Abstract
Protein acetylation, especially histone acetylation, is the subject of both research and clinical investigation. At least four small-molecule histone deacetylase inhibitors are currently in clinical trials for the treatment of cancer. These and other inhibitors also affect microtubule acetylation. A multidimensional, chemical genetic screen of 7,392 small molecules was used to discover “tubacin,” which inhibits α-tubulin deacetylation in mammalian cells. Tubacin does not affect the level of histone acetylation, gene-expression patterns, or cell-cycle progression. We provide evidence that class II histone deacetylase 6 (HDAC6) is the intracellular target of tubacin. Only one of the two catalytic domains of HDAC6 possesses tubulin deacetylase activity, and only this domain is bound by tubacin. Tubacin treatment did not affect the stability of microtubules but did decrease cell motility. HDAC6 overexpression disrupted the localization of p58, a protein that mediates binding of Golgi elements to microtubules. Our results highlight the role of α-tubulin acetylation in mediating the localization of microtubule-associated proteins. They also suggest that small molecules that selectively inhibit HDAC6-mediated α-tubulin deacetylation, a first example of which is tubacin, might have therapeutic applications as antimetastatic and antiangiogenic agents.
Histone deacetylases (HDACs) are zinc-dependent hydrolases that mediate chromatin remodeling and gene expression (1, 2). All 11 known human HDACs have histone deacetylase activity that is inhibited by the small molecule trichostatin A (TSA). Histone hyperacetylation induced by HDAC inhibitors such as TSA correlates with gene expression, cell-cycle arrest, cell differentiation, and cell death (3–8). HDAC inhibitors have been proposed for treatment of cancer as well as neurodegenerative disorders associated with mutations in polyglutamine-encoding tracts (9). In addition, agents already used clinically for other purposes, such as valproic acid, inhibit HDACs and cause histone hyperacetylation in cultured cells (10). Elucidating a functional role for acetylation of proteins other than histones is necessary to understand better the physiological targets of HDACs and the mechanisms by which HDAC inhibitors mediate their spectrum of phenotypic effects (11).
Treatment with TSA increases the cellular acetylation of α-tubulin at lysine 40 (12) (see Fig. 5 A–C, which is published as supporting information on the PNAS web site, www.pnas.org), a protein modification with unknown functional relevance. The effects of TSA on the cytoskeleton indicate a lack of selectivity for nuclear HDACs and confound interpretation of experiments in which this small molecule is used. We set out to identify cell-permeable, selective inhibitors of tubulin deacetylase (TDAC) activity to uncouple α-tubulin acetylation from nuclear consequences of HDAC inhibitors such as TSA.
Toward this end, a multidimensional, high-throughput, cell-based screen of a 7,392-membered deacetylase-biased 1,3-dioxane library prepared by using a one-bead/one-stock solution strategy was performed (see Fig. 5D; refs. 13–17). Two acetylation state-specific antibodies were exploited in cytoblot assays to distinguish between small molecules capable of inducing α-tubulin acetylation (AcTubulin) and histone acetylation (AcLysine). Ability to suppress the induced tubulin-acetylation state with a small-molecule chemical genetic modifier (ITSA1) was also investigated and allowed further classification of the library molecules into functionally related groups (12).
Materials and Methods
Materials.
TSA, nocodazole, paclitaxel (taxol), 5-bromo-2′-deoxyuridine (BrdUrd), α-FLAG M2 agarose affinity gel beads, antiacetylated tubulin (6–11B-1), anti-FLAG M2 antibodies, ribonuclease A, and Harris hematoxylin stain were purchased from Sigma. ITSA1 was purchased from Chembridge (San Diego). Antiacetylated lysine antibody (rabbit) was purchased from Cell Signaling Technology (Beverly, MA). Antiacetyl-histone (K9 and K14) H3 antibody (rabbit) was purchased from Upstate Biotechnology (Lake Placid, NY). Anti-mouse IgG horseradish peroxidase-conjugated and anti-rabbit IgG horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescent mixture (luminol) were purchased from Amersham Pharmacia. Alexa 594 and Alexa 488-conjugated anti-mouse IgG and anti-rabbit IgG antibodies, Hoechst 33342, and propidium iodide were purchased from Molecular Probes. FITC-conjugated anti-mouse IgG (goat) antibody was purchased from Santa Cruz Biotechnology. Microtubule-associated protein (MAP)-rich tubulin and purified tubulin were purchased from Cytoskeleton (Denver). PBS (10 mM, pH. 7.4) (138 mM NaCl/2.7 mM KCl), FBS, penicillin G sodium, streptomycin sulfate, and l-glutamine were purchased from GIBCO/BRL.
Cell Culture.
All cells were cultured at 37°C in a humidified incubator supplemented with 5% carbon dioxide. Human A549 lung carcinoma cells and BSC-1 African green monkey kidney epithelial cells (both from American Type Culture Collection) were cultured in DMEM supplemented with 10% FBS/100 units/ml penicillin G sodium/100 μg/ml streptomycin sulfate/2 mM l-glutamine (DMEM+). Human TAg Jurkat cells and mouse ROSA26 embryonic stem cells (gift of E. Roberston, Harvard University) were cultured as described (12). Mouse NIH 3T3 wild-type (Neo), HDAC6, HDAC6 double-mutant, and HDAC1-overexpressing cells (gift from C. Hubbert, Duke University, Durham, NC) were cultured as described (18).
1,3-Dioxane Library Screen and Cytoblot Assays.
1,3-Dioxanes from a single, polystyrene macrobead were prepared as 1–2 mM stock solutions as described (15). Compounds (2–5 μM) were screened in duplicate in acetylated α-tubulin and acetylated lysine cytoblot assays in A549 cells as described (14). Statistical analyses were performed by using XLSTAT-PRO 5.2. A full description of statistical methods and screen analysis is given in ref. 14. Bead decoding and structure determination were as described (15, 16). Tubacin and niltubacin were synthesized as described (15).
Immunocytochemistry and Fluorescence Microscopy.
Detection of acetylated α-tubulin and acetylated histones by immunofluorescence was performed as described (14). HDAC6 was detected by using a rabbit polyclonal antibody (1:250–1:750) toward the C-terminal 20 aa preceded by an additional cysteine residue (C.M.G., unpublished data) under the conditions used to detect acetylated α-tubulin (14). For determining microtubule stability, A549 cells were treated as indicated, fixed in glutaraldehyde, and stained for acetylated α-tubulin and total α-tubulin in antibody dilution buffer [ADB, Tris (200 mM Tris-Cl)-buffered (pH 7.4) saline (150 mM NaCl) (TBS) with 2% BSA/0.1% Triton X-100]. For cold depolymerization, cells were incubated at 4 or 37°C for an additional 90 min before fixation. For calcium depolymerization, cells were incubated at 37°C for 10 min in a microtubule-stabilizing buffer [0.2% Nonidet P-40/5 mM MgCl2/2 mM ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid/2 M glycerol/100 mM piperazine-1,4-bis(2-ethanesulfonic acid), pH 6.8] with and without the addition of CaCl2 (2 mM) before glutaraldehyde fixation. p58 (1:100) in NIH 3T3 cells was detected after fixation in 3.7% formaldehyde in TBS. Images for these experiments were collected on a Zeiss LSM510 confocal scanning laser microscope at the appropriate wavelengths using the accompanying software and processed with Adobe PHOTOSHOP. Deconvoluted serial images of TSA- and DMSO-treated A549 cells were obtained by taking sections (8-μm) on a Zeiss Axioskop 2 microscope with an AxioCam camera and the accompanying software.
Fluorescence-Activated Cell Sorting.
A549 cells (≈75% confluent) in 10-cm dishes were treated with DMSO (0.1%), TSA (500 nM), or tubacin (2 μM) for 4.5 h, and then either DMSO (0.1%) or ITSA1 (50 μM) was added for an additional 17.5 h. Samples were rinsed in PBS, trypsinized, and fixed for 2 h in 70% ethanol/30% PBS (4°C). Cells then were washed once in PBS and stored overnight in PBS (4°C). To measure BrdUrd incorporation, samples were denatured for 20 min (room temperature) in 2 M hydrochloric acid, quenched in 0.1 M sodium borate (pH 8.5) for 5 min (room temperature), washed twice in TBS with 0.5% Tween-20 and 1% BSA (TBS-TB), and blocked for 45 min in TBS-TB. Samples then were incubated with an anti-BrdUrd antibody (1:1,000) in TBS-TB overnight (4°C). To measure acetylated α-tubulin levels, samples were blocked for 45 min in ADB, aspirated, and incubated in ADB with an antiacetylated α-tubulin antibody (1:500) overnight (4°C). For detection of primary antibodies, samples were washed twice in TBS-TB (or ADB) and incubated with FITC-conjugated anti-mouse IgG (goat; 1:200) in TBST-TB (or ADB) for 1 h (room temperature). After washing twice in TBS, cells were resuspended in 100 μl of ribonuclease A (100 μg/ml) and incubated for 5 min (37°C). To measure the DNA content, 400 μl of propidium iodide (50 μg/ml) was added, and samples were incubated for 1 h (room temperature). Analysis was performed by using a FACScanII flow cytometer (Becton Dickinson) at the Dana Farber Cancer Institute, exciting at 488 nm, and measuring the BrdUrd- or acetylated tubulin-linked FITC fluorescence through a 514-nm filter, and the PI fluorescence was measured through a 600-nm filter. Cell-cycle distributions were calculated from 10,000 cells and modeled by using MODFIT LT 2.0 software.
Cell-Migration Assays.
For Transwell migration assays, wild-type or HDAC6-overexpressing NIH 3T3 cells were seeded (150,000 cells per well) in the upper chamber (uncoated polycarbonate membrane, 6.5 mm, 8 μm) in DMEM+ medium with compounds added as indicated. After incubation (22 h), nonmigrated cells were removed by using cotton swabs, and the remaining cells were stained in Harris hematoxylin solution. For each treatment, cells within a region (spanning the maximum diameter) of three separate membranes were counted under low magnification on a Leitz Laborlux microscope. For DMSO treatments the average count was ≈600 cells per region. Images were obtained by using a Hitachi HV-C12 charge-coupled device camera.
Gene-Expression Analysis.
Gene-expression analysis of tubacin-, TSA-, and DMSO-treated mouse embryonic stem cells was performed according to the Affymetrix protocol by using a murine U74Av2 chip as described in full in ref. 12. Present calls were determined by accompanying software and data analysis including array normalization and clustering performed by using DCHIP software (19, 20).
Transfections, Immunoprecipitations, and Western Blotting.
FLAG-tagged HDAC pBJ5 constructs were transiently transfected by electroporation with 5 μg of DNA for expression of recombinant proteins (6, 21). Cells were mock-transfected without DNA or with a non-FLAG-tagged pCMV-LacZ construct as a negative control. Forty-eight hours after transfection recombinant proteins were immunoprecipitated by using anti-FLAG M2 agarose affinity gel beads as described (6). Western blottiing with antiacetylated α-tubulin (1:1,000), antitubulin (1:1,000), and antiacetylated (K9 and K14) histone H3 (1:1,000) antibodies was performed by using standard methods (12). Tubulin and MAPs were isolated from bovine brain as described (http://mitchison.med.harvard.edu/protocols/tubprep.html). Whole brain and fractions after phosphocellulose chromatography were blotted for HDAC6 and acetylated α-tubulin.
In Vitro HDAC and TDAC Assays.
[3H]Acetate-incorporated histones were isolated from butyrate-treated HeLa cells as described (5, 6). Immunoprecipitates were incubated with acetylated histones, and HDAC activity was determined by scintillation counting as described (5, 6). TDAC assays were performed with immunoprecipitates by using MAP-stabilized microtubules polymerized from MAP-rich tubulin fraction as described (18) or microtubules polymerized in the absence of MAPs. Reactions were incubated (2 h) at 37°C and then incubated on ice (15 min). After centrifugation, the supernatant (diluted to 0.4 μg of tubulin) and α-FLAG M2 agarose affinity gel beads were analyzed by Western blotting with an anti-FLAG M2 antibody (1:1,750) to ensure equal protein levels.
Results and Discussion
Identification of Tubacin, an Inhibitor of α-Tubulin Deacetylation.
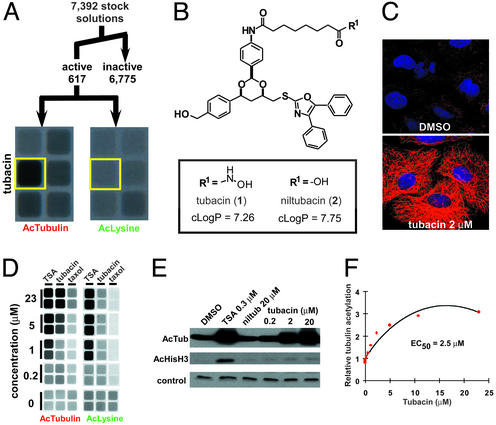
At a threshold of 1.5-fold increase in acetylation levels versus untreated cells, 8.3% (n = 617) of the total library members were active in either the AcTubulin or AcLysine cytoblot assays (Fig. 1A; ref. 14). To verify TDAC-selective inhibitors, stock solutions of compounds active in the AcTubulin and/or ITSA1+AcTubulin cytoblots but not the AcLysine cytoblot were retested in cells by using fluorescence microscopy. Beads corresponding to promising wells then were subjected to gas chromatographic decoding, leading to structure determination and subsequent resynthesis (15, 16). One hydroxamic acid-containing compound, here named tubacin (1) (Fig. 1B), was determined to be a selective (Figs. 1 C–E and 6A, which is published as supporting information on the PNAS web site) and reversible (Fig. 6B) inhibitor of α-tubulin deacetylation.
Figure 1.
Characterization of tubacin, an inhibitor of α-tubulin deacetylation. (A) Summary of multidimensional screen of 7,392 small molecules having 1,3-dioxane diversity and deacetylase-biasing elements (14, 15). An image from wells showing selectivity of tubacin (yellow) in cytoblot assays measuring acetylated α-tubulin (AcTubulin) and acetylated lysine (AcLysine) is shown. (B) Chemical structure of tubacin (1) and an inactive analog niltubacin (2). An estimation of the logarithm of the partition coefficient between octanol and water (cLogP) predicts that both compounds have similar solubility properties. (C) Increased α-tubulin acetylation induced by tubacin in A549 cells detected by immunofluorescence with an antiacetylated α-tubulin antibody (red) and nuclear staining with Hoechst 33342 (blue). (D) Image from cytoblot assays detecting acetylated α-tubulin and acetylated lysine levels in A549 cells treated for 20 h with tubacin. (E) Selectivity of tubacin for inhibiting the TDAC versus HDACs determined by Western blot analysis of acetylated α-tubulin and histone H3 (K9 and K14) in A549 cells (5 h). The control compound niltubacin (2) showed no effect on α-tubulin or histone acetylation levels. A nonspecific band reactive with the acetylated histone H3 antibody was the loading control. (F) Quantification of the mean (n = 6) acetylated α-tubulin level in A549 cells after tubacin treatment using a cytoblot (estimated EC50 of 2.5 μM).
Cellular Effects of Tubacin.
In cultured cells, tubacin (10 μM) induced up to a 3-fold increase in the relative α-tubulin-acetylation level, with a half-maximum effective concentration (EC50) of 2.5 μM (Fig. 1F). The carboxylate analog of tubacin, here named niltubacin (2) (Fig. 1B), had no effect on α-tubulin or histone acetylation and therefore was used as a negative control (Figs. 1E and 6A). Requirement of the hydroxamic acid at position R1 for activity suggests that tubacin targets a metal-dependent hydrolase. Levels of total α-tubulin and α-tubulin containing a C-terminal tyrosine (which is decreased in stabilized microtubules) were unaffected by tubacin (Fig. 6 C and D). Unlike taxol, which increases α-tubulin acetylation by direct stabilization of microtubules, tubacin had no overall effect on the morphology of A549 cells (Fig. 1C). Although α-tubulin acetylation induced by tubacin or TSA was partially suppressed by ITSA1, acetylation augmented by taxol-mediated microtubule stabilization was not suppressed by ITSA1 (Fig. 7 A and B, which is published as supporting information on the PNAS web site). Together, these results suggest that tubacin causes an increase in α-tubulin acetylation without directly stabilizing microtubules.
Tubacin Has No Effect on Gene Expression or Cell-Cycle Progression.
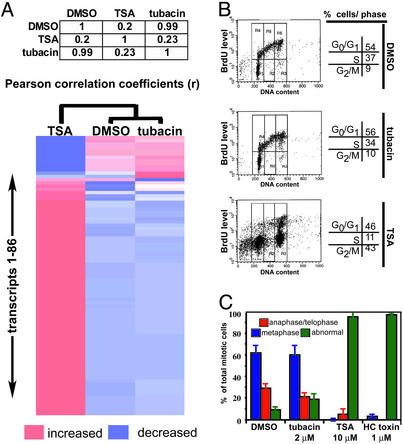
To test the intracellular selectivity of tubacin versus TSA, we compared the genome-wide transcriptional profiles of mouse embryonic stem cells, which exhibited increased sensitivity to HDAC inhibitors compared with tumor cell lines (12). Under conditions where TSA and tubacin increased α-tubulin acetylation significantly (data not shown), TSA altered expression of 232 genes above a 1.3-fold threshold value (12), whereas gene expression was unaltered by tubacin (Fig. 2A). Globally, nearest-neighbor clustering indicated more similarity between profiles of tubacin and the DMSO-treated control than to the TSA profile (Fig. 2A). Thus, TDAC inhibition seems to lack effect or have only minimal effect on gene expression. If cell-cycle arrest induced by TSA-like compounds is linked to gene expression, then tubacin should not affect cell-cycle progression. Indeed, fluorescence-activated cell sorting and BrdUrd labeling indicate that this is the case. Whereas TSA treatment led to a marked loss of a BrdUrd-positive S-phase population and a concomitant increase in sub-G1 and G2 populations, tubacin treatment lacked such effects (Fig. 2B).
Figure 2.
No effect of tubacin on gene expression or cell-cycle progression. (A) Pearson correlation matrix and nearest-neighbor clustering of a subset (86 of the most TSA-sensitive genes) of the average (n = 2) gene-expression data obtained from transcriptional profiling (murine U74Av2 gene chip; DCHIP software) of mouse embryonic stem cells treated (2.5 h) with TSA (300 nM) and tubacin (2 μM). (B) No effect of tubacin on DNA synthesis or cell-cycle distribution as determined by FACS analysis of BrdUrd-labeled and propidium iodide-stained A549 cells after treatment (4.5 h) with TSA (500 nM) and tubacin (2 μM). (C) Mitotic abnormalities in A549 cells treated (24 h) with HDAC inhibitors but not tubacin detected by immunofluorescence. The data are the average (±1 SD) of two treatments (≈100 mitotic cells per treatment).
Besides G1 and G2 cell-cycle arrest, TSA also induces defects in mitotic spindle formation and chromosome orientation. To determine whether these defects result from inhibiting tubulin deacetylation, we quantified abnormal mitotic cells after treatment (18 h) with TSA, tubacin, or HC toxin [a trapoxin-related HDAC inhibitor that, similar to trapoxin, has no effect on α-tubulin acetylation (12)]. Whereas tubacin had neither an effect on the number of mitotic cells nor spindle morphology, both TSA and HC toxin caused a significant number of abnormal mitotic cells, reminiscent of the effects of the small-molecule inhibitor monastrol on Eg5-kinesin (Fig. 2D; ref. 22). Tubacin insensitivity and HC toxin sensitivity suggest that TDAC inhibition is not responsible for mitotic defects caused by HDAC inhibitors. Instead, a deacetylase involved in chromatin remodeling is likely necessary for mitotic gene expression and/or proper kinetochore formation (23). Thus, a chemical genetic approach has allowed the effects of TSA on the TDAC to be uncoupled from its effects on cell-cycle progression.
Inhibition of HDAC6-Mediated α-Tubulin Deacetylation by Tubacin.
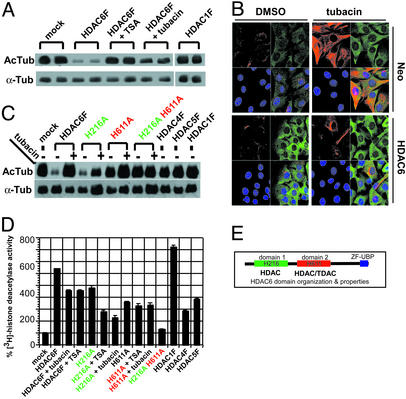
Unique features of class II HDAC6 led us to speculate that it may be the intracellular target of tubacin. HDAC6 contains an internal pair of highly similar catalytic domains, shows largely cytoplasmic localization (Fig. 7 C–F), is resistant to trapoxin/HC toxin (24), and fractionates with MAPs from bovine brain (Fig. 7G). In agreement, while this work was in progress Hubbert et al. (18) described the identification of HDAC6 as a tubulin-associated deacetylase with in vitro TDAC activity. To investigate whether HDAC6 is the TDAC targeted by tubacin, TAg Jurkat cells were transiently transfected with wild-type, FLAG-tagged HDAC6F or HDAC1F expression vectors, and immunoprecipitants assayed for TDAC activity by using MAP-stabilized tubulin as substrate. As shown (Fig. 3A), the TDAC activity of HDAC6F was inhibited by TSA or tubacin but not by niltubacin or trapoxin (Fig. 7H). We then compared the staining pattern of acetylated α-tubulin and HDAC6 in tubacin-treated fibroblasts stably expressing a control vector (Neo), HDAC6, or a catalytically inactive HDAC6 double mutant. As expected, tubacin induced an increase in α-tubulin acetylation in control cells. Overexpression of HDAC6 but not the HDAC6 double mutant (data not shown) resulted in decreased levels of tubacin-induced α-tubulin acetylation (Fig. 3B), consistent with tubacin inhibiting the intracellular deacetylase activity of HDAC6.
Figure 3.
Inhibition of HDAC6 deacetylase activity by tubacin. (A) Western blot of acetylated α-tubulin (AcTub) and total α-tubulin (α-Tub) after incubation of MAP-stabilized tubulin with wild-type HDAC6F and wild-type HDAC1F immunoprecipitated from transfected TAg Jurkat cells. Although HDAC1 showed no TDAC activity, the TDAC activity of HDAC6 was inhibited by TSA (300 nM) or tubacin (2 μM). (B) Decreased α-tubulin acetylation after tubacin (2 μM) treatment (4 h) of HDAC6-overexpressing compared with control (Neo) NIH 3T3 cells detected by immunofluorescence using antiacetylated α-tubulin (red) and anti-HDAC6 (green) antibodies and nuclear staining with Hoechst 33342 (blue). (C) Western blot of acetylated α-tubulin and total α-tubulin after incubation of MAP-stabilized tubulin with immunoprecipitants from transfected TAg Jurkat cells. Mutation of histidine 611 to alanine (H611A) but not histidine 216 to alanine (H216A) was sufficient to abolish the TDAC activity of HDAC6. (D) Effect of TSA and tubacin on the HDAC activity of immunoprecipitants from transfected TAg Jurkat cells. Activities (n = 3; ± mean absolute deviation) were measured by scintillation counting of 3H-labeled acetic acid released from 3H-labeled histones. Wild-type HDAC6F and the HDAC6F H216A mutant were inhibited only partially by TSA or tubacin, and the HDAC6F H611A mutant was not inhibited by either TSA or tubacin. (E) HDAC6 domain organization.
Selectivity of Tubacin for the C-Terminal Deacetylase Domain of HDAC6.
Although both catalytic domains of HDAC6 have histone deacetylase activity (21), their activities toward tubulin deacetylation and sensitivity to tubacin, remained to be determined. To this end, TAg Jurkat cells were transiently transfected with HDAC6F, catalytic subsite mutants (H216A or H611A), or a double mutant (H216A H611A), HDACs1F, -4F, or -5F, and the immunoprecipitants were subjected to in vitro deacetylase assays. With acetylated tubulin as substrate (Fig. 3C), only HDAC6F and the HDAC6F H216A mutant catalyzed deacetylation of α-tubulin, an activity that was inhibited by tubacin and TSA. With [3H]histones as substrate (Fig. 3D), all immunoprecipitants other than the HDAC6 double mutant possessed HDAC activity. Although the HDAC6F H216A mutant was inhibited by tubacin or TSA, the HDAC6F H611A mutant was not inhibited by either small molecule. Thus, the H611 domain alone exhibits TDAC activity and is a target of both tubacin and TSA (Fig. 3E). We speculate that the H216 domain serves as an α-tubulin K40-binding domain that facilitates recruitment and the generation of feedback loops in tubulin function. This two-domain feature is an essential element to histone-modifying enzymes that impart processivity and switch-like behavior (25).
Tubacin Treatment Does Not Stabilize Microtubules to Nocodazole, Calcium, or Cold Depolymerization.
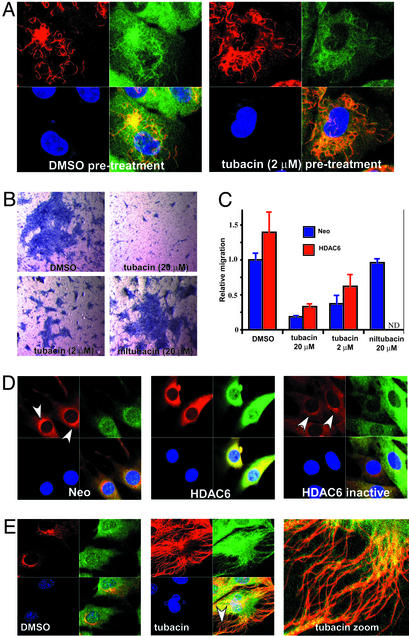
Because α-tubulin acetylation accumulates within stable microtubule populations, particularly in nonmitotic cells (26, 27), we tested whether increasing microtubule acetylation was sufficient to alter microtubule dynamics. A549 cells pretreated with tubacin were treated with the microtubule destabilizer nocodazole, or subjected to these reagents in the reverse order. In separate experiments, cells pretreated with tubacin or taxol were treated with calcium or placed on ice. Although tubacin treatment always increased α-tubulin-acetylation levels, we observed no stabilization of microtubules to any of the depolymerizing conditions tested (Fig. 4A; see also Fig. 8A, which is published as supporting information on the PNAS web site). In addition, neither tubacin nor ITSA1 prevented the polymerization of microtubules after nocodazole was washed out (see Fig. 8B). Thus, although deacetylation may be coupled to depolymerization, deacetylation does not seem necessary for this process. Furthermore, increasing α-tubulin acetylation is not sufficient to stabilize microtubules, a finding that differs from that reported recently by Matsuyama et al. (28). However, because TSA lacks selectivity, and TSA and trapoxin have differential effects on gene expression (12), the reported delay to colcemid-induced microtubule destabilization might result from an effect of TSA on HDACs other than HDAC6. Alternatively, TDAC inhibition by tubacin may affect the stabilization of microtubules beyond the spatial or temporal resolution of our assay.
Figure 4.
Phenotypic effects of tubacin. (A) No effect of tubacin (2 μM) pretreatment (4 h) on the stability of microtubules to nocodazole (332 nM; 2 h)-induced depolymerization determined by immunofluorescence using antiacetylated α-tubulin (red) and anti-α-tubulin (green) antibodies and nuclear staining with Hoechst 33342 (blue) in A549 cells. (B) Hematoxylin staining of NIH 3T3 cells (blue) that have undergone migration in a Transwell assay under the conditions indicated. Tubacin (2–20 μM) but not niltubacin (20 μM) inhibited cell migration. (C) Quantification of the migration of control (Neo) and HDAC6-overexpressing NIH 3T3 cells measured by counting hematoxylin-stained cells. Data are presented as the mean (n = 3; ±1 SD) relative to untreated wild-type (Neo) cells. ND, not determined. (D) Overexpression of HDAC6 but not catalytically inactive HDAC6 in NIH 3T3 cells causes mislocalization of p58 (arrowhead), a Golgi membrane MAP detected by immunofluorescence using anti-p58 (red) and anti-HDAC6 (green) antibodies and nuclear staining with Hoechst 33342 (blue). (E) Increased colocalization of HDAC6 with acetylated α-tubulin in NIH 3T3 cells (Neo) after tubacin (20 μM) treatment (4 h) detected by immunofluorescence using antiacetylated α-tubulin (red) and anti-HDAC6 (green) antibodies and nuclear staining with Hoechst 33342 (blue). The arrowhead indicates a region of higher magnification.
Tubacin Treatment Inhibits Cell Motility and HDAC6 as a MAP.
Because overexpression of wild-type HDAC6 was reported to increase motility of serum-starved NIH 3T3 cells (18), we determined whether tubacin treatment could block cell motility. Tubacin (2 μM) but not niltubacin (20 μM) inhibited the migration of both wild-type and HDAC6-overexpressing cells (Fig. 4 B and C) under normal culture conditions. Because no change in microtubule stability occurred on a commensurate time scale, α-tubulin acetylation may instead affect the activity of MAPs or microtubule motors that regulate cell motility (29). Accordingly, overexpression of HDAC6 but not an inactive mutant disrupted the juxtanuclear localization of p58, a MAP that mediates the binding of Golgi elements to microtubules (Fig. 4D). Microtubules play a role in organization of the Golgi complex through activity of the microtubule-dependent motor dynein (30). Because HDAC6 was reported to colocalize with dynactin (p150/Glued) (18), a processivity factor for dynein, HDAC6 overexpression may alter both the dynamics of α-tubulin acetylation and the activity of dynein. The relationship between HDAC6 and Golgi elements may provide an explanation for the loss of α-tubulin acetylation observed in neuronal cells after disruption of the Golgi complex (31). We observed an increase in the colocalization of HDAC6 along acetylated microtubules in interphase cells after tubacin treatment (Fig. 4E). In mitotic cells (see Fig. 9, which is published as supporting information on the PNAS web site), HDAC6 colocalizes with the highly acetylated spindle microtubules and centrosomes, structures bound by many microtubule motors. These results, along with copurification with MAPs from bovine brain, suggest HDAC6 may have a functional role as a MAP. We speculate that α-tubulin acetylation recruits HDAC6 via its N-terminal domain, causing colocalization with MAPs. Overexpression of HDAC6 then could alter MAP localization, which in turn could affect processes (such as protein trafficking and receptor localization) involved in cell motility (29).
In summary, tubacin is a small-molecule inhibitor selective for α-tubulin deacetylation. With tubacin as a probe, our results show that targeting the TDAC has no effect on cell-cycle progression. Our results highlight the relevance of α-tubulin acetylation in mediating cell motility. Given the dependence of metastasis and angiogenesis on cell movement, increasing α-tubulin acetylation may be an important component to the antimetastatic and antiangiogenic properties of HDAC inhibitors (8, 9). Conversely, decreasing α-tubulin acetylation by HDAC6 overexpression may be the cause of the reduced α-tubulin-acetylation levels observed in neurodegenerative disorders such as Alzheimer's disease (32, 33). In addition, both suberoylanilide hydroxamic acid (12) and butyrate (data not shown), HDAC inhibitors used to suppress neurotoxicity in polyglutamine-repeat disorders (9), also inhibit α-tubulin deacetylation. Besides altering the expression levels of genes important for neuronal viability, the therapeutic effect of HDAC inhibitors in neurodegenerative disorders may involve increased α-tubulin acetylation. By uncoupling the effects of deacetylase inhibitors on the cytoskeleton and chromatin, tubacin allows new applications of deacetylase inhibitors and improved versions of current ones having increased selectivity to be envisioned.
Supplementary Material
Acknowledgments
We thank members of the Schreiber group, Dr. T. Mitchison, Dr. R. Ward, and Z. Perlman, for discussions and comments. We are grateful to Z. Maliga for assistance in tubulin/MAP purification, Dr. C. Li for instruction in using DCHIP software, C. Hubbert and Dr. T. Yao for the gift of the NIH 3T3 stable cell lines, and the National Institute of General Medical Sciences for support of this research. K.M.K. was supported by a Damon Runyon Cancer Research Foundation fellowship. J.C.W. and C.M.G. were supported by National Science Foundation predoctoral fellowships. S.L.S. is an Investigator at the Howard Hughes Medical Institute.
Abbreviations
- HDAC
histone deacetylase
- TSA
trichostatin A
- TDAC
tubulin deacetylase
- AcTubulin
α-tubulin acetylation
- AcLysine
histone acetylation
- MAP
microtubule-associated protein
References
- 1.Grozinger C M, Schreiber S L. Chem Biol. 2002;9:3–16. doi: 10.1016/s1074-5521(02)00092-3. [DOI] [PubMed] [Google Scholar]
- 2.Khochbin S, Verdel A, Lemercier C, Seigneurin-Berny D. Curr Opin Genet Dev. 2001;11:162–166. doi: 10.1016/s0959-437x(00)00174-x. [DOI] [PubMed] [Google Scholar]
- 3.Yoshida M, Kijima M, Akita M, Beppu T. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 4.Kijima M, Yoshida M, Sugita K, Horinouchi S, Beppu T. J Biol Chem. 1993;268:22429–22435. [PubMed] [Google Scholar]
- 5.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 6.Hassig C A, Tong J K, Fleischer T C, Owa T, Grable P G, Ayer D E, Schreiber S L. Proc Natl Acad Sci USA. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnstone R W. Nat Rev Drug Discovery. 2002;1:287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- 8.Remiszewski S W. Curr Opin Drug Discovery Dev. 2002;5:487–499. [PubMed] [Google Scholar]
- 9.Steffan J S, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol B L, Kazantsev A, Schmidt E, Zhu Y Z, Greenwald M, et al. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 10.Phiel C J, Zhang F, Huang E Y, Guenther M G, Lazar M A, Klein P S. J Biol Chem. 2001;276:36734–36741. doi: 10.1074/jbc.M101287200. [DOI] [PubMed] [Google Scholar]
- 11.Polevoda B, Sherman F. Genome Biol. 2002;3:0006.1–0006.6. doi: 10.1186/gb-2002-3-5-reviews0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koeller, K. M., Haggarty, S. J., Perkins, B. D., Leykin, I., Wong, J. C., Kao, M. C. & Schreiber, S. L. (2003) Chem. Biol., in press. [DOI] [PubMed]
- 13.Stockwell B R, Haggarty S J, Schreiber S L. Chem Biol. 1999;6:71–83. doi: 10.1016/S1074-5521(99)80004-0. [DOI] [PubMed] [Google Scholar]
- 14. Haggarty, S. J., Koeller, K. M., Wong, J. C., Butcher, R. A. & Schreiber, S. L. (2003) Chem. Biol., in press. [DOI] [PubMed]
- 15.Sternson S M, Wong J C, Grozinger C M, Schreiber S L. Org Lett. 2001;3:4239–4242. doi: 10.1021/ol016915f. [DOI] [PubMed] [Google Scholar]
- 16.Blackwell H E, Perez L, Stavenger R A, Tallarico J A, Cope-Eatough E, Foley M A, Schreiber S L. Chem Biol. 2001;8:1167–1182. doi: 10.1016/s1074-5521(01)00085-0. [DOI] [PubMed] [Google Scholar]
- 17.Clemons P A, Koehler A N, Wagner B K, Sprigings T G, Spring D R, King R W, Schreiber S L, Foley M A. Chem Biol. 2001;8:1183–1195. doi: 10.1016/s1074-5521(01)00086-2. [DOI] [PubMed] [Google Scholar]
- 18.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang X F, Yao T P. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 19.Li C, Wong W H. Genome Biol. 2001;2:0032.1–0032.11. [Google Scholar]
- 20.Schadt E E, Li C, Ellis B, Wong W H. J Cell Biochem Suppl. 2001;37:120–125. doi: 10.1002/jcb.10073. [DOI] [PubMed] [Google Scholar]
- 21.Grozinger C M, Hassig C A, Schreiber S L. Proc Natl Acad Sci USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayer T U, Kapoor T M, Haggarty S J, King R W, Schreiber S L, Mitchison T J. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 23.Taddei A, Maison C, Roche D, Almouzni G. Nat Cell Biol. 2001;3:114–120. doi: 10.1038/35055010. [DOI] [PubMed] [Google Scholar]
- 24.Furumai R, Komatsu Y, Nishino N, Khochbin S, Yoshida M, Horinouchi S. Proc Natl Acad Sci USA. 2001;98:87–92. doi: 10.1073/pnas.011405598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schreiber S L, Bernstein B E. Cell. 2002;111:771–778. doi: 10.1016/s0092-8674(02)01196-0. [DOI] [PubMed] [Google Scholar]
- 26.Piperno G, LeDizet M, Chang X J. J Cell Biol. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Black M M, Baas P W, Humphries S. J Neurosci. 1989;9:358–368. doi: 10.1523/JNEUROSCI.09-01-00358.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, Seigneurin-Berny D, Osada H, Komatsu Y, Nishino N, Khochbin S, et al. EMBO J. 2002;21:6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palazzo A, Ackerman B, Gundersen G G. Nature. 2003;421:230. doi: 10.1038/421230a. [DOI] [PubMed] [Google Scholar]
- 30.Thyberg J, Moskalewski S. Exp Cell Res. 1999;1:263–279. doi: 10.1006/excr.1998.4326. [DOI] [PubMed] [Google Scholar]
- 31.Elyaman W, Yardin C, Hugon J. J Neurochem. 2002;81:870–880. doi: 10.1046/j.1471-4159.2002.00838.x. [DOI] [PubMed] [Google Scholar]
- 32.Hempen B, Brion J P. J Neuropathol Exp Neurol. 1996;55:964–972. doi: 10.1097/00005072-199609000-00003. [DOI] [PubMed] [Google Scholar]
- 33.Saragoni L, Hernandez P, Maccioni R B. Neurochem Res. 2000;25:59–70. doi: 10.1023/a:1007587315630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.