Abstract
Plant viruses have a significant impact on agronomic losses worldwide. A new strategy for engineering virus-resistant plants by transgenic expression of a dominant interfering peptide is presented here. This peptide of 29 aa strongly interacts with the nucleocapsid proteins (N) of different tospoviruses. Transgenic Nicotiana benthamiana lines expressing the peptide fused to a carrier protein were challenged with five different tospoviruses that have a nucleocapsid protein interacting with the peptide. In the transgenic plants, strong resistance to tomato spotted wilt virus, tomato chlorotic spot virus, groundnut ring spot virus, and chrysanthemum stem necrosis virus was observed. This therefore demonstrates the feasibility of using peptide “aptamers” as an in vivo tool to control viral infection in higher plants.
Viral diseases cause major crop losses worldwide. Actual losses due to plant viruses are difficult to assess, but some estimates indicate total economic damage as high as several billion U.S. dollars per year. Among the most detrimental viruses, tomato spotted wilt virus (TSWV), the type species of the tospovirus genus in the family Bunyaviridae, alone causes crop loss of more than one billion dollars per year (1). For crop protection, so far antiviral chemicals are not available. Therefore, the control of viral diseases employs different strategies, such as integrated vector management, crop rotation, and production of pathogen-free plant stocks and seeds. In addition, natural resistance genes have been widely used to create varieties resistant to one or more viruses. However, the transfer of resistance characters from a wild plant species or a nonadapted variety without losing important qualities is a lengthy and difficult process.
Transgenic technologies have provided new methods for creating virus-resistant plants. The most successful examples rely on the pathogen-derived resistance concept of Sanford and Johnson (2) who proposed to achieve effective resistance by transgenic expression of genes derived from the pathogen itself. The proof of this concept was provided by expressing tobacco mosaic virus (TMV) coat protein resulting in resistant tobacco plants (3). Since then, the same approach was adopted to protect a number of different plant species from several plant viruses (review refs. 4 and 5).
RNA silencing or posttranscriptional gene silencing (PTGS) has been recently uncovered as an important mechanism involved in pathogen-derived resistance. RNAs with homology to the transgenically expressed viral sequences are specifically degraded, with the translation of the gene product being dispensable for virus resistance. Resistance phenotypes based on posttranscriptional gene silencing are usually strong, but because of sequence specificity, they are restricted only to the virus species the transgene was derived from or to closely related isolates (6, 7).
Broad-spectrum virus resistance has been obtained by expressing specific proteins. Here, presence of the viral gene product in inappropriate amounts, form or time, is thought to interfere with viral infection. However, in some cases it is difficult to distinguish between an RNA- and protein-mediated resistance (8, 9). Unambiguous examples of protein-mediated virus resistance mainly concern the expression of viral coat proteins (refs. 3 and 10; review ref. 4), but cases of protein-dependent pathogen-derived resistance due to the expression of viral movement proteins or replicases are also known (9, 11, 12). In some instances, resistance is based on the expression of intact, functional proteins; in others the expression of the intact protein leads only to weak resistance or even to enhanced susceptibility. In contrast, expression of a dysfunctional protein may lead to strong resistance (12, 13). Despite the number of successful examples, the molecular basis of protein-mediated virus resistance is, in most cases, not understood. This is why a rational construction of resistance inducing proteinaceous agents is still not practicable. Also, the rapid evolution of resistance-breaking virus strains calls for the development of new concepts in resistance engineering.
To devise a new knowledge-based strategy to engineer virus-resistant plants, we investigated the molecular properties and functions of TSWV proteins. Consistent with the general notion of the multifunctionality of viral coat proteins (14), the TSWV N protein was found to be involved in a number of molecular interactions, such as homopolymerization (15), packaging of the viral genomic RNA (16), and interaction with NSm, the TSWV movement protein (17). The molecular dissection of homotypic interaction of TSWV N protein revealed the existence of two interaction domains and identified amino acids conserved among the tospoviruses that are essential for the interaction (15).
The isolation of a peptide “microdomain” interacting with TSWV N protein and the development of a strategy to induce virus resistance in higher plants is described. The strategy exemplifies a promising new concept that uses target-specific peptides (“aptamers”), selected with the yeast two-hybrid system, to modulate/inhibit protein functions in vivo (18, 19).
Materials and Methods
Construction of a Protein-Specific Peptide Library and Yeast Two-Hybrid Vectors.
The PCR-amplified TSWV N ORF was digested with DNaseI. Fragments were cloned into the pCRII-Blunt vector (Invitrogen). Subcloned fragments were isolated and cloned into the EcoRI site of the yeast two-hybrid shuttle vector pACT2 (CLONTECH).
N protein-coding ORFs of tomato chlorotic spot virus (TCSV), groundnut ring spot virus (GRSV), chrysanthemum stem necrosis virus (CSNV), impatiens necrotic spot virus (INSV), iris yellow spot virus (IYSV), physalis severe mottle virus (PSMV), and watermelon silver mottle virus (WSMV) were amplified by RT-PCR with gene-specific primer sets adding a NcoI site containing the native start codon and XhoI and SalI sites after the stop codon. The fragments were subcloned in pAS2–1 (CLONTECH) between the NcoI and SalI site as a translational fusion with the Gal4 DNA-binding domain. Proteins with C-terminal deletions were constructed similarly, but with 3′ primers including a stop codon after amino acid 232 of TCSV, GRSV, CSNV, and amino acid 236 of INSV, IYSV, and WSMV and 237 of PSMV N, respectively. Binding domain fusions in pAS2–1 of TSWV N as well as the TSWV N C-terminal deletion (amino acids 1–232, lacking 26 aa at the C terminus = CΔ26) and the C-terminal double mutant F242A/F246A are described in Uhrig et al. (15).
Preparation of β-Glucuronidase (GUS) Fusions.
The protein-coding region of the uidA gene (20), which encodes the β-glucuronidase of Escherichia coli, was modified to include translational peptide fusions with the GUS ORF. One internal BspHI site of the GUS-coding region was eliminated by fusion of an N-terminal fragment of the uidA gene, amplified with primers Gus_5_BspHI, EcoRI (CCCGGGCTCATGAGGGAATTCATGTTACGTCCTGTAGAAACC) and Gus_3_NcoI (GCATCTCCATGGCGACCAAAG), and cut with NcoI, to a second fragment, amplified with primer Gus_5_BspHI (CTTTGGTCGTCATGAAGATGC) and primer Gus_3_NcoI, SalI, BamHI (CGCGGATCCTCAGTCGACCCCGGGGCCCATGGGTTGTTTGCCTCCCTGC) and cut with BspHI. The resulting sequence includes the GUS-coding region without the internal BspHI site, but with additional BspHI and EcoRI sites at the 5′ end and NcoI, SalI, and BamHI sites at the 3′ end. This fragment was cut with BspHI and BamHI and ligated in the vector NcoI and BamHI sites of pEntr4 (Gateway System, Invitrogen), resulting in plasmid pEntry4-Gus-Fus.
The artificial sequence coding for a peptide was created annealing primer T-Pep-52 AGGGTTCTGTGGCAATGGAGCATTATTCTGAGACATTGAATAAATTTTACGAGATGTTTG to primer T-Pep-3 GGGCAATTGCCCCCGCTCGAGAACACCAAACATCTCGTAAAATTTATTCAATGTCTCAGA and blunt ending with Pwo polymerase (Roche Molecular Biochemicals). PCR amplification with primer T-Pep-5 CCGGAATTCGCCATGGCTTCTAGTTCTAACCCAAACGCAAAGGGTTCTGTGGCAATGGAG and primer T-Pep-3 resulted in the final coding region of T-Pep. The fragment was cut with NcoI and XhoI and ligated in plasmid pEntry4-Gus-Fus cut with NcoI and SalI, generating plasmid pEntry4-GusT. For yeast two-hybrid tests, the Gus and GusT ORFs were cloned, as a translational fusion with the Gal4 activation domain, by using a Gateway LR reaction on a pACT2 vector containing the Gateway cassette. This was prepared by ligation of the Gateway cassette B in a blunted pACT2 vector cut with NcoI and XhoI. This reaction gave rise to two vectors called pACT-Gus and pACT-GusT, respectively.
Yeast Protein Interaction Assays.
All yeast two-hybrid interaction assays were carried out by transforming plasmids containing protein fusions to the Gal4 DNA binding domain (pAS2–1) and plasmids containing protein or peptide fusions with the Gal4 DNA activation domain (pACT2), into the yeast strain PJ69-4A as described by Gietz et al. (21). For isolation of TSWV N interacting peptides, the TSWV N double mutant F242A/F246A described in ref. 15 was used as bait. Transformed yeast cells were grown 3 to 5 days at 30°C on solid medium lacking leucine and tryptophan (SD-LW), as well as on solid medium lacking leucine, tryptophan, and histidine (SD-LWH) supplemented with 3 mM 3-aminotriazole to test for protein interactions. Growing colonies were tested for β-galactosidase activity (22).
Plant Transformation.
The plant transformation vectors were prepared based on the LR reaction of the Gateway system (Invitrogen) and the destination vector pLX222-Gateway. Plasmid pLX222-Gateway is based on the pLX222 vector (23) containing a HindIII fragment of vector pRT104 (24) with an additional Gateway destination cassette cloned in the blunted XhoI and XbaI sites such that expression is controlled by the CaMV 35S promoter of plasmid pRT104. Plasmids pLX-Gus and pLX-GusT were constructed by LR recombination of pEntry4-Gus-Fus and pEntry4-GusT into pLX222-Gateway, respectively, and transformed in the Agrobacterium tumefaciens strain LBA4404 (25) and used to transform Nicotiana benthamiana via leaf disk transformation as described (26). Regenerated shoots were tested for expression of recombinant protein through GUS assays (27). Transformed plants were selfed, and homozygous transformants were selected by a kanamycin resistance test and tested again for recombinant gene expression. Seeds obtained from these plants were used to grow plants for the virus resistance tests.
Kanamycin Resistance Test.
Seeds of primary transformants were grown on 3× MS medium containing 100 μg/liter kanamycin. Kanamycin resistance was observed as a measure of root length 14 days after germination. No roots are indicative for kanamycin sensitivity. Candidates segregating with 3 to 1 were used to produce seeds. From these, homozygous plants were generated and chosen for further analysis.
Virus Maintenance.
TSWV (DSM no. PV-0182; DSM, German collection of microorganisms), TCSV (DSM no. PV-0390), GRSV (DSM no. PV-0205), CSNV (DSM no. PV-0529), INSV (DSM no. PV-0280), WSMV (DSM no. PV-0283), as well as the Physalis isolate PSMV (28) were mechanically passaged on N. benthamiana every 3–4 weeks.
Virus Resistance Test.
Young plants grown to leaf stage 3–4 were mechanically inoculated on the third and fourth leaf with 50 μl of 5-fold diluted plant sap prepared from clearly symptomatic leafs with 0,1 M Na2/KH2PO4 buffer, pH 7.0, containing 2% PEG 10000 and 0.2% Na3SO3. Plants were assayed for disease symptoms, and plant height was measured twice a week until 23 days after infection.
GUS Staining.
Total protein extracts from leaves were separated on a nondenaturing PAA gel, and GUS activity was detected by incubating the gel overnight in 10 mM EDTA/0.05% Triton X-100/100 mM NaPi (pH 8.0), containing 0.05% X-Gluc.
Results
Isolation of Peptides Interacting with Tospovirus N Proteins.
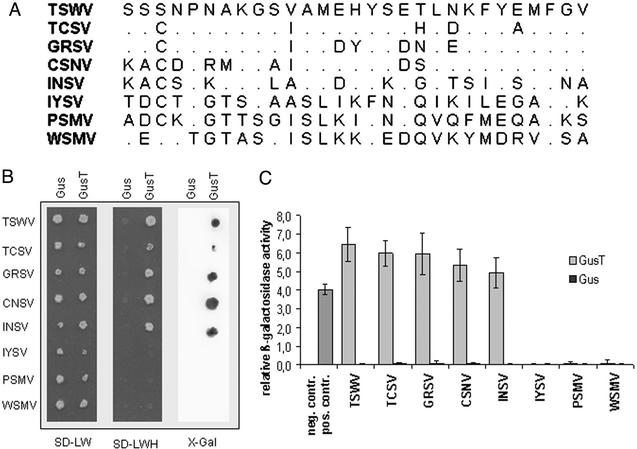
To dissect the functional interaction domains of the TSWV N protein, a library of random fragments of the TSWV N gene was constructed, and fragments were cloned into a yeast two-hybrid activation domain vector. This library of ≈7,000 independent clones was cotransformed with yeast two-hybrid vectors expressing Gal4-DNA binding domain fusions of TSWV N protein carrying two mutations (F242A/F246A). The mutations inactivate the C-terminal interaction domain, allowing the study of the dimerization between bait and prey in the absence of homopolymerization of the bait proteins (15). Two screening rounds led to the isolation of seven transformants with prototrophic growth on medium without histidine. The isolated clones were sequenced, and a minimal peptide sequence was deduced representing amino acids 220–248 of the TSWV N protein (= T220/248; Fig. 1A). The peptide was fused to β-glucuronidase (GusT) and tested in the yeast two-hybrid system (Fig. 1B). Strong interaction of GusT with C-terminally truncated N proteins not only from TSWV but also TCSV, GRSV, CSNV, and INSV was observed. More distantly related tospoviral N proteins, however, did not interact with T220/248 (Fig. 1B; IYSV, PSMV, and WSMV). The interaction pattern correlates with the phylogenetic clustering of N protein sequences in American and Eurasian subgroups (29). The relative interaction strength was evaluated by a quantitative β-galactosidase assay (15) and found to be stronger than the control interaction (CΔ26 as bait, TSWV N protein as prey), with slightly decreased strength in the case of INSV N protein, the protein with the lowest sequence conservation within the European subgroup (Fig. 1C). The GUS protein without the peptide extension fused to the Gal4 activation domain did not show any interaction with tospovirus N proteins.
Figure 1.
Properties of T220/248. (A) Amino acid sequence of the peptide from the TSWV N protein in comparison to homologous sequences of other tospovirus N proteins. (B) Interactions between GusT peptide fusion and GUS, respectively, and C-terminal deletions of different tospovirus N proteins. Interactions were investigated in the yeast two-hybrid system by plating double-transformed yeast cells on SD-LW medium to test for double transformation and on SD-LWH medium for protein interactions. As a second reporter of the interaction, β-galactosidase was detected by using a filter lift assay. (C) Relative interaction strength is measured by a quantitative β-galactosidase activity assay. Yeast cells transformed with the empty vectors pAS2-1 and pACT2 were used as negative control (no column), whereas yeast cells expressing the CΔ26 protein interacting with TSWV N protein were the positive control. GUS protein did not interact with C-terminal deletions from N proteins of different origin.
Plant Transformation and Detection of Native GUS-Peptide Fusion Protein.
Two constructs (pLX-Gus and pLX-GusT, respectively) were produced, the first expressing GUS under control of the CaMV 35S promoter and the second T220/248 C-terminally fused to GUS. To minimize the extent of identity between the coding sequence of T220/248 and the viral genomic RNA, the coding sequence was assembled from oligonucleotides incorporating an altered codon usage optimized for tobacco. Thus, nucleotide sequence identity was consequently reduced to 61%. N. benthamiana leaf disks were transformed by using A. tumefaciens mediated transformation. Nine independent transformants were planted in soil, tested for recombinant GUS expression, and grown to set seed. Transformants showed no morphological abnormalities and were indistinguishable from control plants. Progeny of homozygous T1 plants were tested for TSWV resistance.
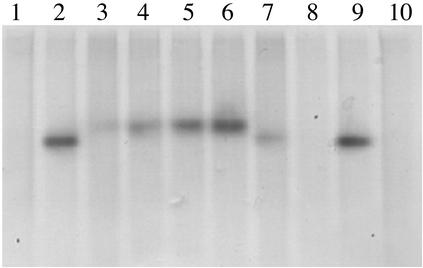
Four homozygous GusT-expressing lines (GusT 1.5, GusT 5.1, GusT 6.2, and GusT 8.1) and one control line expressing GUS (Gus 5.a) were used in further experiments. Native GUS or GusT expression was detected by in-gel staining of GUS activity (Fig. 2). Single bands representing the active tetrameric form of GUS were detected in protein extracts from GUS or GusT-expressing lines, whereas no activity was detected in nontransformed plants. Compared with native GUS, the bands of GusT-expressing lines had a slightly higher molecular weight, as expected because of the additional ≈20 kDa contributed by the peptide fusions. These results supported a correct, native expression of the GUS-peptide fusion proteins (Fig. 2).
Figure 2.
Detection of β-glucuronidase in Gus and GusT-transformed N. benthamiana plants. Twenty micrograms of plant protein extracts were separated under nondenaturing conditions, and gels were stained for β-glucuronidase activity. Lanes 1, 8, and 10, protein extract of nontransformed (wild-type) plants; lanes 2 and 9, protein extract of wild-type plants supplemented with 100 ng of β-glucuronidase purified from E. coli; lanes 3–7, protein extracts from lines GusT 1.5, GusT 5.1, GusT 6.2, GusT 8.1, and Gus 5.a, respectively.
Resistance to TSWV.
Forty-eight T2 plants of homozygous GusT-expressing lines (GusT 1.5, GusT 5.1, GusT 6.2, and GusT 8.1), as well as 12 nontransformed (wild type) plants and 12 plants of a GUS-expressing line (Gus 5.a) were challenged with TSWV. All inoculated individuals of the control lines developed symptoms within 7–10 days postinoculation (dpi), stopped growth at the same time (plant height of ≈4–5 cm), and eventually died. All GusT-expressing lines exhibited strong resistance phenotypes (Figs. 3 and 4 and Table 1). Compared with noninfected wild-type plants, the lines GusT 1.5, GusT 5.1, and GusT 6.2 developed no symptoms and showed no growth reduction. In the line GusT 8.1, 11 plants showed no symptoms, and 1 plant had mild symptoms after 13–17 dpi, a significant delay compared with control lines.
Figure 3.
TSWV inoculated plants evaluated 23 days after inoculation. (A) Wild-type. (B) Gus 5.a expressing unmodified glucuronidase. (C) GusT 8.1 expressing the β-glucuronidase peptide fusion.
Figure 4.
Average plant height of transgenic and wild-type N. benthamiana plant lines after inoculation with different Tospovirus species (23 dpi). Mean values of 12 individual plants per line for TSWV, TCSV, and CSNV and for 24 individual plants per line for GRSV and INSV are displayed. For CSNV infection of Gus 5.a and wild-type plants, the mean values of 6 and 10 plants are displayed, respectively.
Table 1.
Development of viral infection
| Transformants | Virus inoculated
|
||||
|---|---|---|---|---|---|
| TSWV | TCSV | GRSV | CSNV | INSV | |
| GusT 1.5 | 0/12 | 13/18 | 1/24 | 0/12 | 30/30 |
| GusT 5.1 | 0/12 | 17/18 | 0/24 | 0/12 | 29/30 |
| GusT 6.2 | 0/12 | 16/18 | 5/24 | 0/12 | 29/30 |
| GusT 8.1 | 1/12 | 9/18 | 0/24 | 0/12 | 29/30 |
| Gus 5.a | 12/12 | 18/18 | 22/24 | 6/6 | 29/30 |
| Wild type | 12/12 | 18/18 | 24/24 | 10/10 | 30/30 |
Number of symptomatic plants and inoculated plants 23 days after inoculation are reported for each type of inoculation.
Resistance to Other Tospovirus Species.
The potential resistance against other tospovirus species was tested in 12–30 GusT homozygous plants for each GusT 1.5, GusT 5.1, GusT 6.2, and GusT 8.1 line. The candidates and the GUS-expressing and nontransformed controls were inoculated at the third to fourth leaf stage with virus-infected plant sap. Plants were challenged with tospovirus species from the American subgroup expressing N proteins interacting with the TSWV N peptide fused to GUS. Plant height and symptom development was recorded over a period of 23 dpi. No resistance was observed with nontransformed or GUS-expressing plants. In the case of inoculation with TCSV, GRSV, and INSV, systemic symptoms developed within 6–8 dpi, and plants stopped their growth at the height of ≈4 cm, respectively (Fig. 4). In the case of infection with CSNV, control plants developed systemic symptoms ≈17 dpi, growing to a height of ≈23 cm. All four GusT-expressing transgenic lines showed significant resistance against the species TSWV, TCSV, GRSV, and CSNV (Fig. 4). In the cases of TSWV, GRSV, and CSNV, a strong resistance was observed: only one individual carried symptoms on TSWV infection (2% of all tested) and six on GRSV infection (6.25%), whereas after CSNV infection, symptom development was not observed in any plant. A weaker but still significant resistance was observed on TCSV infection. Seventeen of 48 plants (35%) remained free of systemic symptoms (line GusT 8.1 showed no systemic symptoms in 50%; Table 1). Significant resistance was not observed after inoculation with INSV, the most distantly TSWV-related tospovirus species within the American subgroup. In this case, however, in contrast to the controls that stopped growing at 3–5 cm in height, the GusT-expressing lines exhibited a slightly delayed disease development, with 20% of the inoculated plants producing new shoots after 13–17 dpi; some of the plants produced flowers (Fig. 4; data not shown).
Discussion
In this study, we report the development of a knowledge-based method for engineering virus-resistant plants. A detailed analysis of the interaction properties of TSWV N protein allows the adoption of transdominant genetics, a general concept in which proteins, peptides, or RNAs are expressed in cells to generate mutant phenocopies or to dominantly interfere with specific protein functions in vivo (30). Peptides interfering with protein interactions in vivo (peptide aptamers; ref. 18) have successfully been used in yeast cells, E. coli, Drosophila, and cultured human cells. They are a useful tool for basic research, but they also represent a new approach in drug target validation and drug discovery (18, 19, 31–34). The application of dominant negative peptide aptamers to inhibit protein functions in vivo in a living multicellular organism has been reported so far only for Drosophila (19, 35).
We used a random fragment library of TSWV N protein and the yeast two-hybrid system to select peptides strongly interacting with TSWV N protein. The yeast-based screening has the advantage of selecting molecules functional under intracellular conditions (reviewed in refs. 36–38). Overlapping peptides covering a C-terminal region of TSWV N protein were isolated. This region was previously characterized as one of the two interaction domains (15).
The shortest peptide of 29 aa was fused to β-glucuronidase and interacted strongly not only with the TSWV N protein but also with a number of different tospoviral N proteins, indicating that despite sequence variation, the structural basis of homotypic interaction of tospoviral N proteins is evolutionary conserved. Expression of the GUS-peptide fusions in transgenic N. benthamiana resulted in strong resistance not only to TSWV but also to the other tospovirus species TCSV, GRSV, and CSNV. The results provide an example of a successful application of dominant transacting peptides in higher plants to specifically target and inhibit protein functions in vivo. Furthermore, they indicate that homotypic interaction of tospoviral N proteins is essential for the viral life cycle and that the targeting of evolutionary conserved functions of viral proteins in planta can be used to engineer broad-spectrum virus resistance.
The approach presented in this paper targets specifically a well characterized function of a viral protein. In contrast, in most other cases plant disease resistance is obtained by expressing viral genes targeted against structures/functions completely unknown, and in a number of cases even a clear distinction between protein-dependent and RNA-dependent mechanisms is not described. An exception is the coat protein-mediated TMV resistance, which relies on altered interaction properties of the transgenically expressed coat protein interfering with the disassembly of TMV capsids (39). Interestingly, for TSWV, both types of resistance were reported to be induced by the same nucleocapsid transgene: one that requires the expression of the protein and gives low level but fairly broad-spectrum resistance, and the second that is RNA-mediated and provides high-level and highly specific resistance (8, 40).
Our experimental approach aimed at an unambiguous protein-mediated effect. A minimal stretch of 23 nucleotides identical to the target sequence has been shown to be required to trigger posttranscriptional gene silencing (41, 42). We therefore expressed the TSWV-derived peptide from an artificial nucleotide sequence with altered codon usage. This construct shares only 61% sequence identity with the TSWV N gene with a maximum of 14 successive identical nucleotides. Furthermore, the fact that GUS activity was measured from systemic leaves of TSWV-infected resistant plants (data not shown) was considered additional evidence for the absence of posttranscriptional gene silencing in our experiments.
Because peptide aptamers have properties similar to antibodies (18), the approach adopted resembles the engineering of virus resistance by expressing single-chain antibodies directed against viral proteins (43, 44). The aptamer strategy, however, offers additional possibilities, because the application of the antibody approach is limited by the difficulties associated with the expression of antibodies in the plant cytoplasm (45).
The straightforward functional selection of peptides with the yeast two-hybrid system allows to specifically select molecules interacting with essential parts of the target protein. Because such molecular interactions require precisely organized structures, they impose constraints on sequence evolution (46–48). Therefore, the specific targeting of interaction domains with dominant interfering agents should delay the occurrence of resistance-breaking virus strains, thus allowing to engineer long-lasting resistance phenotypes.
An additional advantage of using short sequences to engineer resistance is to minimize unpredictable or even deleterious effects observed in several cases after the expression of functional viral proteins (49, 50). Expression of only a short peptide, as described in this study, or artificial peptides that have been selected from random libraries not only minimizes the potential deleterious effect on the plant cell but also prevents other and possibly undesirable consequences. One such example concerns the evolution of new viruses by recombination with the transgene or by transcapsidation, possibilities frequently discussed in connection with transgenic pathogen-derived resistance, and which can virtually be excluded when an interfering but otherwise nonhomologous and nonfunctional molecule is expressed in the plant.
Peptides selected for a specific function (e.g., interaction) can be considered an extreme form of the dominant negative concept introduced by Herskowitz (51). Here, single functions of multifunctional/multidomain proteins are deleted to give rise to dominant negative proteins. In contrast, in the peptide approach, only one single functional domain is retained to interfere with the target functions. In this sense, our results demonstrate the applicability of such dominant negatively acting peptides targeted to specific protein functions in a higher plant. The finding supports the potential value of these concepts in virus-resistance applications.
Acknowledgments
We thank J. Schell and F. Salamini for constant generous support, J. Hackbusch for pLX222-Gateway, I. Cicerello for help in the greenhouse, and R. Kormelink for PSMV virus isolate.
Abbreviations
- CSNV
chrysanthemum stem necrosis virus
- dpi
days postinoculation
- GRSV
groundnut ring spot virus
- GUS
β-glucuronidase
- INSV
impatiens necrotic spot virus
- PSMV
physalis severe mottle virus
- TCSV
tomato chlorotic spot virus
- TSWV
tomato spotted wilt virus
- WSMV
watermelon silver mottle virus
References
- 1.Goldbach R, Peters D. In: The Bunyaviridae. Elliott R M, editor. New York: Plenum; 1996. pp. 129–157. [Google Scholar]
- 2.Sanford J C, Johnson S A. J Theor Biol. 1985;113:395–405. [Google Scholar]
- 3.Abel P P, Nelson R S, De B, Hoffmann N, Rogers S G, Fraley R T, Beachy R N. Science. 1986;232:738–743. doi: 10.1126/science.3457472. [DOI] [PubMed] [Google Scholar]
- 4.Lomonossoff G P. Annu Rev Phytopathol. 1995;33:323–343. doi: 10.1146/annurev.py.33.090195.001543. [DOI] [PubMed] [Google Scholar]
- 5.Beachy R N, Loesch-Fries S, Tumer N E. Annu Rev Phytopathol. 1990;28:451–474. [Google Scholar]
- 6.Prins M, Resende R O, Anker C, van Schepen A, de Haan P, Goldbach R. Mol Plant Microbe Interact. 1996;9:416–418. doi: 10.1094/mpmi-9-0416. [DOI] [PubMed] [Google Scholar]
- 7.van den Boogaart T, Wen F J, Davies J W, Lomonossoff G P. Mol Plant Microbe Interact. 2001;14:196–203. doi: 10.1094/MPMI.2001.14.2.196. [DOI] [PubMed] [Google Scholar]
- 8.Pang S Z, Slightom J L, Gonsalves D. Biotechnology. 1993;11:819–824. doi: 10.1038/nbt0793-819. [DOI] [PubMed] [Google Scholar]
- 9.Goregaoker S P, Eckhardt L G, Culver J N. Virology. 2000;273:267–275. doi: 10.1006/viro.2000.0430. [DOI] [PubMed] [Google Scholar]
- 10.Ling K, Namba S, Gonsalves C, Slightom J L, Gonsalves D. Biotechnology. 1991;9:752–758. doi: 10.1038/nbt0891-752. [DOI] [PubMed] [Google Scholar]
- 11.Beck D L, Van Dolleweerd C J, Lough T J, Balmori E, Voot D M, Andersen M T, O'Brien I E, Forster R L. Proc Natl Acad Sci USA. 1994;91:10310–10314. doi: 10.1073/pnas.91.22.10310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tacke E, Salamini F, Rohde W. Nat Biotechnol. 1996;14:1597–1601. doi: 10.1038/nbt1196-1597. [DOI] [PubMed] [Google Scholar]
- 13.Cooper B, Lapidot M, Heick J A, Dodds J A, Beachy R N. Virology. 1995;206:307–313. doi: 10.1016/s0042-6822(95)80046-8. [DOI] [PubMed] [Google Scholar]
- 14.Callaway A, Giesman-Cookmeyer D, Gillock E T, Sit T L, Lommel S A. Annu Rev Phytopathol. 2001;39:419–460. doi: 10.1146/annurev.phyto.39.1.419. [DOI] [PubMed] [Google Scholar]
- 15.Uhrig J F, Soellick T R, Minke C J, Philipp C, Kellmann J W, Schreier P H. Proc Natl Acad Sci USA. 1999;96:55–60. doi: 10.1073/pnas.96.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richmond K E, Chenault K, Sherwood J L, German T L. Virology. 1998;248:6–11. doi: 10.1006/viro.1998.9223. [DOI] [PubMed] [Google Scholar]
- 17.Soellick T, Uhrig J F, Bucher G L, Kellmann J W, Schreier P H. Proc Natl Acad Sci USA. 2000;97:2373–2378. doi: 10.1073/pnas.030548397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colas P, Cohen B, Jessen T, Grishina I, McCoy J, Brent R. Nature. 1996;380:548–550. doi: 10.1038/380548a0. [DOI] [PubMed] [Google Scholar]
- 19.Kolonin M G, Finley R L., Jr Proc Natl Acad Sci USA. 1998;95:14266–14271. doi: 10.1073/pnas.95.24.14266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jefferson R A, Burgess S M, Hirsh D. Proc Natl Acad Sci USA. 1986;83:8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gietz R D, Schiestl R H. Methods Mol Cell Biol. 1995;5:255–269. [Google Scholar]
- 22.Bartel P L, Fields S. Methods Enzymol. 1995;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- 23.Landsmann J, Llewellyn D, Dennis E S, Peacock W J. Mol Gen Genet. 1988;214:68–73. doi: 10.1007/BF00340181. [DOI] [PubMed] [Google Scholar]
- 24.Töpfer R, Matzeit V, Gronenborn B, Schell J, Steinbiss H. Nucleic Acids Res. 1987;15:5890. doi: 10.1093/nar/15.14.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoekema A, Hirsch P R, Hooykaas P J J, Schilperoort R A. Nature. 1983;303:179–180. [Google Scholar]
- 26.Benvenuto E, Ordas R J, Tavazza R, Ancora G, Biocca S, Cattaneo A, Galeffi P. Plant Mol Biol. 1991;17:865–874. doi: 10.1007/BF00037067. [DOI] [PubMed] [Google Scholar]
- 27.Morita M, Asami K, Tanji Y, Unno H. Biotechnol Prog. 2001;17:573–576. doi: 10.1021/bp010018t. [DOI] [PubMed] [Google Scholar]
- 28.Cortez I, Saaijer J, Wongjkaew K S, Pereira A M, Goldbach R, Peters D, Kormelink R. Arch Virol. 2001;146:265–278. doi: 10.1007/s007050170174. [DOI] [PubMed] [Google Scholar]
- 29.Silva M S, Martins C R, Bezerra I C, Nagata T, de Avila A C, Resende R O. Arch Virol. 2001;146:1267–1281. doi: 10.1007/s007050170090. [DOI] [PubMed] [Google Scholar]
- 30.Kamb A, Caponigro G. Curr Opin Chem Biol. 2001;5:74–77. doi: 10.1016/s1367-5931(00)00160-5. [DOI] [PubMed] [Google Scholar]
- 31.Norman T C, Smith D L, Sorger P K, Drees B L, O'Rourke S M, Hughes T R, Roberts C J, Friend S H, Fields S, Murray A W. Science. 1999;285:591–595. doi: 10.1126/science.285.5427.591. [DOI] [PubMed] [Google Scholar]
- 32.Butz K, Denk C, Ullmann A, Scheffner M, Hoppe-Seyler F. Proc Natl Acad Sci USA. 2000;97:6693–6697. doi: 10.1073/pnas.110538897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blum J H, Dove S L, Hochschild A, Mekalanos J J. Proc Natl Acad Sci USA. 2000;97:2241–2246. doi: 10.1073/pnas.040573397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tao J, Wendler P, Connelly G, Lim A, Zhang J, King M, Li T, Silverman J A, Schimmel P R, Tally F P. Proc Natl Acad Sci USA. 2000;97:783–786. doi: 10.1073/pnas.97.2.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolonin M G, Finley R L., Jr Dev Biol. 2000;227:661–672. doi: 10.1006/dbio.2000.9916. [DOI] [PubMed] [Google Scholar]
- 36.Phizicky E M, Fields S. Microbiol Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colas P, Brent R. Trends Biotechnol. 1998;16:355–363. doi: 10.1016/s0167-7799(98)01225-6. [DOI] [PubMed] [Google Scholar]
- 38.Colas P. Curr Opin Chem Biol. 2000;4:54–59. doi: 10.1016/s1367-5931(99)00051-4. [DOI] [PubMed] [Google Scholar]
- 39.Bendahmane M, Fitchen J H, Zhang G, Beachy R N. J Virol. 1997;71:7942–7950. doi: 10.1128/jvi.71.10.7942-7950.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaira A M, Semeria L, Crespi S, Lisa V, Allavena A, Accotto G P. Mol Plant Microbe Interact. 1995;8:66–73. doi: 10.1094/mpmi-8-0066. [DOI] [PubMed] [Google Scholar]
- 41.Thomas C L, Jones L, Baulcombe D C, Maule A J. Plant J. 2001;25:417–425. doi: 10.1046/j.1365-313x.2001.00976.x. [DOI] [PubMed] [Google Scholar]
- 42.Sijen T, Wellink J, Hiriart J B, Van Kammen A. Plant Cell. 1996;8:2277–2294. doi: 10.1105/tpc.8.12.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tavladoraki P, Benvenuto E, Trinca S, De Martinis D, Cattaneo A, Galeffi P. Nature. 1993;366:469–472. doi: 10.1038/366469a0. [DOI] [PubMed] [Google Scholar]
- 44.Zimmermann S, Schillberg S, Liao Y C, Fischer R. Mol Breeding. 1998;4:369–379. [Google Scholar]
- 45.Cattaneo A, Biocca S. Trends Biotechnol. 1999;17:115–121. doi: 10.1016/s0167-7799(98)01268-2. [DOI] [PubMed] [Google Scholar]
- 46.Zuckerkandl E. J Mol Evol. 1976;7:167–183. doi: 10.1007/BF01731487. [DOI] [PubMed] [Google Scholar]
- 47.Dickerson R E. J Mol Evol. 1971;1:26–45. doi: 10.1007/BF01659392. [DOI] [PubMed] [Google Scholar]
- 48.Fraser H B, Hirsh A E, Steinmetz L M, Scharfe C, Feldman M W. Science. 2002;296:750–752. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- 49.Herbers K, Tacke E, Hazirezaei M, Krause K P, Melzer M, Rohde W, Sonnewald U. Plant J. 1997;12:1045–1056. doi: 10.1046/j.1365-313x.1997.12051045.x. [DOI] [PubMed] [Google Scholar]
- 50.Prins M, Storms M M, Kormelink R, de Haan P, Goldbach R. Transgenic Res. 1997;6:245–251. [Google Scholar]
- 51.Herskowitz I. Nature. 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]