Abstract
Bruck syndrome is characterized by the presence of osteoporosis, joint contractures, fragile bones, and short stature. We report that lysine residues within the telopeptides of collagen type I in bone are underhydroxylated, leading to aberrant crosslinking, but that the lysine residues in the triple helix are normally modified. In contrast to bone, cartilage and ligament show unaltered telopeptide hydroxylation as evidenced by normal patterns of crosslinking. The results provide compelling evidence that collagen crosslinking is regulated primarily by tissue-specific enzymes that hydroxylate only telopeptide lysine residues and not those destined for the helical portion of the molecule. This new family of enzymes appears to provide the primary regulation for controlling the different pathways of collagen crosslinking and explains why crosslink patterns are tissue specific and not related to a genetic collagen type. A genome screen identified only a single region on chromosome 17p12 where all affected sibs shared a cluster of haplotypes identical by descent; this might be the BS (Bruck syndrome) locus and consequently the region where bone telopeptidyl lysyl hydroxylase is located. Further knowledge of this enzyme has important implications for conditions where aberrant expression of telopeptide lysyl hydroxylase occurs, such as fibrosis and scar formation.
Collagen fibrils are important for the mechanical strength of bone (1–2). The tensile properties of fibrils result from intermolecular crosslinks connecting the nonhelical ends of a collagen molecule (telopeptides) with the triple helical part of an adjacent molecule (2–3). More than ten different collagen crosslinks are known; their structure, number, and location are highly tissue specific and not related to a specific collagen type (4–8). Stereochemical and x-ray diffraction studies revealed that differences in molecular packing of collagen within fibrils are associated with differences in crosslink profiles (9–12). Proper mineralization probably depends on a correct alignment of collagen molecules, as nucleation of calcium apatite crystals starts in the gap region, i.e., in the area adjacent to the crosslink site (9). Alterations in crosslink patterns associated with changes in the molecular packing are, therefore, expected to result in aberrant mineralization.
Residues involved in crosslinking are mainly the amino acids lysine (Lys) or hydroxylysine (Hyl) (4–8). The enzyme lysyl hydroxylase (LH) (procollagen-lysine, 2-oxoglutarate 5-dioxygenase; EC 1.14.11.4) catalyzes the conversion of Lys into Hyl (13–15). Crosslinking is initiated only after specific Lys or Hyl residues of the telopeptides are converted extracellularly by the enzyme lysyl oxidase into the aldehydes allysine and hydroxyallysine, respectively (4–8). The aldehydes subsequently react with Lys, Hyl, or histidyl, residues of the triple helix to give characteristic di-, tri-, and tetrafunctional crosslinks. Two related routes for the formation of crosslinks have been described, based on whether the aldehyde-forming residue in the telopeptide is a lysine (allysine route) or an Hyl (hydroxyallysine route). Each route results in chemically distinct crosslinks (4–5, 8) (Fig. 1). Collagen type I in bone and collagen type II in cartilage are predominantly stabilized by crosslinks from the hydroxyallysine route (i.e., where the telopeptides are hydroxylated), such as hydroxylysylpyridinoline (HP) and lysylpyridinoline (LP). In contrast, the crosslinks in collagen type I in skin, cornea, and certain tendons are mainly derived from the allysine route (4–8). Despite the large differences in lysyl hydroxylation of the telopeptides of collagen type I between skin and bone, for instance, less marked differences are seen in the hydroxylation of lysine residues in the triple helical part of the collagen molecule. To explain this difference, it has been proposed that two forms of LH exist: one that specifically hydroxylates Lys in the α-helix (we propose the term “helical lysyl hydroxylase,” HLH), and another capable of hydroxylating Lys residues in the telopeptides (telopeptide lysyl hydroxylase, TLH) (16–21).
Figure 1.
Simplified scheme to show the origin of the determined crosslinks. Telopeptide Lys is converted to Hyl by TLH. The first step in crosslinking is the oxidation of telopeptide Lys or Hyl to aldehydes by lysyl oxidase. There are two pathways of formation of crosslinks, depending on whether the residue in the telopeptide is a Lys (allysine route) or a Hyl (hydroxyallysine route). The aldehydes react with specific Lys or Hyl residues in the triple helical domain on juxtaposed neighboring molecules to form difunctional intermediates that mature into trifunctional crosslinks. The two pathways lead to different types of crosslinks. Only the difunctional crosslinks that result from the hydroxyallysine route are able to mature into the trifunctional crosslinks HP and LP. Abbreviations: see legend of Fig. 3.
Bruck syndrome [BS; Online Mendelian Inheritance in Man (OMIM) #259450] is an autosomal recessive disease discovered a century ago (22). Patients with BS are characterized by fragile bones with congenital joint contractures. A reduction of mineral content and an increase in size of the hydroxyapatite crystals were observed (23). Worldwide only nine families (comprising 14 patients) have been reported (22–27). Although an abnormality in the bone collagen network in BS was assumed, the underlying defect remained unknown: collagen in bone and the collagen synthesized by cultured skin fibroblasts showed none of the changes commonly found in osteogenesis imperfecta (23, 26–27). Here we report that the molecular defect underlying BS is a deficiency of bone-specific TLH, which results in aberrant crosslinking of bone collagen. A mapping approach based on homozygosity by descent (28) on a consanguineous BS family with three affected and four healthy siblings (27) revealed evidence that the locus responsible for Bruck syndrome (BS) maps to an 18-cM interval on chromosome 17p12.
EXPERIMENTAL PROCEDURES
Patients.
A Kurdish family with three affected (2, 8, and 13 yr old) and four unaffected children was studied (Fig. 3). The parents, who are consanguineous (first cousins), did not show fractures or joint contractures nor did any of their relatives. Trabecular bone and articular cartilage were obtained from the 8-yr-old girl (patient 505; case 2 of ref. 27); cortical bone was obtained from the 13-yr-old girl (patient 507; case 1 of ref. 27). In addition, cortical bone and ligament were obtained from a 4-yr-old boy (patient 3; case 8 of ref. 27) of an unrelated Kurdish family with two affected children. All biochemical studies were carried out on surplus material from orthopedic operations with informed consent of the parents. A detailed description of the affected siblings has been published elsewhere (27).
Figure 3.
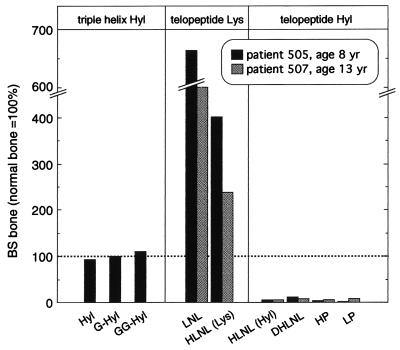
Posttranslational modification levels of type I collagen in BS bone normalized against normal bone. Bar graph shows that (glycosylated) Hyl levels of the triple helix are normal, which is in contrast to that of the telopeptides, where hardly any Hyl is seen. The lack of telopeptide Hyl-derived crosslinks is compensated by increased levels of crosslinks derived from the telopeptide Lys route. HLNL (Lys), hydroxylysinonorleucine derived from telopeptidyl lysine; HLNL (Hyl), hydroxylysinonorleucine derived from telopeptidyl Hyl.
Collagen Analysis.
Bone was demineralized at 4°C with 0.5 M EDTA/0.05 M Tris⋅HCl, pH 7.5, for 2 weeks. Normal human dermis and demineralized BS bone were incubated for 24 h at 4°C with 0.5 M acetic acid (HOAc) or pepsin (enzyme weight:dry tissue weight = 1:10) in 0.5 M HOAc. Aliquots of solubilized collagen were run on 7% SDS/PAGE gels and stained with Coomassie Brilliant Blue.
Hyl and Crosslink Analysis.
Measurements were carried out with cortical BS bone (patient 505), trabecular BS bone, articular cartilage (patient 507), and normal cortical bone (male, 14 yr). Hyl and its glycosides were determined after derivatization with 9-fluorenylmethyl chloroformate with RP-HPLC in acid and alkali hydrolysates, respectively (29–30). Pyridinium crosslinks (HP and LP) were quantified in acid hydrolysates of unreduced tissue samples by RP-HPLC (31–32). The difunctional crosslinks dihydroxylysinonorleucine (DHLNL), hydroxylysinonorleucine (HLNL), and lysinonorleucine (LNL) were determined after reduction of the samples with NaB3H4 as described (7). Hydroxyproline values were used to calculate the amounts of crosslinks per collagen molecule (assuming 300 hydroxyproline residues per molecule). To identify the chemical nature of HLNL, the collected radioactive crosslink was subjected to Smith degradation (6–7, 33). The amounts of radiolabeled proline and lysine, which reflect the proportions of HLNL derived from the telopeptide Hyl or telopeptide lysine route, respectively, were measured chromatographically.
Marker Analysis.
A genome-wide screen on DNA extracted from blood samples of the family described earlier (27) was performed by using 244 markers (average spacing 10 cM; average heterozygosity 0.75) from the Marshfield screening set, version 6 (http://www.marshmed.org). Reverse primers were labeled with either 6-FAM, HEX, or TET fluorescent dyes. PCR reactions and marker analysis were carried out as described previously (34).
Homozygosity Mapping.
A total genome screen using 244 markers was performed on DNA from five members of the BS family (501, 502, 505, 507, 509; Fig. 3). For each marker we determined whether the three affected sibs (505, 507, and 509) were heterozygous or homozygous for the same marker alleles. Regions in which two or more consecutive markers showed homozygosity by state were studied further by (i) screening the four healthy sibs (503, 506, 508, 510) with the same markers, and (ii) haplotype analysis to determine whether the regions were also identical by descent. To determine the region of homozygosity on chromosome 17 more precisely, two additional markers were used: D17S1843 and D17S2196. Multipoint lod scores were calculated with conservative allele frequencies of 0.33 for all alleles using linkage, version 5.03 (35).
RESULTS
Analysis of BS Bone Collagen.
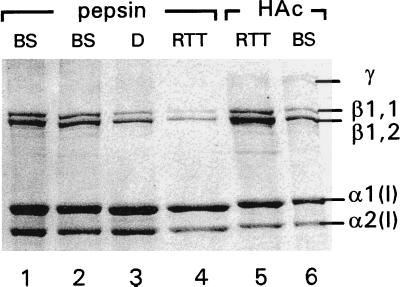
Collagens of BS bone solubilized by 0.5 M acetic acid or pepsin showed a normal migration pattern on SDS/PAGE gels, indicating that the triple helix in BS is not overmodified (Fig. 2). In osteogenesis imperfecta patients such overhydroxylation of the α-chain is frequently seen because of a delayed triple helix formation caused by a mutation in one of the α-chains (36–37). Such a mutation can be excluded in BS. The normal migration of acid-soluble collagen on SDS/PAGE implies the absence of both propeptides. Thus, the N- and C-terminal propeptides were properly processed by their respective peptidases. Furthermore, the collagen molecules are of normal heterotrimeric origin, as a normal α1(I):α2(I) ratio of 2:1 was found. In addition to the monomeric α-bands, dimeric β- and trimeric γ-bands with normal migration behavior were observed. The prominent presence of these crosslinked bands implies that the lysine or Hyl in the telopeptides is effectively oxidized to aldehydes. Therefore, alterations in the enzyme that initiates crosslinking, lysyl oxidase, can be excluded.
Figure 2.
SDS/PAGE of BS bone collagen type I, either solubilized by pepsin (lanes 1 and 2) or extracted with 0.5 M HOAc (lane 6), show no differences compared with pepsinized collagen from normal human dermis (D, lane 3) or rat tail tendon (RTT, lane 4), or with HOAc extracted RTT (lane 5). Collagens solubilized with acid contain intact telopeptides, accounting for the slightly slower electrophoresis migration rate compared with pepsinized collagen with partly degraded telopeptides. Lanes 1 and 6: cortical BS bone, patient 505; lane 2: trabecular BS bone, patient 507.
(Glycosylated) Hyl Levels in Bone, Ligament, and Cartilage.
The amount of Hyl, reflecting the lysyl hydroxylation of the triple helix, was normal in collagen type I from joint ligament (Table 1) and bone (Table 2; Fig. 3). The same was observed with respect to the level of the two glycosylated derivatives of Hyl, galactosyl-Hyl (G-Hyl), and glucosylgalactosyl-Hyl (GG-Hyl). These findings are in agreement with the normal migration pattern of collagen type I on SDS/PAGE (Fig. 2). Thus, in BS, HLH and hydroxylysyl glycosyl transferases are normal. All these findings indicate that the collagen triple helix in BS bone is no different from that of normal bone (Fig. 3). Normal Hyl and glycosylation levels were also observed in collagen type II of cartilage (Table 1).
Table 1.
Lysylhydroxylation and crosslinking in ligament and cartilage
| Residues per collagen molecule
| ||
|---|---|---|
| Amino acid | Ligament (collagen type I) | Cartilage (collagen type II) |
| Bruck/control | Bruck/control | |
| Hyl | 20.5/21.3 | 42.4/43.7 |
| G-Hyl | Not determined | 8.2/9.6 |
| GG-Hyl | Not determined | 12.5/10.4 |
| HP | 0.28/0.41 | 1.2/1.6 |
| LP | 0.018/0.020 | 15.6/20.5 |
Table 2.
Lysylhydroxylation and crosslinking in bone (collagen type I)
| Residues per collagen molecule
| |||
|---|---|---|---|
| Amino acid | Patient 505 | Patient 507 | Control |
| Hyl | 13.7 | 13.1 | 14.4 |
| G-Hyl | <0.8 | <0.8 | <0.8 |
| GG-Hyl | 4.3 | 3.9 | 3.7 |
| HP | 0.011 | 0.021 | 0.35 |
| LP | 0.002 | 0.007 | 0.09 |
| DHLNL | 0.10 | 0.08 | 0.92 |
| HLNL | 0.55 | 0.33 | 0.52 |
| LNL | 0.20 | 0.14 | 0.03 |
| % HLNL (Hyl) | 4% | 6% | 75% |
Crosslinking in Bone.
As shown in Table 2 and Fig. 3, collagen type I from BS bone (patient 505 and 507) showed strongly reduced levels of both pyridinium crosslinks, HP and LP (<8% of normal levels), which are derived from telopeptide Hyl residues. The levels of the difunctional crosslink DHLNL, another crosslink that originates from hydroxylated telopeptides, were <10% of normal values. These findings indicate that hydroxylation of the lysine residues in the telopeptides is very low (Fig. 3). Normal amounts of HLNL were found; this crosslink can result from both telopeptidyl Lys and Hyl. To corroborate further that the telopeptides in BS contain little Hyl, the origin of HLNL was determined by Smith degradation (6–7, 33). The percentage of HLNL derived from hydroxylated telopeptides in BS was around 5%; in normal bone this is around 75%. The sum of telopeptidyl Hyl-derived crosslinks per collagen molecule (HP, LP, DHLNL, part of the HLNL pool) in BS bone was 0.14, whereas in normal bone this is around 2. The low hydroxylation level of the telopeptides was further substantiated by the observed 5-fold elevation over normal levels for LNL, a crosslink derived from telopeptide lysyl residues. The presence of the allysine crosslinking pathway in BS bone was not accompanied by maturation to the nonreducible crosslink histidinohydroxylysinonorleucine (HHL) as in skin and cornea collagen (10–11): only marginal amounts of HHL were detected in affected bone (<0.05 HHL/triple helix). Pyridinium crosslink levels in bone of patient 3 showed, like patient 505 and 507, a 10-fold decrease compared with control bone (data not shown); other crosslinks were not determined.
Crosslinking in Ligament and Cartilage.
In contrast to collagen type I from bone, collagen type I from ligament (patient 3) and collagen type II from cartilage (patient 507) showed normal pyridinium (HP and LP) levels (Table 1). Thus, aberrant crosslinking is restricted to bone.
Genome Screen.
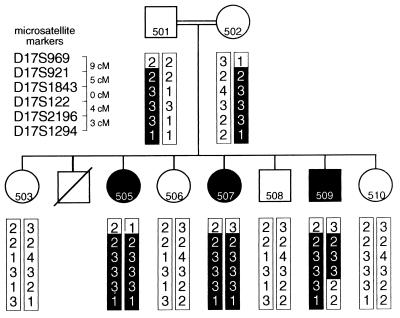
The existence of affected inbred BS individuals made the localization of the gene underlying this disorder a suitable target for homozygosity mapping (Fig. 4). This mapping approach is based on the inheritance of two identical copies of the disease locus by the affected inbred child from a common ancestor, i.e., homozygosity by descent (28). Of 244 markers, 34 showed homozygosity by state in all three affected sibs; they were scattered throughout the genome. Clusters of two or more adjacent homozygous markers were identified on chromosomes 1, 2, 8, and 17 and were considered putative BS candidate regions needing further investigation. Haplotype analysis excluded the chromosomes other than 17 as a candidate region for the BS locus since the three affected sibs did not show identity by descent (data not shown). The affected individuals were homozygous for D17S921 and D17S122 and haplotype analysis showed that the affected individuals, in contrast to the four healthy sibs, inherited two identical copies of that chromosomal region (i.e., homozygous by descent). Further investigation with two additional markers refined the position of the BS further. The telomeric border was defined by D17S969; the recombination event in patient 509 localized the BS locus to a region of maximally 18 cM at 17p12 (Fig. 3). This chromosomal location by haplotype analysis was in agreement with the results of the lod score analysis. The peak lod score was obtained at D17S1843 (Z = 2.5).
Figure 4.
Pedigree and genotypes in the region of homozygosity on 17p12. The regions of homozygosity in the affected children are boxed. The distances between the DNA markers are indicated next to the markers. These distances were extracted from the Marshfield genetic maps (http://www.marshmed.org).
DISCUSSION
BS Provides Genetic Evidence for the Existence of a Tissue-Specific TLH.
In BS bone, substantial amounts of difunctional crosslinks were found, indicating that collagen in the extracellular matrix met most of the spatial requirements for crosslinking. However, the relative abundance of the different di- and trifunctional crosslinks was completely different from that observed in normal bone: there was a preponderance of crosslinks derived from lysine in the telopeptides with very low amounts of telopeptide Hyl-derived crosslinks (Fig. 3). The normal levels of Hyl in the triple helix indicate that the hydroxylation defect is limited to the telopeptides. The most likely explanation for these results is that BS is caused by the lack of an enzyme that is capable of hydroxylating telopeptide lysyl residues (TLH). The normal crosslinking in ligament (collagen type I) and cartilage (collagen type II) indicates a normal telopeptide lysyl hydroxylation in these tissues. Thus, BS not only provides evidence for the existence of a TLH; it also shows the presence of tissue-specific forms: in BS only bone is affected.
Collagen Crosslinking and Mineralization of Bone.
Major changes occur in the crosslink profile of collagen type I during the transition from nonmineralized to mineralized tissue (9, 19). These crosslink profiles in mineralized tissue are different from those of other type I collagen matrices and are characterized by the high amount of difunctional crosslinks, the relatively low HP content, and the presence of significant amounts of LP. These findings suggest that correct crosslinking is critical for proper mineralization. Indeed, inhibition of crosslinking with lathyritic agents results in decreased mineral density of bone (38). It is therefore not surprising that in BS bone, in view of the almost complete absence of HP and LP, a reduction of mineral in relation to the organic matrix is seen, as well as an aberrant size of the apatite crystals (23). The bone brittleness observed in BS is most likely caused by a combination of diminished crosslinking of the telopeptide Hyl route and defective mineralization.
Ehlers–Danlos Syndrome Type VI (EDS-VI) and BS: LH Disorders.
In HP and LP, the amino acid of the telopeptides involved in crosslinking is Hyl. The difference between HP and LP is that the triple helical residue in HP is an Hyl; it is a Lys in LP (4–6, 8). In patients with EDS-VI, defects in a LH gene has been demonstrated. These defects result in a replacement of HP by LP (18, 21), indicating that the telopeptides are normally hydroxylated but that the triple helix contains less Hyl than normal. Thus, EDS-VI provides evidence that the cloned LH gene (13–15) is actually an HLH (18, 21). Ihme et al. (39) showed in a particular individual with EDS-VI that the triple helix of collagen type I from skin and bone was Hyl-deficient, whereas the triple helix of collagen type I from tendon and lung was not. In addition, the triple helix of collagen type V from bone was, in contrast to collagen type I from bone, normally hydroxylated. These data indicate that a family of HLH genes exist, each with its own tissue specificity and collagen type specificity. Evidence for the presence of LH isoenzymes was recently provided at the gene level (40–43). The three LH genes (PLOD1, PLOD2, and PLOD3) can, however, be excluded as candidate genes of BS, as they are localized on chromosome 1, 3, and 7 respectively (42, 43), whereas we localized the BS locus by homozygosity mapping to an 18-cM region of the p12 region on chromosome 17 between the markers D17S969 and D17S2196. Confirmation and further delimitation of this region will require more families (the major limiting factor) and the use of additional polymorphic DNA markers. In summary, the combined findings in EDS-VI and BS show that regulation of lysyl hydroxylation, a key posttranslational event in collagen biosynthesis, is highly complex: telopeptide and triple helical lysine residues are hydroxylated by different enzymes; both enzymes show tissue-specific forms; and, at least in the case of HLH, collagen type-specific forms exist as well.
Implications of Tissue-Specific TLHs.
Growth, development, and aging involve major changes in crosslinking, even within the same tissue (9, 19, 44–46). Despite its obvious importance, the mechanism of tissue specificity of crosslinking remained elusive. Our findings for BS provide compelling evidence that the type of crosslinking, as well as its abundance, is regulated primarily by tissue-specific enzymes that specifically hydroxylate telopeptide Lys residues, explaining why crosslink patterns are tissue specific and not necessarily related to a genetic collagen type. Notably, a switch in the telopeptide-lysyl-crosslink pathway toward the telopeptide-hydroxylysyl-pathway is seen in injured tissues, fibrotic tissues, scars, and sclerosis (12, 20, 47–48). This switch is considered to be involved in the accumulation of collagen in, e.g., fibrosis (20, 48). Therefore, molecules capable of regulating (i.e., suppressing) the expression or enzyme activity of TLH provide an exciting avenue of novel antifibrotic drugs. Presently, suppression of excessive deposition of collagen, the hallmark of fibrosis, focuses on inhibition of collagen processing enzymes such as prolyl hydroxylase or lysyl oxidase. This results in unphysiological collagen that is underhydroxylated or lacks crosslinking. In contrast, inhibition of TLH does not affect the normal processing of collagen molecules; it only reduces the aberrant crosslinking typical of fibrotic pathology. Identification of the gene, its transcriptional/translational control, and the biochemical properties of TLH is an important challenge that merits further investigation.
Acknowledgments
We are indebted to Dr. M. Yamauchi (University of North Carolina, Chapel Hill) for providing a HHL standard, to Geja J. Oostingh for running SDS gels, to Bob Beekman for art work, and to Lodewijk A. Sandkuijl for statistical analysis. S.P.R. is indebted to the Scottish Office of Agriculture, Environment and Fisheries Department for support.
ABBREVIATIONS
- BS
Bruck syndrome
- LH
lysyl hydroxylase
- HLH
helical lysyl hydroxylase
- G-Hyl
galactosyl-hydroxylysine
- GG-Hyl
glucosylgalactosyl-hydroxylysine
- LNL
lysinonorleucine
- DHLNL
dihydroxylysinonorleucine
- HLNL
hydroxylysinonorleucine
- HP
hydroxylysylpyridinoline
- LP
lysylpyridinoline
- Hyl
hydroxylysine
- TLH
telopeptide lysyl hydroxylase
- HOAc
acetic acid
- HHL
histidinohydroxylysinonorleucine
- EDS-VI
Ehlers–Danlos syndrome type VI
References
- 1.Einhorn T. Calcif Tissue Int. 1992;51:333–339. doi: 10.1007/BF00316875. [DOI] [PubMed] [Google Scholar]
- 2.Oxlund, H., Barckman, M., Ortoff, G. & Andreassen, T. T. (1995) Bone 17, Suppl., 365S–371S. [DOI] [PubMed]
- 3.Knott L, Whitehead C C, Fleming R H, Bailey A J. Biochem J. 1995;310:1045–1051. doi: 10.1042/bj3101045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eyre D R, Paz A, Gallop P M. Annu Rev Biochem. 1984;53:717–748. doi: 10.1146/annurev.bi.53.070184.003441. [DOI] [PubMed] [Google Scholar]
- 5.Reiser K, McCormick R J, Rucker R B. FASEB J. 1992;6:2439–2449. doi: 10.1096/fasebj.6.7.1348714. [DOI] [PubMed] [Google Scholar]
- 6.Herbage D, Le Lous M, Cohen-Solal L, Bazin S. Front Matrix Biol. 1985;10:59–91. [Google Scholar]
- 7.Robins S P. Methods Biochem Anal. 1982;28:330–379. doi: 10.1002/9780470110485.ch8. [DOI] [PubMed] [Google Scholar]
- 8.Yamauchi M, Mechanic G L. In: Collagen. Vol. 1: Biochemistry. Nimni M E, editor. Boca Raton, FL: CRC; 1988. pp. 157–172. [Google Scholar]
- 9.Yamauchi M, Katz E P. Connect Tissue Res. 1993;29:81–98. doi: 10.3109/03008209309014236. [DOI] [PubMed] [Google Scholar]
- 10.Mechanic G L, Katz E P, Henmi M, Noyes C, Yamauchi M. Biochemistry. 1987;26:3500–3509. doi: 10.1021/bi00386a038. [DOI] [PubMed] [Google Scholar]
- 11.Yamauchi M, Chandler G S, Tanzawa H, Katz E P. Biochem Biophys Res Commun. 1996;219:311–315. doi: 10.1006/bbrc.1996.0229. [DOI] [PubMed] [Google Scholar]
- 12.Brinckmann J, Açil Y, Tronnier M, Notbohm H, Bätge B, Schmeller W, Koch M H J, Müller P K, Wolff H H. J Invest Dermatol. 1996;107:589–592. doi: 10.1111/1523-1747.ep12582991. [DOI] [PubMed] [Google Scholar]
- 13.Hautala T, Byers M G, Eddy R L, Shows T B, Kivirikko K I, Myllylä R. Genomics. 1992;13:62–69. doi: 10.1016/0888-7543(92)90202-4. [DOI] [PubMed] [Google Scholar]
- 14.Heikkinen J, Toppinen T, Yeowell H, Krieg T, Steinmann B, Kivirikko K I, Myllylä R. Am J Hum Genet. 1997;60:48–56. [PMC free article] [PubMed] [Google Scholar]
- 15.Yeowell H N, Walker L C. Proc Assoc Am Physicians. 1997;109:383–396. [PubMed] [Google Scholar]
- 16.Royce P M, Barnes M J. Biochem J. 1985;230:475–480. doi: 10.1042/bj2300475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerriets J E, Curwin S L, Last J A. J Biol Chem. 1993;268:25553–25560. [PubMed] [Google Scholar]
- 18.Steinmann B, Eyre D R, Shao P. Am J Hum Genet. 1995;57:1505–1508. [PMC free article] [PubMed] [Google Scholar]
- 19.Knott L, Tarlton J F, Bailey J F. Biochem J. 1997;322:535–542. doi: 10.1042/bj3220535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerriets J E, Reiser K M, Last J A. Biochim Biophys Acta. 1996;1316:121–131. doi: 10.1016/0925-4439(96)00019-1. [DOI] [PubMed] [Google Scholar]
- 21.Pasquali M, Still M J, Vales T, Rosen R I, Evinger J D, Dembure P P, Longo N, Elsas L J. Proc Assoc Am Physicians. 1997;109:33–41. [PubMed] [Google Scholar]
- 22.Bruck A. Dtsch Med Wochenschr. 1897;23:152–155. [Google Scholar]
- 23.Brenner R E, Vetter U, Stöss H, Müller P K, Teller W M. Eur J Pediatr. 1993;152:505–508. doi: 10.1007/BF01955060. [DOI] [PubMed] [Google Scholar]
- 24.Sharma N L, Anand J S. Indian Med J. 1964;53:124–126. [PubMed] [Google Scholar]
- 25.Viljoen D, Versfeld G, Beighton P. Clin Genet. 1989;36:122–126. doi: 10.1111/j.1399-0004.1989.tb03174.x. [DOI] [PubMed] [Google Scholar]
- 26.McPherson E, Clemens M. Am J Med Genet. 1997;70:28–31. [PubMed] [Google Scholar]
- 27.Breslau-Siderius E J, Engelbert R H H, Pals G, Van der Sluijs J A. J Pediatr Orthop. 1998;B 7:35–38. [PubMed] [Google Scholar]
- 28.Lander E S, Botstein D. Science. 1987;236:1567–1570. doi: 10.1126/science.2884728. [DOI] [PubMed] [Google Scholar]
- 29.Bank R A, Jansen E J, Beekman B, TeKoppele J M. Anal Biochem. 1996;240:167–176. doi: 10.1006/abio.1996.0346. [DOI] [PubMed] [Google Scholar]
- 30.Bank R A, Beekman B, Tenni R, TeKoppele J M. J Chromatogr. 1997;B:703. doi: 10.1016/s0378-4347(97)00392-7. , 267–272. [DOI] [PubMed] [Google Scholar]
- 31.Bank R A, Beekman B, Verzijl N, De Roos J A D M, Sakkee A N, TeKoppele J M. J Chromatogr. 1997;B 703:37–44. doi: 10.1016/s0378-4347(97)00391-5. [DOI] [PubMed] [Google Scholar]
- 32.Robins S P, Duncan A, Wilson N J, Evans B J. Clin Chem. 1996;42:1621–1626. [PubMed] [Google Scholar]
- 33.Robins S P, Bailey A J. Biochem J. 1975;149:381–385. doi: 10.1042/bj1490381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giltay J C, Brunt T, Beemer F A, Wit J M, Ploos van Amstel H K, Pearson P L, Wijmenga C. Am J Hum Genet. 1998;62:937–940. doi: 10.1086/301796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lathrop G M, Lalouel J M, Julier C, Ott J. Am J Hum Genet. 1985;37:428–498. [PMC free article] [PubMed] [Google Scholar]
- 36.Engel J, Prockop D J. Annu Rev Biophys Biophys Chem. 1991;20:137–152. doi: 10.1146/annurev.bb.20.060191.001033. [DOI] [PubMed] [Google Scholar]
- 37.Raghunath M, Bruckner P, Steinmann B. J Mol Biol. 1994;236:940–949. doi: 10.1006/jmbi.1994.1199. [DOI] [PubMed] [Google Scholar]
- 38.Lees S, Barnard S M, Churchill D. Ultrasound Med Biol. 1987;139:19–24. doi: 10.1016/0301-5629(87)90156-6. [DOI] [PubMed] [Google Scholar]
- 39.Ihme A, Krieg T, Nerlich A, Feldmann U, Rauterberg J, Glanville R W, Edel G, Müller P K. J Invest Dermatol. 1984;83:161–165. doi: 10.1111/1523-1747.ep12263502. [DOI] [PubMed] [Google Scholar]
- 40.Yeowell H N, Ha V, Clark W, Marshall M K, Pinnell S R. J Invest Dermatol. 1994;102:382–384. doi: 10.1111/1523-1747.ep12371799. [DOI] [PubMed] [Google Scholar]
- 41.Valtavaara M, Papponen H, Pirttilä, Hiltunen K, Helander H, Myllylä R. J Biol Chem. 1997;272:6831–6834. doi: 10.1074/jbc.272.11.6831. [DOI] [PubMed] [Google Scholar]
- 42.Szpirer C, Szpirer, Rivière M, Vanvooren P, Valtavaara M, Myllylä R. Mamm Genome. 1997;8:707–708. doi: 10.1007/s003359900549. [DOI] [PubMed] [Google Scholar]
- 43.Valtavaara M, Szpirer C, Szpirer J, Myllyläla R. J Biol Chem. 1998;273:12881–12886. doi: 10.1074/jbc.273.21.12881. [DOI] [PubMed] [Google Scholar]
- 44.Yamauchi M, Woodley D T, Mechanic G L. Biochem Biophys Res Commun. 1988;152:898–903. doi: 10.1016/s0006-291x(88)80124-4. [DOI] [PubMed] [Google Scholar]
- 45.Barnes M J, Constable B J, Morton L F, Royce P M. Biochem J. 1974;139:461–468. doi: 10.1042/bj1390461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robins S P, Shimokomaki M, Bailey A J. Biochem J. 1973;131:771–780. doi: 10.1042/bj1310771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bailey A J, Bazin S, Sims T J, Le Lous M, Nicoletis C, Delauny A. Biochim Biophys Acta. 1975;405:412–421. doi: 10.1016/0005-2795(75)90106-3. [DOI] [PubMed] [Google Scholar]
- 48.Bailey A J, Light N D. Ciba Found Symp. 1985;114:80–96. doi: 10.1002/9780470720950.ch6. [DOI] [PubMed] [Google Scholar]