chen K, Torres R, Thomson C T, Sacchettini J C, DePinho R A. Oncogene. 1994;9:3167–3177. [PubMed] [Google Scholar]
- 48.Boyd J M, Gallo G J, Elangovan B, Houghton A B, Malstrom S, Avery B J, Ebb R G, Subramanian T, Chittenden T, Lutz R J, et al. Oncogene. 1995;11:1921–1928. [PubMed] [Google Scholar]
- 49.Boyd J M, Malstrom S, Subramanian T, Venkatesh L K, Schaeper U, Elangovan B, D’Sa-Eipper C, Chinnadurai G. Cell. 1994;79:341–351. doi: 10.1016/0092-8674(94)90202-x. [DOI] [PubMed] [Google Scholar]