Abstract
Background
This study was aimed primarily at testing in the liver of brown trout (Salmo trutta) spectrophotometric methods previously used to measure the activities of catalase and hydrogen peroxide producing oxidases in mammals. To evaluate the influence of temperature on the activities of those peroxisomal enzymes was the second objective. A third goal of this work was the study of enzyme distribution in crude cell fractions of brown trout liver.
Results
The assays revealed a linear increase in the activity of all peroxisomal enzymes as the temperature rose from 10° to 37°C. However, while the activities of hydrogen peroxide producing oxidases were strongly influenced by temperature, catalase activity was only slightly affected. A crude fraction enriched with peroxisomes was obtained by differential centrifugation of liver homogenates, and the contamination by other organelles was evaluated by the activities of marker enzymes for mitochondria (succinate dehydrogenase), lysosomes (aryl sulphatase) and microsomes (NADPH cytochrome c reductase). For peroxisomal enzymes, the activities per mg of protein (specific activity) in liver homogenates were strongly correlated with the activities per g of liver and with the total activities per liver. These correlations were not obtained with crude peroxisomal fractions.
Conclusions
The spectrophotometric protocols originally used to quantify the activity of mammalian peroxisomal enzymes can be successfully applied to the study of those enzymes in brown trout. Because the activity of all studied peroxisomal enzymes rose in a linear mode with temperature, their activities can be correctly measured between 10° and 37°C. Probably due to contamination by other organelles and losses of soluble matrix enzymes during homogenisation, enzyme activities in crude peroxisomal fractions do not correlate with the activities in liver homogenates. Thus, total homogenates will be used in future seasonal and toxicological studies of brown trout peroxisomes.
Background
A literature survey on animal peroxisomes reveals that most studies are confined to laboratory mammals and humans. The scarcity of knowledge on other vertebrate and invertebrate species, and the need for more comparative studies has been recently emphasised [1-3]. Nevertheless, it is known that fish peroxisomes have enzymatic systems similar to those reported in mammals. Catalase, D-amino acid oxidase, urate oxidase, allantoinase, allantoicase, L-α-hydroxyacid oxidase, alanine:glyoxylate aminotransferase, fatty acyl-CoA oxidase, enoyl-CoA hydratase and carnitine acetyltransferase are among the enzymes that were detected in fish peroxisomes [3-10]. It is also known that peroxisomal metabolism is lower in fish than in mammals. For example, in the liver of rainbow trout (Onchorynchus mykiss) and Japanese medaka (Oryzias latipes) peroxisomal enzyme activities are at least four folds lower than in rodent liver. These results were either assigned to a reduced number of peroxisomes per hepatocyte, or a lower amount of peroxisomal enzymes, or both [9].
The research in fish revealed that environmental factors like water temperature, salinity, season and feeding habits, exerted changes in peroxisomal enzyme activities that, additionally, vary greatly among species [1]. It was also discovered that season, age and gender affect the morphology of fish liver peroxisomes. In the gray mullet (Mugil cephalus) hepatic peroxisomes tend to be larger in summer and in aged animals [11]. Moreover, in the liver of brown trout (Salmo trutta) both the individual dimensions of peroxisomes and their total volume per hepatocyte, but not their number, change during the annual breeding cycle in both genders [12]. Yet, in the latter species hepatic peroxisomes are significantly smaller in females than in males during the breeding season [12]. These observations raised the question about the kind of biochemical changes that may be associated with those morphological variations detected in fish liver peroxisomes. To measure some peroxisomal enzyme activities in brown trout liver, a method originally applied for mammalian peroxisomal hydrogen peroxide producing oxidases [13] was tested for the first time in fish. Catalase, a well-known peroxisomal marker enzyme [14], was also included in this work. Since the temperature is an important factor that affects enzyme activities, its influence on several trout peroxisomal enzymes was investigated. This work also aimed at studying the distribution of several peroxisomal enzymes in crude cell fractions. To control the amount of cross contamination in the crude peroxisomal fraction by other organelles, standard spectrophotometric procedures were used in order to measure the activity of marker enzymes of mitochondria (succinate dehydrogenase), lysosomes (aryl sulphatase) and microsomes (NADPH cytochrome c reductase). Finally, this paper reports the first data about the activities of peroxisomal enzymes in the liver of brown trout.
Results
Temperature and sample concentration tests
Since protein content and enzyme activities in brown trout liver were much different from those reported in rodent liver, several sample dilutions were tested in order to determine the best proportion between protein concentration and the concentration of all other components in the incubation medium. Therefore, after the dilution of homogenates and fractions the following final protein concentrations were used: 2 and 4 μg ml-1 for catalase, 45 and 90 μg ml-1 for D-aminoacid oxidase and fatty acyl-CoA oxidase, 10 and 20 μg ml-1 for urate oxidase, 0.15 and 0.3 mg ml-1 for glycolate oxidase and succinate dehydrogenase, 0.2 and 0.4 mg ml-1 for both aryl sulphatase and NADPH cytochrome c reductase. With these two dilutions for each enzyme, activities were linear in time and a good proportionality between enzymatic activity and the amount of protein was obtained. These sample concentrations also provided results with low variability between the two dilutions used (Coefficient of Variation ≤ 0.05) and high reproducibility between replicates of the same dilution (CV ≤ 0.02).
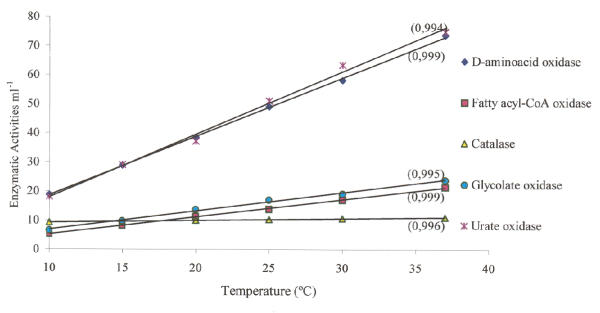
In brown trout liver homogenates, a linear relationship between temperature and enzyme activity was found for the five peroxisomal enzymes tested (Fig. 1). The activities of peroxisomal oxidases were highly influenced by temperature whereas catalase was only slightly affected. In fact, the activity of urate oxidase was strongly dependent on temperature, since its activity at 37°C was about five times higher than at 10°C. Also between 10 and 37°C, fatty acyl-CoA oxidase and D-aminoacid oxidase activities raised about four times. Glycolate oxidase, also known as L-α-hydroxyacid oxidase A, seems to be slightly less influenced by temperature since its activity showed an increment of 3.4 times within this temperature range. In contrast, catalase activity increased only 1.2 times between 10 and 37°C.
Figure 1.
Temperature influence on the activity of peroxisomal enzymes. The regression lines express the relationship between temperature and activity (s-1 ml-1 for catalase and nmol min-1 ml-1 for the others) for five peroxisomal enzymes in brown trout liver homogenates. For each line the regression coefficient is showed between brackets. Data was obtained from 4 fishes. Assays were carried out as described under Methods.
Measurement of enzyme activities
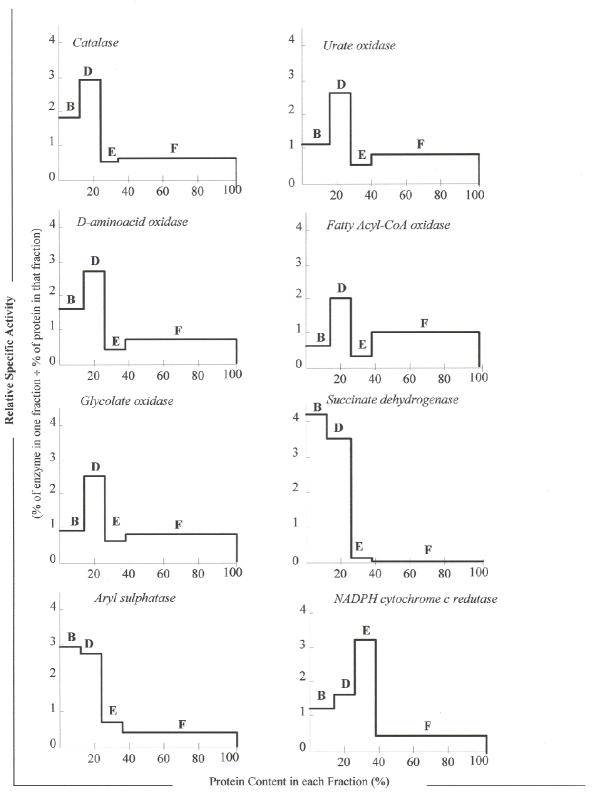
Catalase is the peroxisomal enzyme with the highest activity in brown trout liver. Urate and D-aminoacid oxidases were, among the hydrogen peroxide producing oxidases, those with the highest activities in liver homogenates (A fraction) and in the crude peroxisomal fraction (D fraction). Such differences in the activity of peroxisomal oxidases, well noticed at the physiological temperature of 10–15°C, become more pronounced when the assays are made at higher temperatures. Specific activities of peroxisomal enzymes were about twice as higher in D fraction than in A fraction (Table 1), as expected for a crude peroxisomal fraction. All the peroxisomal enzymes had the same pattern of distribution in liver fractions, with high relative specific activities in D fraction, and their pattern of distribution is clearly different from the marker enzymes of other organelles (Fig. 2). Moreover, the cytosolic F fraction had high distribution percentages for these enzymes too (Table 2). Presumably, part of the peroxisomal soluble matrix enzymes ended in the supernatant as a result of membrane damage during homogenisation. However, due to the high amount of protein in F fraction, both the specific activities and the relative specific activities were low in this fraction (Fig. 2 and Table 1).
Table 1.
Specific and per g enzyme activities in brown trout liver
| Enzymes |
Activity per g of liver s-1 g-1 liver (*) or nmol min-1 g-1 liver |
Specific activity s-1 mg-1 protein (*) or nmol min-1 mg-1 protein |
||||
| A | B | D | E | F | ||
| Peroxisomal markers | ||||||
| Catalase (*) | 95.9 (0.4) | 1.3 (0.2) | 2.0 (0.3) | 3.5 (0.3) | 0.4 (0.3) | 0.6 (0.2) |
| Urate oxidase | 790.4 (0.6) | 10.3 (0.6) | 13.0 (0.7) | 22.6 (0.6) | 4.7 (0.8) | 8.8 (0.8) |
| D-aminoacid oxidase | 471.0 (0.6) | 6.0 (0.4) | 8.9 (0.5) | 14.4 (0.1) | 1.4 (0.4) | 3.5 (0.4) |
| Fatty acyl-CoA oxidase | 73.3 (0.5) | 1.0 (0.4) | 0.4 (0.5) | 1.5 (0.3) | 0.3 (0.8) | 1.0 (0.5) |
| Glycolate oxidase | 60.2 (0.5) | 0.8 (0.3) | 0.8 (0.4) | 2.3 (0.4) | 0.5 (0.5) | 0.7 (0.4) |
| Mitochondrial marker | ||||||
| Succinate dehydrogenase | 310.7 (0.5) | 3.6 (0.3) | 29.1 (0.5) | 27.8 (0.5) | 1.4 (0.7) | 0.2 (0.6) |
| Lysosomal marker | ||||||
| Aryl sulphatase | 275.0 (0.2) | 3.2 (0.4) | 10.4 (0.5) | 10.6 (0.5) | 2.4 (0.4) | 1.4 (0.4) |
| Microsomal marker | ||||||
| NADPH cytochrome c reductase | 2520 (0.4) | 32.5 (0.3) | 29.2 (0.4) | 41.8 (0.3) | 99.0 (0.5) | 11.5 (0.3) |
|
Protein per g of liver (mg g-1 liver) |
Total protein per fraction (mg) |
|||||
| 77.8 (0.2) | 225.6 (0.2) | 26.4 (0.5) | 22.3 (0.3) | 25.4 (0.6) | 128.4 (0.2) | |
Activities per g of liver and specific activities were measured at 37°C in liver homogenates and cell fractions. Protein per mg of liver and in cell fractions is also given. A – liver homogenates; B – crude mitochondrial/lysosomal fraction; D – crude peroxisomal fraction; E – crude microsomal fraction; F – cytosolic fraction. Presented data are a mean from 6 fishes and the coefficient of variation is given in brackets. (*) – Catalase activity is expressed by the first-order rate constant (s-1) and all other enzyme activities are express in nmol min-1, per mg of protein or per g of liver.
Figure 2.
Relative specific activities of marker enzymes in cell fractions. Distributions of several marker enzymes from the liver of brown trout in B, D, E and F fractions. For details of centrifugation conditions see Methods.
Table 2.
Percentage distribution of the marker enzymes
| Enzymes | Percentage Distribution (%) | |||
| B | D | E | F | |
| Peroxisomal markers | ||||
| Catalase | 23.0 (0.3) | 34.2 (0.2) | 4.6 (0.8) | 38.2 (0.2) |
| Urate oxidase | 14.2 (1.0) | 28.7 (0.4) | 6.0 (0.7) | 51.1 (0.2) |
| D-aminoacid oxidase | 21.2 (0.5) | 30.5 (0.2) | 4.2 (1.0) | 44.1 (0.3) |
| Fatty acyl-CoA oxidase | 7.1 (0.7) | 22.7 (0.3) | 3.7 (0.6) | 66.5 (0.2) |
| Glycolate oxidase | 11.6 (0.7) | 27.3 (0.2) | 7.7 (0.6) | 53.5 (0.2) |
| Mitochondrial marker | ||||
| Succinate dehydrogenase | 56.0 (0.1) | 40.4 (0.2) | 1.9 (0.5) | 2.2 (0.8) |
| Lysosomal marker | ||||
| Aryl sulphatase | 37.4 (0.4) | 30.5 (0.3) | 7.9 (0.7) | 24.4 (0.2) |
| Microsomal marker | ||||
| NADPH cytochrome c reductase | 15.5 (0.6) | 18.2 (0.5) | 38.5 (0.4) | 27.9 (0.5) |
| Protein | 12.9 (0.4) | 11.1 (0.2) | 12.0 (0.4) | 64.0 (0.1) |
Percentage distribution of enzymes and protein in brown trout liver fractions. B – crude mitochondrial / lysosomal fraction; D – crude peroxisomal fraction; E – crude microsomal fraction; F – cytosolic fraction. Data are a mean from 6 fishes and the coefficients of variation are given in brackets.
Fraction D also showed high percentage distributions (Table 2) and elevated specific activities for the enzymatic markers of both mitochondria (succinate dehydrogenase) and lysosomes (aryl sulphatase) (Table 1). Moreover, the relative specific activities of succinate dehydrogenase and aryl sulphatase showed that they were more or less equally distributed both in B and D fractions (Fig. 2). Nonetheless, percentage distribution, specific activity and relative specific activities of these two enzymes were lower in E and F fractions than in the other fractions (Fig. 2, Tables 1 and 2). Considering the microsomal marker enzyme NADPH cytochrome c reductase, it was observed that in D fraction its activity was less than half of that determined in E fraction (Table 1).
The percentage distribution of peroxisomal enzymes in D fraction (Table 2) ranged from 22.7 for fatty acyl-CoA oxidase to 34.2 for catalase, but according to ANOVA such differences have no statistical significance. The same was verified in both B and E fractions. However, in F fraction the percentage distribution of catalase (38.2) is significantly different from the percentage distribution of fatty acyl-CoA oxidase (66.5), although no other statistical differences among peroxisomal enzymes were found within this fraction (Table 2).
Statistical analyses also revealed that specific activities of peroxisomal enzymes in D fraction did not have any explicit pattern of correlation with the specific activities in fraction A, neither with the activities per g of liver or per liver. Contrastingly, in fraction A it was observed that for each peroxisomal enzyme there was a strong and positive correlation between the specific activity and the activity per g of liver: urate oxidase (r = 0.96; p = 0.002); D-aminoacid oxidase (r = 0.99; p = 0.0002); fatty acyl-CoA oxidase (r = 0.96; p = 0.002); glycolate oxidase (r = 0.95; p = 0.003); catalase (r = 0.89; p = 0.02). In this fraction, the specific activity of each oxidase was also highly correlated with the total activity per liver: urate oxidase (r = 0.97; p = 0.001); D-aminoacid oxidase (r = 0.86; p = 0.03); fatty acyl-CoA oxidase (r = 0.97; p = 0.001); glycolate oxidase (r = 0.94; p = 0.005).
Discussion
One of the purposes of this study was to test in brown trout liver the spectrophotometric method originally used by Cablé et al. [13] for measurement of peroxisomal oxidase activities in mammalian tissues. However, some modifications were introduced and the method was extended to urate and glycolate oxidases. To calculate the amounts of H2O2 produced by these enzymes, instead of using the extinction coefficient previously published [13], a calibration line was made with known amounts of H2O2. Others also made similar calibration lines, but when using luminometric methods [15] and fluorometric techniques [16].
Since the physiological temperature is much lower in brown trout than in mammals, temperature influence in the activity of five peroxisomal enzymes was investigated. The results showed that the activities of these enzymes rose in a linear mode from 10 to 37°C, but while the activity of hydrogen peroxide producing oxidases were strongly influenced by temperature, catalase activity was only slight affected. This last observation is in total accordance with previous data regarding catalase activity in mammals [14]. Therefore, these enzymatic activities can be measured at whatever temperature within the above-referred range. However, at 37°C activities are higher and therefore easier to measure than at the physiological temperatures of brown trout (10 – 15°C). In fact, other authors have used a wide range of temperatures in studies involving fish peroxisomal enzymes. For example, in carp (Cyprinus carpio) catalase activity was determined at 25°C whereas urate oxidase, fatty acyl-CoA oxidase and glycolate oxidase were measured at 37°C [4,5,17] and in rainbow trout (O. mykiss) catalase activity was determined on ice whereas fatty acyl-CoA oxidase was measured at 15°C [18].
Some similarities are evident when crude cell fractions of rat liver [19] are compared with those obtained from brown trout liver. In both species relative specific activities of catalase and hydrogen peroxide producing oxidases are higher in the D fraction obtained using very similar centrifugation procedures. Moreover, catalase and glycolate oxidase (or L-α-hydroxyacid oxidase) present a high percentage distribution in the cytosolic fraction of rat and brown trout, has expected for soluble matrix enzymes released from peroxisomes due to membrane damage during homogenisation. On the other hand, urate oxidase is not detectable in the supernatant of rat liver [19] while in brown trout about 50% of urate oxidase activity can be found in the supernatant. This discrepancy is due to a fundamental difference between brown trout and mammalian urate oxidase, which behaves exactly like a soluble matrix enzyme in the former but is a crystallised core enzyme in the latter. Indeed cores were neither found in brown trout liver peroxisomes [12] or in other fish species [3]. Although urate oxidase was described as a membrane-linked enzyme in carp liver peroxisomes [17] in other fish species was considered a soluble matrix enzyme [6] as it seems to be in brown trout liver peroxisomes.
Differences between rat and brown trout crude liver fractions are also evident in the microsomal fraction, in which the percentage distributions of peroxisomal enzyme activities are higher in rat than in brown trout [19].
The absence of correlations between D fractions and total liver homogenates showed that enzyme activities in peroxisomal enriched fractions are not proportional to the activities in homogenates. The contamination of crude peroxisomal fractions with other organelles and the substantial loss of soluble matrix enzymes can explain this result. Because of these facts, and knowing that in rainbow trout the gradient centrifugation of the crude peroxisomal fraction produced a further decrease in the activities of some oxidases [18], in future seasonal and toxicological studies about brown trout peroxisomes total homogenates will be used instead of cell fractions. Moreover, variation coefficients suggest that enzyme activities have relevant individual variability in brown trout. Thus, very subtle differences due to experiments or natural events may pass unnoticed when working with a small number of animals.
Conclusions
In conclusion: 1) the spectrophotometric protocols used for measurement of mammalian peroxisomal enzymes can be successfully applied to brown trout; 2) the enzymatic activities of the peroxisomal enzymes studied by us can be correctly measured between 10° and 37°C, this because their activity rose in a linear mode with temperature, with catalase activity increasing only slightly; 3) for peroxisomal enzymes in brown trout liver homogenates there is a clear correlation between specific activity and activity per g of liver or per liver; 4) during preparation of cell fractions enriched with peroxisomes a significant amount of matrix enzymes are loosed, and the remaining enzyme activities are not correlated with the activities in liver homogenates.
Methods
Fish
Two-year-old brown trout males (n = 6) were collected by random net fishing from aquaculture pools (Posto Aquícola do Torno – Amarante, Portugal) in late February. Fish of 188 g (coefficient of variation, CV = 0.10) in weight and 26 cm (CV = 0.04) in length were kept in dechlorinated water (pH 6–7, at 8–10°C) for 1–2 days before sacrifice. Mean liver weight was 3.0 g (CV = 0.30).
Chemicals
Cofactors, substrates for enzyme assays and bovine serum albumin (BSA) were purchased from Sigma Chemical Co. (Poole, Dorset, U.K). Other chemicals were obtained from Merck (Darmstadt, Germany).
Preparative procedures
Animals were anaesthetised by bathing in a solution of ethylene glycol monophenyl ether (0.4 ml l-1) and then perfused at 5.6 ml min-1 kg-1 via the portal venous system for 4–5 min with an isosmotic buffer for salmonids [20]. The liver was weighted just after removal, immediately minced in chilled homogenization buffer with a pH of 7.4 (250 mM sucrose, 5 mM MOPS, 1 mM EDTA and 0.1% ethanol saturated with phenyl-methyl-sulfonyl-fluoride PMSF) [17] and then homogenized in the same buffer using a Potter-Elvehjem homogenizer at 1000 rpm, held in an ice-water-bath. After sedimentation of unhomogenized material (50 g, 10 min, 4°C) according to Cajaraville et al. [21], supernatant volume was adjusted to 5 ml g-1 of liver using ice-cold homogenisation buffer (A fraction). A previously described method [17] was used for differential pelleting of the A fraction in order to produce a B fraction containing mainly mitochondria and lysosomes (2,000 g, 10 min, 4°C), a D fraction enriched with peroxisomes (20,000 g, 30 min, 4°C), a E fraction enriched with microsomes (100,000 g, 60 min, 4°C) and a cytosolic F fraction (supernatant of 100,000 g, 60 min, 4°C). Pellets (B, D and E) were rinsed once by resuspension in an appropriate volume of buffer using a glass-rod and recentrifuged under the same conditions. Before enzymatic assay, all fractions were treated with Triton X-100 at a final concentration of 0.5% (v/v) in homogenization buffer. Proteins were assayed according to Lowry et al. [22] using BSA standards and results are expressed in BSA equivalents (Table 1).
Enzyme assays
Enzymatic assays were carried out in a spectrophotometer connected to a circulating water system for temperature regulation in the cuvette compartment. Enzyme activities linear in time and proportional to the amount protein in the assays were obtained using an appropriate sample dilution. Two different dilutions of each sample were used to produce a mean value of enzymatic activities. Temperature effect on peroxisomal enzyme activities was evaluated by measurements made at 10, 15, 20, 25, 30 and 37°C.
Catalase
For catalase the methodology previously described by Aebi [14] was used. Incubation medium contained 50 mM sodium phosphate buffer (pH 7), 10 mM hydrogen peroxide (H2O2) and a diluted sample. Consumption of H2O2 was followed by the decrease in absorbance at 240 nm for 30 seconds and the activity is expressed by the first-order rate constant (k) for degradation of H2O2. k = (1 ÷ Δt) × ln (c1 ÷ c2) where c1 and c2 correspond to H2O2 concentrations at t = 0 and t = 30 seconds, respectively [14].
Peroxisomal oxidases
Measurement of peroxisomal oxidase activities was based on the production of H2O2 and followed the procedure of Cablé et al. [13] with some modifications. Incubation medium contained 50 mM potassium phosphate buffer (pH 8.3), 0.082 mM 4-amino-antipyrine, 1 mM phenol and 2 IU ml-1 of horseradish peroxidase. To avoid the interference of catalase, 10 mM of azide was added to the medium, according to Leupold et al. [15]. For glycolate oxidase assay, 0.01 mM flavin mononucleotide (FMN) was added in the medium [23]. For other oxidases, 0.01 mM flavin adenine dinucleotide (FAD) was included in the medium [13,23]. Substrates were used in the following concentrations: 20 mM D-alanine, 1 mM uric acid, 0.1 mM palmitoyl-CoA (C:16) and 20 mM sodium glycolate. Each enzymatic reaction was started by addition of 25 μl of a diluted sample to 650 μl of incubation medium. Absorbance increase was measured at 500 nm for 10 min. For urate oxidase, a baseline was made with the complete medium without sample, which was thereafter subtracted from the absorbance increase in each assay. This procedure was not necessary for all other assayed oxidases, because non-specific reactions were not detected in other cases.
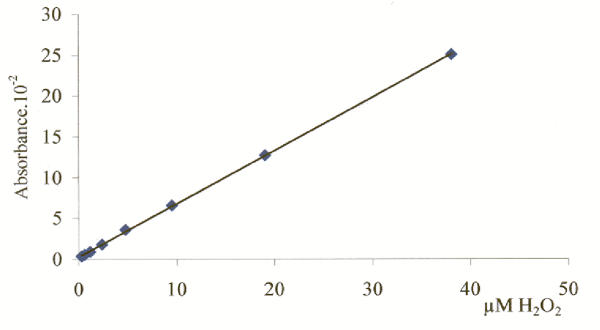
The amount of H2O2 (in μM) produced by peroxisomal oxidases was calculated from the equation of the calibration line: [H2O2] = 185.07 × Absorbance at 500 nm. This line (Fig. 3) showed a regression coefficient of 1.0 and was constructed using several standards of H2O2, which were added to the incubation medium without sample. The concentration of the H2O2 standards was calculated from the absorbance at 240 nm (ε = 39.4 M-1 cm-1) [24].
Figure 3.
Calibration line for peroxisomal oxidases assays. This plot shows the linear relationship between Absorbance and the concentration of several H2O2 standards. Absorbance was measured at 500 nm as described in Methods.
Succinate dehydrogenase
Succinate dehydrogenase activity was estimated according to Schoner et al. [25]. Incubation medium contained 50 mM potassium phosphate buffer (pH 7.5), 5 mM sodium succinate, 1 mM potassium cyanide, 0.1 mM 2,6-dichlorophenol indophenol (DPIP), and a diluted sample. The extinction coefficient of DPIP at 600 nm, determined with several standard solutions ranging from 0 to 100 μM of DPIP (ε = 17.3 mM-1 cm-1), was used to calculate the activity [25].
Aryl sulphatase
Measurements of aryl sulphatase activity followed the method of Worwood et al. [26]. Incubation medium contained 0.5 M acetate buffer (pH 5.6), 20 mM nitrocatechol sulphate and a diluted sample. The reaction was stopped by addition 1 ml of 1 M sodium hydroxide and the extinction coefficient of nitrocatechol solutions at 515 nm (ε = 12.4 mM-1 cm-1) was used to calculate the activity [26].
NADPH cytochrome c reductase
Measurements of NADPH cytochrome c reductase activity followed the method of Johannesen et al. [27]. Incubation medium contained 0.1 M TRIS-HCl buffer (pH 7.6), 10 mM potassium cyanide, 0.4 mM oxidised cytochrome c, 3 mM NADPH and a diluted sample. This reaction was started by addition of NADPH. Activity calculations were based upon the extinction coefficient of a reduced cytochrome c solution at 550 nm (ε = 19.6 mM-1 cm-1) [28].
Calculations, units and statistical analysis
The percentage distribution [(protein or enzyme activity in one fraction ÷ Σ of protein or enzyme activity in fractions B to F) × 100] and the relative specific activity [percentage distribution of the enzyme in one fraction ÷ percentage distribution of protein in that fraction] were calculated as in other papers [5]. Catalase activity is expressed by the first-order rate constant in s-1 g-1 of liver and s-1 mg-1 of protein, for all others, results are given in nmol min-1 g-1 of liver and nmol min-1 mg-1 of protein.
Data are reported as means per group of animals, followed by the respective coefficients of variations (CV = standard deviation ÷ mean). Correlation analysis was used to establish several relationships considered biologically or technically relevant. Results were judged significant when p ≤ 0.05 as reported by Pearson's coefficient of correlation (r). A linear regression analysis was made to study enzyme activities at six different chosen temperatures.
Authors' contributions
Prof. Maria João Rocha participated in study design and collection of samples, performed most of the experimental work related with enzyme assays and data analysis, wrote and edited the manuscript. Prof. Eduardo Rocha participated in study design and collection of samples, made the statistical analysis and edited the manuscript. Dr. Albina D. Resende participated in the collection of samples, and performed experimental assays related with the temperature effects on enzyme activities and related data analysis. Prof. Alexandre Lobo-da-Cunha conceived the study, participated in its design and collection of samples, performed experimental work, data analysis, and edited the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
The fish were generously provided by the "Divisão de Caça e Pesca nas Águas Interiores – Direcção Regional de Agricultura de Entre-Douro e Minho", under an official cooperation protocol. The commitment of Eng. Álvaro Gonçalves and Eng. Augusto Maia is particularly appreciated. This study was financially supported by the pluriannual program of the CIIMAR and by the "Fundação para a Ciência e a Tecnologia" (research contract n° PRAXIS/C/BIA/14268/1998).
Contributor Information
Maria João Rocha, Email: mjrocha@netc.pt.
Eduardo Rocha, Email: erocha@icbas.up.pt.
Albina D Resende, Email: dolores@icbas.up.pt.
Alexandre Lobo-da-Cunha, Email: alcunha@icbas.up.pt.
References
- Fahimi HD, Cajaraville MP. Induction of peroxisome proliferation by some environmental pollutants and chemicals in animal tissues. In: Cajaraville MP, editor. Cell Biology in Environmental Toxicology. Basque Country Press Service, Bilbo; 1995. pp. 221–255. [Google Scholar]
- Lobo-da-Cunha A. The peroxisomes of the hepatopancreas in two species of chitons. Cell Tissue Res. 1977;290:655–664. doi: 10.1007/s004410050971. [DOI] [PubMed] [Google Scholar]
- Cancio I, Cajaraville MP. Cell biology of peroxisomes and their characteristics in aquatic organisms. Int Rev Cytol. 2000;199:201–293. doi: 10.1016/s0074-7696(00)99005-3. [DOI] [PubMed] [Google Scholar]
- Kramar R, Goldenberg H, Böck P, Klobucar N. Peroxisomes in the liver of the carp (Cyprinus carpio L.) electron microscopic cytochemical and biochemical studies. Histochem. 1974;40:137–154. doi: 10.1007/BF00495962. [DOI] [PubMed] [Google Scholar]
- Goldenberg H, Hüttinger M, Kampfer P, Kramar R. Preparation of peroxisomes from carp liver by zonal rotor density gradient centrifugation. Histochem J. 1978;10:103–113. doi: 10.1007/BF01003417. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Takada Y, Fujiwara S. Degradation of uric acid to urea and glyoxylate in peroxisomes. J Biol Chem. 1979;254:5272–5275. [PubMed] [Google Scholar]
- Baldwin LA, Calabrese EJ, Kostecki PT, Yang J-H. Isolation of peroxisomal enoyl-CoA hydratase in rainbow trout and immunochemical identification with the bifuntional enzyme. Fish Physiol Biochem. 1990;8:347–351. doi: 10.1007/BF00003430. [DOI] [PubMed] [Google Scholar]
- Braunbeck T, Völkl A. Induction of biotransformation in the liver of eel (Anguilla anguilla) by sublethal exposure to dinitro-o-kresol: an ultrastructural and biochemical study. Ecotoxicol Environ Saf. 1991;21:109–127. doi: 10.1016/0147-6513(91)90014-g. [DOI] [PubMed] [Google Scholar]
- Scarano LJ, Calabrese EJ, Kostecki PT, Baldwin LA, Leonard DA. Evaluation of a rodent peroxisome proliferator in two species of freshwater fish: rainbow trout (Onchorynchus mykiss) and Japanese medaka (Oryzias latipes). Ecotoxicol Environ Saf. 1994;29:13–19. doi: 10.1016/0147-6513(94)90026-4. [DOI] [PubMed] [Google Scholar]
- Sakuraba H, Fujiwara S, Noguchi T. Metabolism of glyoxylate, the en product of purine degradation in liver peroxisomes of fresh water fish. Biochem Biophys Res Commun. 1996;229:603–606. doi: 10.1006/bbrc.1996.1850. [DOI] [PubMed] [Google Scholar]
- Orbea A, Beier K, Völkl A, Fahimi HD, Cajaraville MP. Ultrastructural, immunocytochemical and morphometric characterisation of liver peroxisomes in gray mullet, Mugil cephalus. Cell Tissue Res. 1999;297:493–502. doi: 10.1007/s004410051376. [DOI] [PubMed] [Google Scholar]
- Rocha E, Lobo-da-Cunha A, Monteiro RAF, Silva MW, Oliveira MH. A stereological study along the year on the hepatocytic peroxisomes of brown trout (Salmo trutta). J Submicr Cytol Pathol. 1999;31:91–105. [Google Scholar]
- Cablé S, Kedinger M, Dauça M. Peroxisomes and peroxisomal enzymes along the crypt-villus axis of the rat intestine. Differentiation. 1993;54:99–108. doi: 10.1111/j.1432-0436.1993.tb00712.x. [DOI] [PubMed] [Google Scholar]
- Aebi H. Catalase in vitro. Method Enzymol. 1984;105:121–126. doi: 10.1016/s0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- Leupold C, Völkl A, Fahimi HD. Luminometric determination of oxidase activity in peroxisomal fractions of rat liver: Glycolate oxidase. Anal Biochem. 1985;151:63–69. doi: 10.1016/0003-2697(85)90053-3. [DOI] [PubMed] [Google Scholar]
- Walusimbi-Kisitu M, Harrison EH. Fluorometric assay for rat liver peroxisomal fatty acyl-coenzyme A oxidase activity. J Lipid Res. 1983;24:1077–1084. [PubMed] [Google Scholar]
- Goldenberg H. Organisation of purine degradation in the liver of a teleost (carp; Cyprinus carpio L.). A study of its subcellular distribution. Molec Cell Biochem. 1977;16:17–21. doi: 10.1007/BF01769834. [DOI] [PubMed] [Google Scholar]
- Henderson RJ, Sargent JR. Chain-length specificities of mitochondrial and peroxisomal β-oxidation of fatty acids in livers of rainbow trout (Salmo gardneri). Comp Biochem Physiol. 1985;82:79–85. doi: 10.1016/0305-0491(85)90131-2. [DOI] [PubMed] [Google Scholar]
- Völkl AE, Fahimi HD. Isolation and characterisation of peroxisomes from the liver of normal untreated rats. Eur J Biochem. 1985;149:257–265. doi: 10.1111/j.1432-1033.1985.tb08920.x. [DOI] [PubMed] [Google Scholar]
- Rocha E, Monteiro RAF, Pereira CA. The liver of the brown trout, Salmo trutta fario: a light and electron microscope study. J Anat. 1994;185:241–249. [PMC free article] [PubMed] [Google Scholar]
- Cajaraville MP, Völkl A, Fahimi HD. Peroxisomes in digestive gland cells of the mussel Mytilus galloprovincialis Lmk. Biochemical, ultrastructural and immunocytochemical characterisation. Eur J Cell Biol. 1992;59:255–264. [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Vamecq J. Fluorometric assay of peroxisomal oxidases. Anal Biochem. 1990;186:340–349. doi: 10.1016/0003-2697(90)90092-n. [DOI] [PubMed] [Google Scholar]
- Nelson DP, Kiesow LA. Enthalpy of decomposition of hydrogen peroxide by catalase at 25°C (with molar extinction coefficients of H2O2 solutions in the UV). Anal Biochem. 1972;49:472–478. doi: 10.1016/0003-2697(72)90451-4. [DOI] [PubMed] [Google Scholar]
- Schoner W, von Ilberg C, Kramer R, Seubert W. On the mechanism of Na+- and K+-stimulated hydrolysis of adenosine triphosphate. 1. Purification and properties of a Na+- and K+-activated ATPase from ox brain. Euro J Biochem. 1967;1:334–343. doi: 10.1007/978-3-662-25813-2_45. [DOI] [PubMed] [Google Scholar]
- Worwood M, Dodgson KS, Hook GER, Rose FA. Problems associated with the assay of arylsulphatases A and B of rat tissues. Biochem J. 1973;134:183–190. doi: 10.1042/bj1340183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesen K, Depierre JW, Bergstrand A, Dallner G, Ernster L. Preparation and characterisation of total, rough and smooth microsomes from the lung of control and methylcholanthrene-treated rats. Biochem Biophys Acta. 1977;496:115–135. doi: 10.1016/0304-4165(77)90120-9. [DOI] [PubMed] [Google Scholar]
- Livingstone DR, Moore MN, Widdows J. Ecotoxicology: biological effects measurements on molluscs and their use in impact assessment. In: Salomons W, Duvrsma EK, Bayne BL, Forstner U, editor. Pollution of the North Sea: an assessment. Spriger-Verlag, Berlin, Heidelberg, New York; 1988. pp. 624–637. [Google Scholar]