Abstract
White–opaque switching in the human fungal pathogen Candida albicans is an alternation between two distinct types of cells, white and opaque. White and opaque cells differ in their appearance under the microscope, the genes they express, their mating behaviors, and the host tissues for which they are best suited. Each state is heritable for many generations, and switching between states occurs stochastically, at low frequency. In this article, we identify a master regulator of white–opaque switching (Wor1), and we show that this protein is a transcriptional regulator that is needed to both establish and maintain the opaque state. We show that in opaque cells, Wor1 forms a positive feedback loop: It binds its own DNA regulatory region and activates its own transcription leading to the accumulation of high levels of Wor1. We further show that this feedback loop is self-sustaining: Once activated, it persists for many generations. We propose that this Wor1 feedback loop accounts, at least in part, for the heritability of the opaque state. In contrast, white cells (and their descendents) lack appreciable levels of Wor1, and the feedback loop remains inactive. Thus, this simple model can account for both the heritability of the white and opaque states and the stochastic nature of the switching between them.
Keywords: phenotypic switching, transcriptional regulation, WOR1
In this article, we examine the interconversion between two distinctive types of cells in the human fungal pathogen Candida albicans. This interconversion, which plays important roles in both pathogenesis and mating, exemplifies two characteristics shared by many examples of cell differentiation: The conversion from one cellular state to another is stochastic, and each state, once formed, is heritable for many generations.
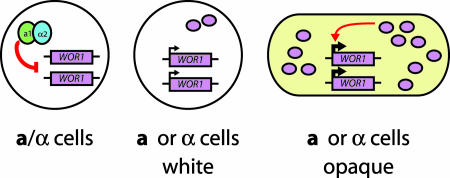
The property of C. albicans we investigate is called white–opaque switching, and it refers to an alternation between two distinctive types of cells, white and opaque (1). White cells generally give rise to white cell progeny, but, approximately every 10,000 generations, a white cell spontaneously switches to the opaque form, which then will produce opaque cell progeny for many generations (2, 3). Conversely, an opaque cell can spontaneously switch back to the white form, and the progeny of this cell will remain in the white form for many generations. Any molecular mechanism for white–opaque switching therefore must account for the ability of each state to stochastically convert to the other as well as the heritability of each state, once formed.
White and opaque cells of C. albicans differ in many features (for reviews, see refs. 4–6). They are easily distinguished under the microscope, with white cells appearing nearly spherical and opaque cells appearing larger and more elongated. When grown on agar plates, white cells form white, dome-shaped colonies, whereas opaque colonies are darker and lie flatter against the agar. Many, if not all, of the differences between white and opaque cells are due to differences in gene expression: For example, mRNAs from ≈400 genes (≈7% of the genome) are present in significantly different levels in white compared with opaque cells (7, 8). These genes cover a wide range of functions including adhesion, drug resistance, metabolism, virulence, and mating.
Although the full range of biological roles for white–opaque switching are only beginning to be appreciated, a few specific examples are well documented. C. albicans can colonize many different niches in the mammalian host, and white and opaque cells differ significantly in this regard. Although white cells are more suited for bloodstream infections, opaque cells are better at colonizing skin surfaces (9–11). Thus, white–opaque switching provides C. albicans with two distinctive types of cells that interact differently with the host. White–opaque switching also has a key role in the mating of C. albicans. White cells mate poorly (if at all), whereas opaque cells mate with high efficiency (12). The key role of white–opaque switching in mating also is reflected by the fact that the mating-type locus of C. albicans controls white–opaque switching: Whereas a and α cells (the mating forms) are permissive for switching, a/α cells cannot switch and remain locked in the white form. This block to white–opaque switching in a/α cells is mediated through the a1-α2 heterodimer, a transcriptional repressor (12, 13).
In this article, we investigate the molecular mechanism of white–opaque switching. We begin by identifying a master regulator of white–opaque switching, the WOR1 gene. We show that strains deleted for WOR1 are locked in the white form and that ectopic expression of WOR1 in white cells converts the population en masse to opaque cells. Ectopic WOR1 expression also can override, at least partially, the a1-α2 block to white–opaque switching. We show that Wor1 protein is normally present at very low levels in white cells but accumulates to high levels in opaque cells through the action of a positive feedback loop: Wor1 binds to its own promoter and activates its own synthesis. Finally, we show that a pulse of ectopically expressed Wor1 in white cells converts the entire population to opaque cells and that these opaque cells continue to give rise to opaque progeny for many generations after the ectopic construct has been turned off. Based on these results, we propose a simple model, a self-sustaining transcriptional feedback loop present in opaque but not white cells, that can account for the stochastic nature of white–opaque switching and for the heritability of each of the two states.
Results
Identification of WOR1 as a Regulator of White–Opaque Switching.
To identify genes controlling white–opaque switching, we based our strategy on the observation that the a1-α2 heterodimer blocks white–opaque switching (12). We therefore considered all genes repressed by a1-α2 in white cells as candidate regulators of white–opaque switching. Six a1-α2 repressed genes [CEK2 (orf19.460), STE2 (orf19.696), FGR23 (orf19.1616), FAR1 (orf19.7105), CAG1 (orf19.4015), and WOR1 (orf19.4884)] were identified in our previous microarray analysis (8), and we constructed two independent knockout strains for each gene in an a cell background. All of the mutant strains underwent white-to-opaque switching at normal frequencies (14), as monitored by sectored colony formation (typically 2–5% of colonies show opaque sectors), except for the two strains deleted for the WOR1 gene (based on the work in this article, WOR1 was named as White–Opaque Regulator 1). These strains failed to switch; that is, they appeared locked in the white phase. In the course of this work, we examined >6,000 colonies of the wor1Δ strains and never observed an opaque colony or sector. Note that C. albicans is diploid and that construction of a knockout strain requires sequential disruption of both gene copies. For convenience, we will denote strains deleted for both copies of the WOR1 gene simply as wor1Δ mutants.
WOR1 codes for a class of conserved fungal proteins that have been implicated in several biological processes but whose precise biochemical function is not known (Fig. 1). For example, Schizosaccharomyces pombe has two proteins closely related to WOR1: GTI1 regulates alternative sugar uptake (16), and PAC2 regulates sexual development (17). The C. albicans WOR1 gene was identified (as EAP2) by its ability to enhance adhesiveness to polystyrene when introduced into Saccharomyces cerevisiae, but the basis of this effect has not been investigated (18). We show in this article that Wor1 is a transcriptional regulator, a result strongly suggesting that all members of this protein class share this function.
Fig. 1.
Alignment of Wor1 homologs across fungal species. Protein sequences were aligned by using ClustalW (15), which identified a highly conserved region at the N terminus of each protein (blue). For instance, C. albicans Wor1 and Sc. pombe Gti1 are 53% identical across the conserved region. C. albicans Wor1 protein is 785 aa in length, and the other proteins are drawn to scale.
Ectopic Expression of WOR1 Converts an Entire Population of White Cells to Opaque Cells.
As described in the Introduction, white a or α cells typically switch to the opaque form once every ≈10,000 generations. When a copy of the WOR1 coding region was placed under control of the MET3 promoter (19) and its expression was induced in white a cells, the entire population of white cells was converted to opaque cells (Fig. 2). We identify these cells as bona fide opaque cells by four criteria: They have the cell shape characteristic of opaque cells (Fig. 2 A and B), they form colonies with the highly characteristic morphology of those formed by normal opaque cells (data not shown), they up-regulate transcription of opaque-specific genes and down-regulate white-specific genes (Fig. 2C), and they respond to mating pheromone in the way that only opaque cells do: by forming highly characteristic mating projections (ref. 14; Fig. 2D). Control experiments demonstrate that the pMET3-WOR1 construct has no significant effect on white–opaque switching unless it is induced (Fig. 2A); in addition, the nutritional conditions used to regulate the MET3 promoter have no effect on white–opaque switching in cells that lack the pMET3-WOR1 construct (data not shown). We considered the possibility that Wor1 also could regulate hyphal growth in C. albicans, thereby confusing our identification of opaque cells by their increased length-to-width ratios. To test this possibility, we monitored hyphal growth in the wor1Δ strains on spider medium and on YEPD + 10% serum and found it to be normal (data not shown). Thus, WOR1 is necessary for opaque cell formation but not for hyphal cell formation.
Fig. 2.
Ectopic expression of WOR1 in white cells drives the cells to the opaque phase. (A and B) Ectopic expression of WOR1 causes white cells to resemble opaque cells in appearance. Cell dimensions were measured in differential interference contrast images (B), and populations of cells were compared based on the distribution of length/width ratios for 50 cells per strain for each condition (A). (C) Ectopic expression of WOR1 in white cells causes them to express genes characteristic of opaque cells. Quantitative RT-PCR was used to monitor transcription of the white-specific genes WH11 and EFG1 and the opaque-specific genes SAP1 and OP4. All values were normalized to PAT1, a transcript that is not regulated by white–opaque switching. (D) Ectopic expression of WOR1 in white cells renders them sensitive to the mating pheromone α-factor. This specialized property of true opaque cells is visualized by the formation of mating projections on the ends of the cells (14). Cells were treated with α-factor (10 μg/ml in DMSO) or an equivalent amount of DMSO as a control. In A and B, “ON” and “OFF” indicate the expression of the pMET3-WOR1 construct, as controlled by media conditions. For strains that lack the pMET3-WOR1 construct, media conditions are designated by “ON” or “OFF.” For experiments shown in C and D, strains were grown in media that induces pMET3-WOR1 expression. All strains are a strains.
Wor1 Activates Its Own Transcription and Binds to Its Own Promoter.
When WOR1 is expressed ectopically in white cells, transcription from the endogenous copies of WOR1 is strongly induced, demonstrating that Wor1 activates its own transcription (Fig. 3A). As indicated in the figure, the endogenous transcript is considerably larger than that produced from the pMET3-WOR1 construct and is easily distinguished from it.
Fig. 3.
Northern and Western blot analysis of WOR1 expression. (A) Northern blot analysis of total RNA isolated from strains grown under conditions that induce the pMET3-WOR1 construct. The relevant genotypes are indicated above each lane (all strains are a strains). RPL5 serves as a loading control. (B) Immunoblot analysis of Wor1 protein levels in white and opaque cells of several different strains. Wor1 was detected in WCE by using an antibody (α-Wor1) generated against a peptide portion of Wor1. Strain 1 (a, wor1Δ) controls for nonspecific binding of the antibody. Strain 2 (CAF2-1, a, white) and strain 3 (CAF2-1, a, opaque) show the differential expression of Wor1 between white and opaque cells. Strain 4 (a, pMET3-WOR1), strain 5 (a, wor1Δ + pMET3-WOR1), and strain 6 (a/α, pMET3-WOR1) show that the pMET3-WOR1 construct is tightly regulated by media conditions and that the protein is not grossly overexpressed. Strain 7 (SN87, a, white) and strain 8 (SN87, a, opaque) again show the differential expression of Wor1 in white versus opaque cells. The blot was stripped and reprobed with α-Tub1 as a loading control. “ON” and “OFF” indicate media conditions used to regulate the pMET3-WOR1 construct, as described in Fig. 2.
The simplest model for Wor1 activating its own transcription predicts that Wor1 binds to its own DNA regulatory region. Inspection of the Wor1 amino acid sequence indicated that it lacks all of the conventional motifs associated with sequence-specific DNA binding or other aspects of transcriptional regulation. We therefore experimentally tested the idea that Wor1 is a transcriptional regulator of its own gene by ChIP. Protein–DNA complexes were cross-linked in opaque cells, sheared, and precipitated by using affinity-purified antibodies directed against a Wor1 peptide. As shown in Fig. 4, the Wor1 protein specifically occupies several discrete positions upstream of its gene. For this experiment, all values for immunoprecipitated DNA were normalized to an ADE2 control; the peaks of Wor1 occupancy are ≈10-fold above both the ADE2 values and those of the “troughs” in the WOR1 upstream region. Control experiments demonstrate that no significant precipitation of the WOR1 control region is observed in white a, a/α, or a wor1Δ cells (Fig. 4).
Fig. 4.
Wor1 protein is bound to the region upstream of its gene. ChIP was performed with α-Wor1 antibodies in wild-type a opaque, wild-type a white, wild-type a/α, and wor1Δ a strains. Wor1 ChIP enrichment was detected by quantitative PCR at ≈250-bp intervals across the 10.3-kb intergenic region. Shown are enrichment values at each position upstream of WOR1 relative to a control gene (ADE2) that is not regulated by white–opaque switching.
Although we have not formally shown that Wor1 binds DNA directly, binding seems likely given the multiple discrete sites of occupancy in the WOR1 upstream region. If true, this binding could mean that Wor1 exemplifies a previously undescribed motif for sequence-specific DNA recognition.
Wor1 Protein Accumulates to High Levels in Opaque Cells.
The results of the ChIP experiments described above demonstrate that Wor1 occupies its own DNA regulatory region in opaque but not in white cells of the same genotype (Fig. 4). A simple explanation for this finding is that Wor1 is present at a much higher concentration in opaque cells than in white cells, thereby driving its DNA occupancy. As shown in Fig. 3A, the WOR1 transcript is present at higher levels in opaque cells than white cells, a result also consistent with previous microarray experiments (7, 8). In the experiment of Fig. 3B, we monitored levels of Wor1 protein by Western blot analysis of crude extracts prepared from a variety of white and opaque strains. Consistent with the mRNA regulation, the results clearly show that the Wor1 protein is present in much higher concentrations in opaque cells compared with white cells. The immunoblot also shows that, when induced, the pMET3-WOR1 construct does not grossly under- or overexpress the Wor1 protein. This experiment also includes a series of control experiments that unambiguously establish that the antibodies specifically recognize the Wor1 protein; these same antibodies were used in the ChIP experiments of Fig. 4.
A Pulse of WOR1 Expression Is Sufficient to Stably Convert White Cells to Opaque Cells.
Our results indicate that Wor1 turns on its own transcription, resulting in high levels of the protein in opaque cells. In contrast, the protein is expressed at very low levels in white cells. These observations suggest a simple model for white–opaque switching based on a positive feedback loop: In white cells, levels of Wor1 protein are below the threshold needed to activate its own synthesis; switching to opaque cells occurs when this threshold is exceeded and the feedback loop is activated. According to this model, the positive feedback loop should be self-sustaining; that is, once excited, it should persist for many generations. In this regard, the model makes two important predictions, which we test in turn.
First, if the model is correct, ectopic expression of WOR1 in white cells should be needed only transiently to convert the population to stable opaque cells. In other words, ectopic expression of WOR1 should be needed to initially excite the feedback loop, but continued ectopic expression should not be required to maintain it. This prediction was borne out by the experiment of Table 1 (lines 1 and 2). Here, ectopic expression of WOR1 was induced in white cells for several generations, converting the population to opaque cells, and then shut off. Many generations later (enough to form a colony from a single cell), these cells were still in the opaque form, and they continued to give rise to opaque progeny cells. Thus, the cells retained a memory of the pulse of ectopic WOR1 expression, faithfully maintaining the opaque state for many generations after the original stimulus had been removed.
Table 1.
Transient ectopic expression of WOR1 forms stable opaque colonies in a strains containing the endogenous WOR1 genes
| Strain | Mating type | Phenotype on inducing media | % opaque colonies when replated to repressing media | n |
|---|---|---|---|---|
| pMET3 control | a | White | 0.38 | 789 |
| pMET3-WOR1 | a | Opaque | 97 | 443 |
| wor1Δ + pMET3 control | a | White | <0.44 | 228 |
| wor1Δ + pMET3-WOR1 | a | Opaque-like | <0.34 | 295 |
| pMET3 control | a/α | White | <0.13 | 770 |
| pMET3-WOR1 | a/α | Opaque-like | <0.13 | 768 |
Ectopic expression of pMET3-WOR1 was induced by growth on appropriate media, and strains formed opaque or opaque-like colonies as described in Results. When replated onto media that represses expression of the pMET3-WOR1 construct, only the a strains that contain the endogenous copies of WOR1 were able to maintain the opaque state.
A second prediction of the feedback loop model is that continued expression of Wor1 should be needed to maintain the opaque state; that is, Wor1 should be needed not only to establish but also to maintain the opaque state. To test this prediction, we deleted the endogenous copies of WOR1 and introduced the pMET3-WOR1 construct. When the construct was induced in white cells, the population converted to a mix of cells types, with many resembling true opaque cells but others showing a less elongated shape. However, when the pMET3-WOR1 construct was turned off, all of the cells, including those resembling true opaques, reverted to white cells as judged by both cell appearance and colony morphology (Table 1, lines 3 and 4). This experiment shows that, unless the endogenous copies of WOR1 are present, a pulse of Wor1 expression from the ectopic construct is not sufficient to generate a heritable opaque state. Thus, Wor1 is necessary for both the establishment and the maintenance of the opaque state.
Ectopic Expression of WOR1 Can Override the a1-α2 Block to White–Opaque Switching.
As discussed in the Introduction, white–opaque switching can occur in a cells and α cells, but is blocked in a/α cells by the transcriptional repressor a1-α2. We present four lines of evidence that argue that the a/α block to white–opaque switching is due to repression of WOR1 transcription by a1-α2. First, as discussed above, WOR1 is absolutely required for white–opaque switching, hence, its repression would be sufficient to block switching. Second, our previous work (8) showed that WOR1 transcription is indeed repressed by a1-α2. Third, a1-α2 binding sites are highly conserved between C. albicans and Sa. cerevisiae (20), and we found a close match (TTGATGTGATTTTTAACACG) to the composite consensus sequence in the WOR1 upstream region. To provide a fourth test of the idea that the a/α block to white–opaque switching is due to repression of WOR1, we introduced the pMET3-WOR1 construct into a/α cells and induced expression of WOR1. We observed conversion en masse of the population to an opaque-like form: Many, but not all, of the cells resembled true opaque cells under the microscope (Fig. 5A); the colonies resembled, but were not identical to, those formed by true opaque cells (not shown); and opaque-specific genes were induced, and white specific genes were repressed (Fig. 5B). As for the case of the wor1Δ strain, continued expression of the pMET3-WOR1 construct was required to maintain this state: When the construct was turned off in the a/α strain, the cells reverted to the white form (Table 1, lines 5 and 6). This experiment shows that ectopic expression of WOR1 partially can override the a/α block to white–opaque switching and, in combination with the other observations cited above, demonstrates that repression of WOR1 by a1-α2 is sufficient to explain why a/α cells cannot undergo white–opaque switching. The experiment also confirms that expression of the endogenous copies of WOR1 is necessary to maintain the heritability of the opaque state once the ectopic construction is turned off.
Fig. 5.
Ectopic expression of WOR1 in a/α cells induces opaque-like characteristics. Ectopic expression of WOR1 was regulated by using the pMET3-WOR1 construct in a/α strains. (A) Ectopic expression of WOR1 causes white cells to resemble opaque cells in appearance. Cell dimensions were measured in differential interference contrast images, and populations of cells were compared based on the distribution of length/width ratios for 50 cells per strain for each condition. “ON” and “OFF” indicate media conditions used to regulate the pMET3-WOR1 construct, as described in Fig. 2. (B) Ectopic expression of WOR1 in white cells causes them to express genes characteristic of opaque cells. Quantitative RT-PCR was used to monitor transcription of the white-specific genes WH11 and EFG1 and the opaque-specific genes SAP1 and OP4 under conditions that induce the pMET3-WOR1 construct. All values were normalized to PAT1, a transcript that is not regulated by white–opaque switching.
Discussion
Discovered nearly 20 years ago, white–opaque switching in C. albicans is an interconversion between two different types of cells, white and opaque. As reviewed in Introduction, white and opaque cells differ in their appearances, the genes they express, the host tissues they are most suited for, and their mating behavior. White–opaque switching in C. albicans embodies two critical features of gene expression that underlie numerous examples of cell differentiation. First, switching is stochastic, occurring on average once per 10,000 cell generations. Second, the two states are heritable; that is, white cells give rise to white progeny and opaque cells to opaque progeny. This inheritance proceeds for many generations until a cell spontaneously switches to the other form.
In this article, we investigate the molecular mechanism of white–opaque switching. We first identify a master regulator of white–opaque switching, the WOR1 gene. We show that this gene is required for white–opaque switching, and that, when ectopically expressed, it converts wholescale a population of white cells to opaque cells. We show that WOR1 forms a positive feedback loop: The protein binds to its own promoter, activates its own transcription, and accumulates to high levels in opaque cells. We demonstrate that this feedback loop is self-sustaining by showing that a pulse of ectopic WOR1 expression is sufficient to convert a whole population of white cells into opaque cells and that these cells continue to generate opaque cell progeny many generations after the pulse was ended. We also provide an explanation for the genetic block to white–opaque switching in a/α cells: The a1-α2 heterodimer represses WOR1 expression. These results show that WOR1 is a master regulator of white–opaque switching and that it is required both to establish and maintain the opaque state. We propose that it does so by forming a self-sustaining positive feedback loop, which produces high levels of the protein in the opaque state. It seems likely that, in addition to turning on its own expression, Wor1 activates a set of opaque-specific genes, whose expression endows the opaque state with its specialized properties.
In its most succinct form, our model for white–opaque switching is given in Fig. 6. a/α cells cannot undergo white–opaque switching because WOR1 transcription is repressed. In a and α white cells, WOR1 is expressed at low levels, below the threshold necessary to excite the WOR1-positive feedback loop. According to the model, the level of WOR1 expression in white cells exhibits cell-to-cell variation (noise) and, in a population, the threshold level of WOR1 will be exceeded in a small number of cells. In these rare cells, the feedback loop will be activated, Wor1 levels will accumulate, and the cells will switch to the opaque form. A similar idea can explain the switch from opaque back to white: If the levels of WOR1 expression drop below the threshold needed for self-activation, the feedback loop will be broken, and cells will revert to the white form. The self-sustaining feedback loop model also explains the heritability of the two states. According to the model, white cells give rise to white cells because their progeny receive only low levels of Wor1 protein. In contrast, opaque cell progeny would receive sufficiently high levels of Wor1 to maintain the feedback loop.
Fig. 6.
A model for a positive Wor1 feedback loop in regulation of white–opaque switching.
Although the simplified model of Fig. 6, in principle, can account for the critical features of white–opaque switching, the actual circuitry probably includes additional features that further stabilize the white and opaque states. For example, Wor1 binds to multiple positions in the WOR1 upstream region, and cooperative effects may well sharpen the switch-like behavior of the white–opaque transition. It is also possible that WOR1 levels in white a and α cells are kept low by a repressor, thereby ensuring a low (but observable) switching rate, and the production of this repressor is antagonized as Wor1 levels increase. It is also possible that the high levels of WOR1 production in opaque cells are limited, perhaps by a negative feedback loop. The structure of the WOR1 gene itself is consistent with additional regulatory inputs: Its mRNA contains extensive untranslated regions, and the DNA control region appears to be on the order of 8 kb in length.
In closing, we note that our experiments establish that white–opaque switching is almost certainly an epigenetic phenomenon; that is, switching creates a heritable change without altering the primary DNA sequence. (Technically, a small, reversible DNA rearrangement cannot be rigorously ruled out, but this idea seems highly unlikely in light of the experiments presented here.) In eukaryotes such as C. albicans, epigenetic changes often are attributed to heritable changes in chromatin structure, but we are proposing a very different type of model for white–opaque switching: the inheritance of a diffusible protein that directs its own production. This idea is reminiscent of the epigenetic alterations between the immune and antiimmune states of Escherichia coli containing derivatives of bacteriophage λ (21). In this case, inheritance of the immune state is based on a diffusible protein (the λ repressor), which activates its own synthesis.
Many challenges remain in understanding white–opaque switching in C. albicans. Given the apparent absence of this type of phenomenon in many other fungi, including Sa. cerevisiae, it is possible that white–opaque switching coevolved during C. albicans’ long association with warm-blooded animals (22). Perhaps producing two distinct types of cells has enabled C. albicans to thrive in the hostile environment created by an evolving innate immune system. The simple mechanism we have proposed suggests that an epigenetic phenomenon like white–opaque switching easily could have evolved from more conventional (that is, nonheritable) types of transcriptional circuits.
Materials and Methods
Media.
Standard laboratory media have been described in ref. 23. Synthetic complete medium plus 2% glucose and 100 μg/ml uridine (SCD+Urd) was used to maintain strains in the white and opaque phases at room temperature. Supplemented Lee’s medium (1) was supplemented further with 70 μg/ml Arg/81 μg/ml Ade/0.023 μg/ml His/100 μg/ml Urd. For the MET3 induction experiments (18), cells were grown in SCD+Urd-lacking Met and Cys (SD-Met-Cys+Urd). To repress the MET3 promoter, SCD+Urd was supplemented with 2.5 mM each of Met and Cys. Strains were grown on solid media for 5–7 days at room temperature before inoculating liquid cultures used in the experiments described below.
Plasmids.
To generate pMET3-WOR1, the WOR1 ORF was amplified from SC5314 genomic DNA by using primers containing BglII and AvaI restriction sites and was cloned into BamHI/BspEI-digested pCaEXP (18), creating plasmid pRZ25.
Strain Construction.
All strains were derived from SC5314. With the exception of the cag1Δ mutant, the a strains were generated by growth on sorbose-containing medium, as described (see ref. 14 and references therein). Mating type was determined by PCR (12).
The cag1Δ mutant was derived from RM1000: Urablaster methods by using pCH152 disrupted the MTLa1 and MTLa2 genes (24, 25), and the two alleles of CAG1 were disrupted by using HIS1 and URA3 markers (26). This strains also contains a maltose-inducible STE3 construct, generated from pAU15 (27).
Deletions of CEK2, FGR23, FAR1, and WOR1 were created from the parent strain RZY47, an a derivative of SN87 (-His -Leu) generated by sorbose selection (28). The target genes were disrupted by using the fusion PCR strategy as described in ref. 28. For each target gene, at least two independent deletion mutants were generated from independent heterozygous mutants.
The fusion knockout strategy also was used to create a wor1Δ mutant in SN78 (a/α -His -Leu -Ura) (28). The strain then was transformed with linearized pRZ25 (containing pMET3-WOR1) to direct integration to the RP10 locus (18). Proper integration was verified by using PCR. This strain was sorbose-selected on media supplemented with 2.5 mM Met and Cys to generate a isolates.
The pRZ25 plasmid also was transformed into CAI4 (-Ura) or RZY9 (a derivative of CAI4). These strains are referenced as pMET3-WOR1 (a/α or a) in the text. As controls, pCaEXP was introduced into CAI4 or RZY9 to create pMET3 control (a/α or a) strains.
DNA sequences of C. albicans genes were obtained from the Candida Genome Database (www.candidagenome.org). Fungal protein sequences were obtained from Proteome Bioknowledge Library (www.proteome.com).
White–Opaque Switching Assays.
White–opaque switching assays were performed as described in ref. 12, with the following modifications. Each strain was streaked onto supplemented Lee’s medium (described above) and grown for 5 days at 25°C. Cells then were plated onto SCD+Urd and grown for 7 days at room temperature, at which time the colonies were monitored for the presence of opaque colonies and sectors. At least two independent isolates of each mutant were used in this assay.
Ectopic Expression of WOR1 and Cell Measurements.
Cells were grown on SD+Urd plus 2.5 mM concentration each of Met and Cys (repressing) media at room temperature for 5 days. For each strain, 5–10 colonies were resuspended in sterile water and plated onto repressing or inducing media (SCD+Urd-lacking Met and Cys). After growth at room temperature for 5 days, colony phenotypes were recorded. Colonies were resuspended in sterile water, and cells were examined by using differential interference contrast microscopy on a Axiovert 200M microscope (Carl Zeiss, Oberkochen, Germany). Cell dimensions were measured by using Zeiss AxioVision software. Additional experiments with independent wild-type and mutant strains were nearly identical to those shown in Figs. 2 and 5 (data not shown). To test the effect of transient ectopic expression of WOR1, colonies grown on inducing media were replated on repressing media (or inducing, as a control). Plates were grown at room temperature for ≈7 days, and colony phenotypes were recorded.
Quantitative RT-PCR.
Cultures were grown in SCD+Urd-lacking Met and Cys at room temperature to mid-log phase, harvested by centrifugation, and frozen in liquid nitrogen. Total RNA was isolated from the cell pellets by using buffered phenol extractions. Total RNA from each sample was linearly reverse-transcribed, and cDNA was amplified by quantitative PCR, as monitored by Sybr Green fluorescence in a MJ Research Opticon instrument (Waltham, MA). Quantitative PCR was performed three times on the same cDNA preparation, and the median value is shown in Figs. 2 and 5. Signal for each gene is normalized to the median PAT1transcript level in the corresponding strain.
Response to Mating Pheromone.
Synthetic α-factor treatment was performed as described in ref. 14. Cells were fixed and observed by differential interference contrast microscopy after 4 h of pheromone treatment.
Northern Blot Analysis.
Five micrograms of total RNA (isolated above) were analyzed by Northern blot analysis. Radiolabeled DNA probes were generated by PCR and purified with Probe Quant G50 Sephadex Columns (Amersham Biosciences, Buckinghamshire, England). Signal was detected by using a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Western Blot Analysis.
C. albicans cultures were grown in repressing or inducing medium and harvested as described above. Whole-cell extracts (WCE) were prepared in urea lysis buffer (29), and 5 μg of WCE from each sample was separated by SDS/PAGE and analyzed by Western blotting. α-Wor1 is an affinity-purified antibody generated against a peptide at the C terminus of Wor1 (DDAVGNSSGSYYTGT) (Bethyl Laboratories, Montgomery, TX). As a loading control, the membrane was stripped and reprobed with rat α-Tub1, raised against Sa. cerevisiae Tub1 (no. ab1616; Abcam, Cambridge, MA).
ChIP Experiments.
Overnight cultures were grown in SCD+Urd for ≈16h at 25°C to an OD600 of 0.4. Cells were formaldehyde cross-linked and lysed by spheroplasting and osmotic lysis. Using 5 μl of α-Wor1 antibody (described above), immunoprecipitation (IP) was performed as described in ref. 30, with modifications. After spheroplasting, micrococcal nuclease digestion was omitted, and spheroplasts were resuspended in lysis buffer (50 mM Hepes-KOH, pH 7.5/140 mM NaCl/1 mM EDTA/1% Triton X-100/0.1% sodium deoxycholate). DNA was sheared by sonication 10 times for 10 s at power setting 2 on a Branson 450 sonicator (Danbury, CT), incubating on ice for 2 min between sonication pulses. Extracts were clarified by centrifugation.
PCR primers were designed at ≈250-bp intervals across the intergenic sequence upstream of the WOR1 ORF. For each ChIP experiment, DNA derived from the WCE and IP eluate was analyzed by quantitative PCR (qPCR). For each primer pair, three independent pairs of WCE and IP qPCRs were run, and median IP/WCE quantity ratios across the three replicates were divided by median IP/WCE quantity ratios across three control reactions run in parallel by using primers to the ADE2ORF. ChIP experiments using an MTLα1ΔMTLα2Δ opaque strain (data not shown) had an enrichment profile that was virtually identical to the a opaque profile shown in Fig. 4.
Supporting Information.
See Tables 2 and 3, which are published as supporting information on the PNAS web site, for a list of strains used in this study and a list of the PCR primers, respectively.
Supplementary Material
Acknowledgments
We thank Matt Miller, Richard Bennett, Brian Tuch, Annie Tsong, and other laboratory members for many valuable discussions and reagents and Marie Bao and Hiten Madhani (University of California, San Francisco, CA) for providing the α-Tub1 antibody. R.E.Z. was supported by an Achievement Rewards for College Scientists (ARCS) Foundation scholarship. D.J.G. was supported by a National Science Foundation predoctoral fellowship. The work was supported by National Institutes of Health Grant R01 AI49187 and an Ellison Foundation grant (to A.D.J.).
Abbreviations
- IP
immunoprecipitation
- SCD+URD
synthetic complete medium plus 2% glucose and 100 μg/ml uridine
- WCE
whole-cell extract.
Footnotes
Conflict of interest statement: No conflicts declared.
See Commentary on page 12659.
References
- 1.Slutsky B., Staebell M., Anderson J., Risen L., Pfaller M., Soll D. R. J. Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergen M. S., Voss E., Soll D. R. J. Gen. Microbiol. 1990;136:1925–1936. doi: 10.1099/00221287-136-10-1925. [DOI] [PubMed] [Google Scholar]
- 3.Rikkerink E. H., Magee B. B., Magee P. T. J. Bacteriol. 1988;170:895–899. doi: 10.1128/jb.170.2.895-899.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soll D. R., Morrow B., Srikantha T. Trends Genet. 1993;9:61–65. doi: 10.1016/0168-9525(93)90189-O. [DOI] [PubMed] [Google Scholar]
- 5.Lockhart S. R., Daniels K. J., Zhao R., Wessels D., Soll D. R. Eukaryot. Cell. 2003;2:49–61. doi: 10.1128/EC.2.1.49-61.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson A. Nat. Rev. Microbiol. 2003;1:106–116. doi: 10.1038/nrmicro752. [DOI] [PubMed] [Google Scholar]
- 7.Lan C.-Y., Newport G., Murillo L. A., Jones T., Scherer S., Davis R. W., Agabian N. Proc. Natl. Acad. Sci. USA. 2002;99:14907–14912. doi: 10.1073/pnas.232566499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsong A. E., Miller M. G., Raisner R. M., Johnson A. D. Cell. 2003;115:389–399. doi: 10.1016/s0092-8674(03)00885-7. [DOI] [PubMed] [Google Scholar]
- 9.Kvaal C. A., Srikantha T., Soll D. R. Infect. Immun. 1997;65:4468–4475. doi: 10.1128/iai.65.11.4468-4475.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kvaal C., Lachke S. A., Srikantha T., Daniels K., McCoy J., Soll D. R. Infect. Immun. 1999;67:6652–6662. doi: 10.1128/iai.67.12.6652-6662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lachke S. A., Lockhart S. R., Daniels K. J., Soll D. R. Infect. Immun. 2003;71:4970–4976. doi: 10.1128/IAI.71.9.4970-4976.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller M. G., Johnson A. D. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- 13.Lockhart S. R., Pujol C., Daniels K. J., Miller M. G., Johnson A. D., Pfaller M. A., Soll D. R. Genetics. 2002;162:737–745. doi: 10.1093/genetics/162.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennett R. J., Uhl M. A., Miller M. G., Johnson A. D. Mol. Cell. Biol. 2003;23:8189–8201. doi: 10.1128/MCB.23.22.8189-8201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caspari T. J. Cell Sci. 1997;110:2599–2608. doi: 10.1242/jcs.110.20.2599. [DOI] [PubMed] [Google Scholar]
- 17.Kunitomo H., Sugimoto A., Wilkinson C. R., Yamamoto M. Curr. Genet. 1995;28:32–38. doi: 10.1007/BF00311879. [DOI] [PubMed] [Google Scholar]
- 18.Li F., Palecek S. P. Biotechnol. Prog. 2005;21:1601–1609. doi: 10.1021/bp050236c. [DOI] [PubMed] [Google Scholar]
- 19.Care R. S., Trevethick J., Binley K. M., Sudbery P. E. Mol. Microbiol. 1999;34:792–798. doi: 10.1046/j.1365-2958.1999.01641.x. [DOI] [PubMed] [Google Scholar]
- 20.Hull C. M., Raisner R. M., Johnson A. D. Science. 2000;289:307–310. doi: 10.1126/science.289.5477.307. [DOI] [PubMed] [Google Scholar]
- 21.Ptashne M. A Genetic Switch: Phage Lambda Revisited. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 22.Lott T. J., Fundyga R. E., Kuykendall R. J., Arnold J. Fungal Genet. Biol. 2005;42:444–451. doi: 10.1016/j.fgb.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 23.Guthrie C., Fink G. R. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic; 1991. [Google Scholar]
- 24.Hull C. M., Johnson A. D. Science. 1999;285:1271–1275. doi: 10.1126/science.285.5431.1271. [DOI] [PubMed] [Google Scholar]
- 25.Fonzi W. A., Irwin M. Y. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson R. B., Davis D., Enloe B. M., Mitchell A. P. Yeast. 2000;16:65–70. doi: 10.1002/(SICI)1097-0061(20000115)16:1<65::AID-YEA508>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 27.Uhl M. A., Johnson A. D. Microbiology. 2001;147:1189–1195. doi: 10.1099/00221287-147-5-1189. [DOI] [PubMed] [Google Scholar]
- 28.Noble S. M., Johnson A. D. Eukaryot. Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ubersax J. A., Woodbury E. L., Quang P. N., Paraz M., Blethrow J. D., Shah K., Shokat K. M., Morgan D. O. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 30.Liu C. L., Kaplan T., Kim M., Buratowski S., Schreiber S. L., Friedman N., Rando O. J. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.