Abstract
Compartmentalization of the cAMP-dependent protein kinase (PKA) is coordinated through association with A-kinase anchoring proteins (AKAPs). A defining characteristic of most AKAPs is a 14- to 18-aa sequence that binds to the regulatory subunits (RI or RII) of the kinase. Cellular delivery of peptides to these regions disrupts PKA anchoring and has been used to delineate a physiological role for AKAPs in the facilitation of certain cAMP-responsive events. Here, we describe a bioinformatic approach that yields an RII-selective peptide, called AKAP-in silico (AKAP-IS), that binds RII with a Kd of 0.4 nM and binds RI with a Kd of 277 nM. AKAP-IS associates with the type II PKA holoenzyme inside cells and displaces the kinase from natural anchoring sites. Electrophysiological recordings indicate that perfusion of AKAP-IS evokes a more rapid and complete attenuation of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor currents than previously described anchoring inhibitor peptides. Thus, computer-based and peptide array screening approaches have generated a reagent that binds PKA with higher affinity than previously described AKAPs.
The intracellular transduction of signals from the plasma membrane to cellular compartments evokes a variety of physiological responses. Perhaps the most rigorously studied signaling pathway utilizes the ubiquitous second messenger cAMP (1). Engagement of heptahelical receptors and the recruitment of intermediary G proteins activate adenylyl cyclases on the inner face of the plasma membrane (2–4). This molecular chain of events triggers an increase of cAMP concentration in certain intracellular compartments, where it activates molecules such as cyclic nucleotide-gated ion channels, guanine nucleotide exchange factors, cyclic nucleotide phosphodiesterases, and cAMP-dependent protein kinases (PKAs). These cAMP-responsive enzymes propagate disparate intracellular events, including the excitation of olfactory neurons, control of certain mitogen-activated protein kinase cascades, and a plethora of phosphorylation events catalyzed by PKA (5–8).
PKA is the predominant intracellular receptor for cAMP. In its dormant form, the PKA holoenzyme consists of two catalytic (C) subunits held in an inactive conformation by a regulatory (R) subunit dimer (9). Multiple C subunits (Cα, Cβ, and Cγ) and R subunits (RIα, RIβ, RIIα, and RIIβ) have been identified (10). Binding of cAMP to the R subunits causes the dissociation of the C subunits and the concomitant phosphorylation of target substrates within the vicinity of the kinase. Several regulatory mechanisms control the spatial and temporal activation of PKA. Elegant fluorescent imaging techniques have detected intracellular gradients and nanocompartments of cAMP, formed by the opposing actions of adenylyl cyclases and phosphodiesterases (11–13). These local fluctuations in cAMP influence where and when the kinase becomes active. Furthermore, spatial restriction of PKA is achieved through association with A-kinase anchoring proteins (AKAPs). AKAPs represent a group of functionally related proteins, classified by their ability to interact with PKA inside cells (14). Early on, most AKAPs were identified by a solid-phase overlay procedure and were thought to interact exclusively with RII (15–19). More recently, two-hybrid screening and affinity purification techniques have identified dual-function anchoring proteins that can interact with RI or RII (20–22). In a few instances, RI-selective AKAPs have been reported (23–25).
A defining characteristic of most AKAPs is a 14- to 18-aa sequence that binds to the R subunit dimer (18, 26–28). Peptides encompassing this region are effective antagonists of PKA anchoring inside cells and are routinely used to demonstrate a role for AKAPs in the coordination of cAMP-responsive events (29–35). Structural studies on two such AKAP peptides indicate that this region folds to form an amphipathic helix that slots into a binding pocket formed by the amino-terminal regions of each RII protomer (36, 37). Nonetheless, individual AKAPs bind RII with dissociation constants (Kd) ranging from 2 to 90 nM, which reflects the diversity of sequences that form these PKA-anchoring regions (18, 38–40). Therefore, we initiated a comprehensive analysis of multiple AKAP sequences in an attempt to define a consensus PKA-anchoring motif. Through a combination of bioinformatics, screening of peptide arrays, and the RII overlay procedure, we have designed a high-affinity RII-selective binding peptide that we have named AKAP-in silico (AKAP-IS).
Experimental Procedures
Autospot Peptide Array.
Peptide arrays were synthesized on cellulose paper by using an Autospot Robot ASP222 (ABiMED, Langenfeld, Germany) as described (41).
RII Overlay.
Proteins were separated by SDS/PAGE and electrotransferred to a nitrocellulose. RII overlays were conducted as described, using 32P-labeled recombinant murine RIIα (42).
meme Software.
MEME software was used for consensus sequence generation (http://meme.sdsc.edu) (43). MEME setting included one motif per sequence, and a motif length of 17 was specified.
Fluorescence Polarization.
FITC-labeled peptides (Cell Essentials, Boston) that were used for fluorescence polarization studies included AKAP-IS (AMAQIEYLAKQIVDNAIQQAKA), scrambled peptide (AMAQDVEIQLKAAYNQKLIAIA), and Ht31 (AADLIEEAASRIVDAVIEQVKA). Peptides (1 nM for RI experiments and 0.1 nM for RII experiments) were suspended to working dilutions in phosphate-buffered saline containing 5 μg/μl BSA, pH 7.0. Increasing concentrations of recombinant bovine RIα or recombinant murine RIIα were added to a PBS solution and mixed with each FITC-labeled peptide. Each sample was incubated for 10 min. Fluorescence polarization was measured on a Beacon 2000 (Panvera, Madison, WI), following the manufacturer's instructions. Saturation binding curves were generated with prism graphing software (GraphPad, San Diego). Dissociation constants (Kd) were calculated from the nonlinear regression curve from averages of three individual experiments.
Molecular Modeling of Peptides.
Predictions of the intrinsic α-helical content of the Ht31 and AKAP-IS peptides were performed with the predictive algorithm agadir (44). The solution structure of the Ht31-RIIα was used as a template for comparative modeling of the structure of the AKAP-IS–RIIα complex (37). The structural model was refined by using the swiss-model and swiss-pdb viewer (Glaxo Wellcome) programs.
Recombinant DNA Constructs.
Oligonucleotides with the following sequences were directionally cloned into the pcDNA3.1 V5/His TOPO vector (CLONTECH). The GFP coding region was cloned into a plasmid containing AKAP-IS and Scramble sequences by using HindIII and KpnI sites. All chimeric cDNA constructs were verified by sequencing. IS (+), 5′-CACCATGGCACAAATCGAATACTTAGCAAAACAAATCGTAGACAACGCAATCCAACAAGCAAAAGCA-3′; IS (−), 5′-TGCTTTTGCTTGTTGGATTGCGTTGTCTACGATTTGT- TTTGCTAAGTATTCGATTTGTGCCATGGTG-3′; Scramble (+), 5′-CACCATGGCACAAGACGTAGAAATCCAACTCAAAGCAGCATACAACCAAAAATTAATCGCAAT- CGCA-3′; and Scramble (−), 5′-TGCGATTGCGATTAATTTTTGGTTGTATGCTGCTTTGAGTTGGATTTCTACGTCTTGTGCCAGGGTG-3′.
Coimmunoprecipitation and PKA Activity Assay.
Cells at 50–80% confluency were transfected by using FuGENE (Roche Biochemicals). Five micrograms of plasmid DNA (GFP-IS-V5His, GFP-Scramble-V5His, GFP-Ht31, or GFP-Ht31PP) was transfected into HEK-293 cells. Cells were lysed 24 h later in 20 mM Hepes, pH 7.5/150 mM NaCl/1 mM EDTA/1% Triton X-100. AKAP-IS or the scrambled peptide was immunoprecipitated with an anti-V5 monoclonal antibody (Invitrogen). Coprecipitation of RIIα, RIIβ, RI, and C subunit were detected with isoform-specific polyclonal antibodies (Santa Cruz Biotechnology). PKA kinase assays were performed by the filter paper assay (45). The protein kinase inhibitor (PKI) residues 5–24 peptide [PKI-(5–24)] was used as a specific inhibitor of the kinase (46).
Confocal Microscopy.
Cells were seeded on glass coverslips and incubated overnight at 37°C, 5% CO2/95% air atmosphere. AKAP-IS or Scrambled was detected by intrinsic GFP fluorescence. Immunocytochemistry was performed as described (47).
Electrophysiology.
HEK293 cells were maintained in DMEM with 10% FBS. Cells were transfected by Fugene (Roche Biochemicals) with 1 μg of plasmids, which contains GluR1flip in pRK5 vector and pEGFP-N1 vector at a ratio of 5:1. One day after transfection, cells were replated onto 35-mm dishes at low density. Recordings were made at room temperature 2 days after transfection. GFP-positive cells were visually selected for recording by the fluorescence. Whole-cell recordings were made with an Axopatch200B amplifier (Axon Instruments, Foster City, CA). Patch pipettes (2–4 MΩ) contained (in mM) 140 cesium methanesulfonate, 5 adenosine triphosphate, 5 MgCl2, 0.2 CaCl2, 1 BAPTA [1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate], and 10 Hepes, pH 7.4. Extracellular solution contained (in mM) 150 NaCl, 5 KCl, 1.8 CaCl2, 10 glucose, 0.1 cyclothiazide, and 10 Hepes, pH 7.4. Solution exchanges were accomplished through a two-barrel pipe controlled by a solution stimulus delivery device, SF-77B (Warner Instruments, Hamden, CT). GluR1 receptor currents were evoked by a 500-ms application of 1 mM glutamate at 30-s intervals. Data were acquired and analyzed by using pclamp software (Axon Instruments). Currents were digitized at 5 kHz and filtered at 1 kHz.
Results
Design of a Consensus RII Binding Sequence.
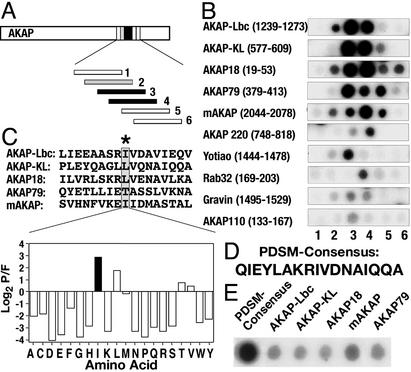
Solid-phase peptide arrays of overlapping 20-mer peptides (offset every three amino acids) that encompassed the PKA-anchoring regions of 10 AKAPs were screened by the RII overlay assay (Fig. 1A). Binding of 32P-radiolabeled RII was detected by autoradiography. The sequence and source of each peptide are included in Fig. 6, which is published as supporting information of the PNAS web site, www.pnas.org. Peptides from AKAP-Lbc, AKAP-KL, AKAP18, AKAP79, and mAKAP consistently bound RII with a higher apparent affinity than peptides from the other anchoring proteins (Fig. 1B). The most favorable RII-binding sequence from each high-affinity AKAP was aligned by using MEME software (Fig. 1C Upper). The MEME algorithm generated a position-dependent scoring matrix (PDSM) by systematically calculating the probability that a side chain would be found at a given position. For example, isoleucine was selected to occupy position 9 (Fig. 1C Lower), although leucine or threonine occupies the corresponding position in some high-affinity AKAPs (Fig. 1C Upper). The end result was a computer-generated PDSM consensus sequence of 17 amino acids (Fig. 1D) that exhibited a higher apparent RII-binding affinity than peptides from the five high-affinity AKAPs (Fig. 1E).
Figure 1.
Derivation of PDSM consensus sequence. (A) Schematic representation of how the RII-binding sequences within AKAPs were defined. Peptide arrays of 20-mer peptides (each offset by three residues) from 10 individual AKAPs were screened for RII binding by the overlay procedure. (B) Autoradiographs of RII-binding peptides from 10 anchoring proteins. The name of each AKAP and segment of sequence analyzed is indicated. (C Upper) Alignment of the five highest-affinity RII-binding sequences by using the MEME software. An AKAP-specific position-dependent scoring matrix (PDSM) was calculated by the log (base 2) of the probability that an amino acid is found at a given position in the alignment divided by the frequency that this amino acid is found in the nonredundant protein database (P/F). (C Lower) Values derived for position 9 in the PDSM sequence represented as a graphical output. Isoleucine (black bar) is the highest-scoring amino acid. Amino acids are indicated by their single-letter codes. (D) The MEME-derived PDSM consensus sequence. (E) RII overlay autoradiograph of a peptide array containing the PDSM consensus sequence and five high-affinity AKAPs (indicated above each lane).
Optimization of the PDSM Sequence.
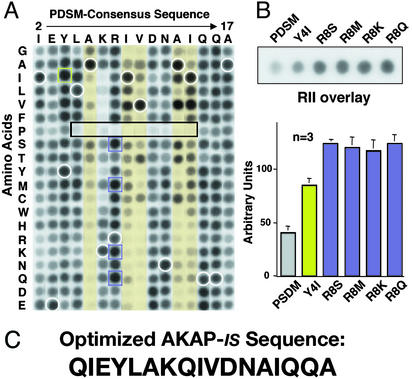
Optimization included screening a two-dimensional array of 320 peptides, where each residue between position 2 and 17 of the PDSM consensus sequence was systematically replaced with residues having every possible side chain (Fig. 2A). Solid-phase binding of 32P-labeled RII was assessed by the overlay assay and detected by autoradiography. The relative binding affinity of each modified peptide was compared with the signal strength of internal control peptides (the original PDSM consensus sequence) distributed throughout the array (Fig. 2A, white circles). Analysis of these data confirmed and extended our previous findings in three ways. First, substitution of polar side chains at positions 6, 9, 10, 13, and 14 generally decreased RII binding (Fig. 2A, yellow columns). This is consistent with our NMR data suggesting that nonpolar side chains at these positions form a hydrophobic face that contacts the RII dimer (37). Second, the substitution of proline anywhere within the core of the PDSM consensus sequence abolished RII binding (Fig. 2A, boxed region). Site-directed mutagenesis experiments on several AKAPs have shown that the introduction of helix-breaking amino acids into PKA-anchoring sites prevents RII binding (17, 26, 48). Third, substitutions at certain positions increased RII binding as evidenced by changing tyrosine-4 to isoleucine (Y4I) (Fig. 2A, green squares) or replacing arginine-8 with serine, methionine, lysine, or glutamine (Fig. 2A, purple squares). In independent experiments, the relative binding affinity of these peptide derivatives was measured by RII overlay (Fig. 2B Upper) followed by densitometry analysis of the autoradiographs (Fig. 2B Lower). The original PDSM-generated consensus sequence bound at a level of 43 ± 3 arbitrary units (n = 3), the Y4I substituted peptide was measured at 85 ± 8 (n = 3), and the arginine-8 substituted peptides ranged from 117 ± 8 (n = 3) for the R8K peptide to 124 ± 13 (n = 3) for the R8Q peptide. Therefore, we incorporated the neutral amino acid glutamine at position 8 (Fig. 2C). Dual substitutions at positions 4 and 8 did not increase the relative RII-binding affinity of the peptides further (data not shown). We named the optimized peptide AKAP-in silico (AKAP-IS) to acknowledge the computer-aided design of this reagent (Fig. 2C).
Figure 2.
Optimization of PDSM consensus sequence. (A) A two-dimensional array of 320 AKAP-IS peptide derivatives was generated where each residue between positions 2 and 17 in the PDSM sequence (above the array) was replaced with residues having every possible side chain (left of the array). Amino acids are indicated by their single-letter codes. Binding of 32P-labeled RII was detected by autoradiography. Peptide derivatives with substitutions at positions 6, 9, 10, 13, and 14 (yellow columns), proline substitutions with reduced RII binding (black rectangle), and internal control peptides of native sequence (white circles) are indicated. AKAP-IS derivatives with higher apparent RII-binding affinity (green and purple squares) are indicated. (B Upper) Solid-phase RII binding of the original PDSM consensus and the five peptides with higher affinity was quantified by densitometry of autoradiographs. Representative data from three individual experiments are presented. (B Lower) The relative binding affinity (arbitrary units) of each peptide is presented in graphic form. (C) The optimized AKAP-IS sequence.
In Vitro Characterization of the AKAP-IS Peptide.
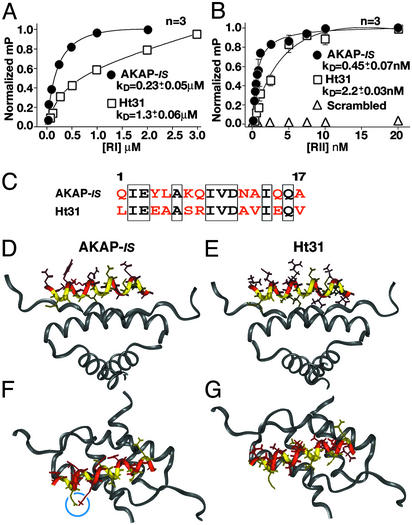
Dissociation constants (Kd) for the optimized AKAP-IS peptide were measured by fluorescence anisotropy by using both R subunit subtypes. The Kd for RIα was 227 ± 55 nM (n = 3), whereas RIIα bound more tightly with a Kd of 0.45 ± 0.07 nM (n = 3) (Fig. 3 A and B). A control peptide of identical amino acid composition but with a scrambled sequence did not interact with either R subunit type (Fig. 3B). Parallel experiments performed with the Ht31 peptide, which is derived from AKAP-Lbc, generated Kd values of 1,277 ± 56 nM (n = 3) for RIα and 2.2 ± 0.03 nM (n = 3) for RIIα (Fig. 3 A and B).
Figure 3.
Biochemical and structural analysis of AKAP-IS. Dissociation constants (Kd) of AKAP-IS (●) and Ht31 (□) peptides were determined by fluorescence polarization. Saturation binding curves were generated with increasing concentrations of RIα (A) or RIIα (B). Polarization values (mP) were determined at equilibrium and normalized to the highest value of saturation. Nonlinear regression analysis was used to derive Kd values from three independent experiments. Interaction of RII was not detected when a scrambled peptide (▵) of amino acid composition identical to that of the AKAP-IS sequence was used. (C) Alignment of the AKAP-IS and Ht31 sequences. Identical (boxed) and dissimilar (red) residues are indicated. (D–G) Molecular modeling of the AKAP-IS–RIIα complex used coordinates from the NMR structure of the Ht31/RIIα complex. The core peptide (yellow) and sites of divergence between AKAP-IS and Ht31 (red) are indicated. A side view reveals a change in RII contact side chains at positions 14 and 17 in AKAP-IS (D) compared with the Ht31 (E). Top view of AKAP-IS (F) reveals the formation of a salt bridge formed by residues E3 and K7 (blue circle) that is not found in Ht31 (G).
Although 8 of 17 residues are identical in the AKAP-IS and Ht31 sequences (Fig. 3C), modeling studies show that the majority of the changes are located on the noncontacting polar face of the amphipathic helix (Fig. 3 C and D). Two factors predict that AKAP-IS is a more helical peptide than Ht31. Alanine residues at positions 13 and 17 in AKAP-IS are predicted to increase the helicity of the peptide (Fig. 3 D and E), and a salt bridge formed between Glu-3 and Lys-7 stabilizes the overall conformation of the peptide (Fig. 3 F and G). These data suggest why AKAP-IS binds to either R subunit more tightly than does Ht31.
Cell-Based Analysis of AKAP-IS.
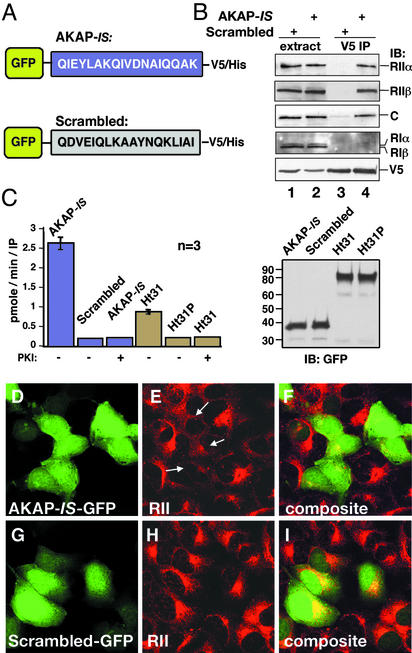
Expression constructs encoding AKAP-IS or the scrambled sequence fused to a V5/His epitope-tagged GFP were transfected into HEK293 cells (Fig. 4A). Three cell-based approaches were used to characterize the AKAP-IS interaction with the PKA holoenzyme. First, immune complexes were isolated with a monoclonal antibody against the V5 epitope, and the coprecipitation of PKA subunits was assessed by immunoblotting using specific antibodies (Fig. 4B). Both type II R subunits (RIIα and RIIβ) and the catalytic (C) subunit were detected in the AKAP-IS-GFP immunoprecipitates (Fig. 4B, top three blots, lane 4) but not found in immune complexes with the Scrambled-GFP fusion protein (Fig. 4B, lane 3). Neither type I R subunit (RIα or RIβ) copurified with AKAP-IS (Fig. 4B, fourth blot, lane 4), which probably reflects their lower binding affinities. Equivalent amounts of AKAP-IS-GFP or the scrambled peptide-GFP fusion proteins were present in the immune complexes as assessed by immunoblotting using V5 monoclonal antibodies (Fig. 4B Bottom). Coomassie blue stains of these samples detected only proteins that migrated with the same mobilities as the R and C subunits of the PKA holoenzyme (data not shown).
Figure 4.
AKAP-IS interacts with PKA inside cells. (A) Schematic representation of GFP fusion proteins with the AKAP-IS (Upper) and scrambled (Lower) peptides used in the cell-based studies. Sequences are given by the single-letter amino acid codes. (B) HEK293 cells were transfected with either construct, and immune complexes were immunoprecipitated from cell extracts (extract) with a V5 antibody (V5 IP). Copurification of PKA holoenzyme subunits was detected by immunoblotting (IB) using antibodies against the RIIα (top blot), RIIβ (second blot), C subunit (third blot), or RI subunits (fourth blot). The GFP fusion proteins were detected by immunoblotting using the V5 antibody (bottom blot). (C) The specific activity (pmol/min per IP) of PKA C subunit coprecipitating with chimeric AKAP fusion proteins (indicated above each column) was measured by a filter paper binding assay using the Kemptide as a substrate. PKA activity was blocked when PKI-(5–24) peptide (10 μM) was added to the reaction mixture. The accumulated data from three independent experiments are shown (Left). Immunoblot shows that equal amounts of the GFP fusion proteins were used in these experiments (Right). (D–I) Cells transiently transfected with plasmids expressing AKAP-IS or the scrambled peptide for 24 h were fixed, and immunocytochemical techniques were used to detect intrinsic GFP fluorescence (green; D and G). The subcellular location of RII (red; E and H) was detected with a monoclonal anti-RII antibody and Texas red-conjugated secondary antibody. Arrows indicate the mislocalization of RII from the Golgi/centrosomal area in AKAP-IS-expressing cells. Composite images (F and I) are presented.
Second, PKA activity coprecipitating with AKAP-IS-GFP or an Ht31-GFP fragment was measured by in vitro kinase assay using Kemptide as a substrate (Fig. 4C). PKA activity was 2.6-fold higher in the AKAP-IS-GFP fraction than in the Ht31-GFP fraction (Fig. 4C Right), although equivalent levels of the fusion proteins were used (Fig. 4C Left). Kinase activity was not copurified with the scrambled peptide or an Ht31P-GFP control, which is unable to anchor PKA (Fig. 4C Right). Control experiments confirmed that all PKA activity was blocked by the PKI-(5–24) inhibitor peptide (Fig. 4C Right).
Third, immunofluorescence techniques demonstrated that overexpression of AKAP-IS-GFP (Fig. 4D) displaced endogenous RII from anchoring sites at the perinuclear regions and Golgi complex of HEK293 cells (Fig. 4 E and F). In contrast, expression of the Scrambled-GFP construct (Fig. 4H) had no effect on the subcellular distribution of RII (Fig. 4 H and I). Similar results were obtained when these experiments were performed in COS7 cells (data not shown). These approaches demonstrate that AKAP-IS is a high-affinity ligand for the type II PKA holoenzyme inside cells.
Functional Analysis of AKAP-IS.
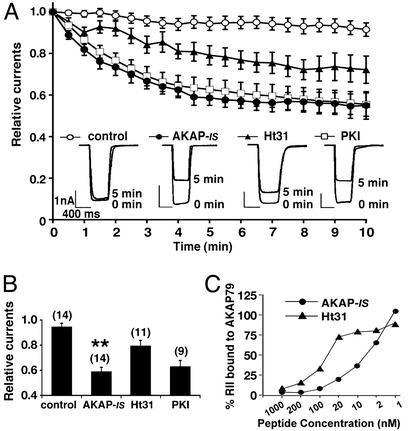
A physiologically relevant model of PKA anchoring is the time-dependent down-regulation (rundown) of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)-responsive currents in hippocampal neurons upon intracellular dialysis of the Ht31 peptide (29). This system was reconstituted in HEK293 cells by coexpressing the neuronal anchoring protein AKAP79 and the AMPA channel subunit GluR1. Whole-cell patch-clamp techniques provided a sensitive means to record GluR1 currents upon the delivery of bioactive peptides through the patch pipette (Fig. 5A). Control recordings demonstrated that GluR1 currents were stable over a 10-min time course (Fig. 5A, ○, and B). Perfusion of AKAP-IS (1 μM) evoked a pronounced and rapid reduction (41.4 ± 3.6%; n = 14) in GluR1 currents that was almost complete within 5 min (Fig. 5A, ●, and B). In contrast, perfusion of the Ht31 peptide (1 μM) evoked a less robust (20.8 ± 4.6%; n = 11) and slower response (Fig. 5A, ▴, and B). Interestingly, the magnitude and time course of the AKAP-IS response was similar to the effect of PKI-(5–24) peptide (10 μM), a potent and specific inhibitor of the kinase (Fig. 5A, □, and B). Collectively, these functional data suggest that AKAP-IS is a potent cell-based antagonist of AKAP79/PKA interaction. Additional support for this notion was provided by in vitro binding assays showing that AKAP-IS (IC50 = 8 nM, n = 5) was more effective at displacing RII from interaction with AKAP79 than was the Ht31 peptide (IC50 = 55 nM, n = 5; Fig. 5C).
Figure 5.
AKAP-IS is a potent antagonist of PKA anchoring. (A) Whole-cell patch-clamp recording techniques were used to measure the effects of AKAP-IS (1 μM, ●), Ht31 (1 μM, ▴), or PKI (10 μM, □) peptides on the time-dependent rundown of GluR1 receptor currents expressed in HEK293 cells. The accumulated data from 9–14 experiments including control currents (○) are presented. (Inset) Representative current trace from 0 and 5 min. Amplitude and time scale bars are presented. (B) Graphical representation of the peak current amplitudes upon glutamate stimulation 5 min after delivery of the peptides (indicated below each column). Each bar is normalized to the peak amplitude found at time 0. Amalgamated data from a number of experiments (indicated above each column) are shown. Asterisks indicate significant difference between AKAP-IS and Ht31 (**, P < 0.01). (C) In vitro competition assays. Recombinant AKAP79 (100 ng) immobilized to glutathione-Sepharose was loaded with recombinant 32P-labeled RII. Decreasing concentrations of AKAP-IS (●) or Ht31 (▴) peptide were added. After washing, the remaining RII was detected by autoradiography. RII binding (% bound) and peptide concentrations (nM) are indicated.
Discussion
Bioinformatics is a powerful methodology in biomedical research. Computational biology approaches are routinely used to search genomes for conserved structural motifs and to chart the evolutionary history of gene superfamilies (49). The MEME software used in this study was originally designed to identify distantly related protein sequences from a user-defined substitution-scoring matrix. This is an ideal system to analyze PKA-anchoring sites on AKAPs, which represent structurally related regions of low sequence identity on otherwise unrelated proteins. Consequently, we have combined this innovative bioinformatic approach with analysis of peptide arrays and a solid-phase RII binding assay to design a potent antagonist of PKA anchoring. Although the latter two techniques allowed us to quickly pare down our initial pool of 60 candidate peptides to five high-affinity AKAP sequences, the most critical element in the design strategy was analysis by the MEME program. Not only did the consensus sequence it derived bind RII more tightly, but the subsequent analysis of 320 derivatives during the optimization procedure prompted only a single substitution in the final AKAP-IS peptide. Thus, our product is a reagent that binds RII with significantly higher affinity than naturally occurring AKAP peptides.
Ht31 is the prototypic AKAP sequence that has been to used to study PKA anchoring for the past decade (26). This 24-residue peptide is derived from human anchoring protein AKAP-Lbc and binds RII with a Kd of 2–4 nM (18, 50). Our biochemical studies show that AKAP-IS binds RII with a 5-fold higher affinity (Kd = 0.4 nM), and our cell-based experiments confirm that it is a more effective inhibitor of PKA anchoring in vivo. Two observations emphasize this latter point: (i) an AKAP-IS-GFP fusion protein more effectively isolated PKA activity from cells than an Ht31 derivative, and (ii) the intracellular dialysis of the AKAP-IS peptide promoted a more rapid and complete attenuation of GluR1 currents than Ht31. In fact, our electrophysiological data indicate that the rundown in GluR1 channels upon AKAP-IS-mediated disruption of PKA anchoring occurs with a rate and magnitude similar to complete inhibition of the kinase by PKI-(5–24). Our evidence that AKAP-IS is as effective as PKI has very important functional implications. First, it suggests that disruption of PKA proximity to its substrates prevents their efficient phosphorylation to the same extent as blocking kinase activity. Second, the time course of the AKAP-IS effect is rapid and mostly complete within 5 min, which is more rapid than the Ht31 peptide. Thus, AKAP-IS has clear advantages over the currently available anchoring inhibitor peptides and should prove to be a superior reagent for disrupting anchored PKA pools in cells, tissues, and possibly whole animals.
In an effort to understand the higher affinity of AKAP-IS for PKA anchoring, we performed modeling studies of the AKAP-IS–RII interaction that used coordinates from the Ht31/RIIα structure as a starting point. The results suggest that most aliphatic side chains that contact the RII protomer are retained in both peptides (37). Interesting exceptions to this observation are the alteration of the RII contact residues Ala-13 and Ala-17 (Fig. 3C). Surprisingly, these changes remove hydrophobic contacts to the RII protomer from the peptide surface and, a priori, would have been predicted to destabilize the complex. However, alanine is the most favored residue for helix formation and, when combined with the introduction of a lysine at position 7 on the solvent-exposed surface of the peptide, serves to increase the predicted helical content of the unbound peptide. This likely involves the formation of an intramolecular salt bridge between Glu-3 and Lys-7 in AKAP-IS (Fig. 3F). However, a helical structure, in and of itself, does not provide sufficient determinants for high-affinity RII interaction, as specific surface contacts must be maintained to achieve binding. Indeed, a structure that is too rigid might hinder binding because of steric clashes with the RII surface. Thus, interplay between structural dynamics and the favorable energetics associated with the presence of specific surface contacts must be considered. The kinetic structural mechanism for AKAP binding is presently unknown, but the following two scenarios exist: (i) the peptide binds to RII then folds into a helical structure (binding-induced folding) or (ii) the peptide folds into an α-helix and then binds to RII (conformational regulation of binding). The former hypothesis is less likely, as the loss of intermolecular hydrophobic interactions would serve to destabilize the complex (Fig. 3D). Alternatively, if the recognition surface of the AKAP is already assembled, then increasing the helical content of the peptide could shift the equilibrium toward a binding-competent form. In this case, the observed on-rate would be maximized under conditions where the helical form predominates. Indirect support for this notion is provided by our electrophysiology data showing that AKAP-IS is a more potent and rapid antagonist of PKA anchoring inside cells.
Although AKAPs were initially thought to interact only with the type II PKA holoenzyme, there is now ample evidence showing that many anchoring proteins also target the type I kinase (20, 24, 51). In fact, substitution of aliphatic side chains in the hydrophobic face of the Ht31 helix increases affinity for RI (52). However, the native RI binding affinities of AKAP79 and AKAP-Lbc are 500-fold less for RI than for RII. This difference suggests that these AKAPs will preferentially associate with the type II PKA inside cells when both kinase subtypes are available (40, 53). Likewise, AKAP-IS exhibits a 500-fold preference for RII over RI in vitro, and its interaction with RI cannot be detected inside cells. This isoform selectivity is not surprising, given that the five AKAP sequences used to generate the progenitor AKAP-IS consensus are recognized as high-affinity RII binding proteins (14). In contrast, anchoring proteins such as D-AKAP-1/sAKAP84/149 and D-AKAP-2 exhibit less selectivity for either R subunit and have been designated dual-function AKAPs (20, 21). Recent evidence suggests that a single nucleotide polymorphism that is identified in the aging population causes a valine to isoleucine mutation in the anchoring helix of D-AKAP-2. This mutation increases RI binding affinity 3-fold but has no effect on the RII/D-AKAP-2 interaction (54). In an accompanying paper, the same authors used peptide array technologies to generate a high-affinity binding peptide with a 100-fold preference for RIα (55). Thus, peptide antagonists are now available to test the hypothesis that isoform-selective kinase anchoring occurs in a cellular context.
Supplementary Material
Acknowledgments
We thank Robert Mouton for preparation of the peptides arrays, Dr. Kjetil Tasken for providing us with RI protein and helpful discussion of this work, and members of the Scott lab for critical evaluation of this manuscript. N.M.A. was supported by National Institutes of Health Grant GM48231, and J.D.S. and P.A.J. were supported in part by National Institutes of Health Grant DK54441.
Abbreviations
- PKA
protein kinase A
- AKAP
A-kinase anchoring protein
- AKAP-IS
AKAP-in silico
- PKI
protein kinase inhibitor
- PDSM
position-dependent scoring matrix
References
- 1.Sutherland E W. Science. 1972;171:401–408. doi: 10.1126/science.177.4047.401. [DOI] [PubMed] [Google Scholar]
- 2.Lefkowitz R J. J Biol Chem. 1998;273:18677–18680. doi: 10.1074/jbc.273.30.18677. [DOI] [PubMed] [Google Scholar]
- 3.Taussig R, Gilman A G. J Biol Chem. 1995;270:1–4. doi: 10.1074/jbc.270.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Smith F D, Scott J D. Curr Biol. 2002;12:R32–R40. doi: 10.1016/s0960-9822(01)00646-7. [DOI] [PubMed] [Google Scholar]
- 5.Kaupp U B, Seifert R. Physiol Rev. 2002;82:769–824. doi: 10.1152/physrev.00008.2002. [DOI] [PubMed] [Google Scholar]
- 6.de Rooij J, Zwartkruis F J, Verheijen M H, Cool R H, Nijman S M, Wittinghofer A, Bos J L. Nature. 1998;396:474–477. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 7.Shabb J B. Chem Rev. 2001;101:2381–2411. doi: 10.1021/cr000236l. [DOI] [PubMed] [Google Scholar]
- 8.Pearson G W, Cobb M H. J Biol Chem. 2002;277:48094–48098. doi: 10.1074/jbc.M208535200. [DOI] [PubMed] [Google Scholar]
- 9.Taylor S S, Buechler J A, Yonemoto W. Annu Rev Biochem. 1990;59:971–1005. doi: 10.1146/annurev.bi.59.070190.004543. [DOI] [PubMed] [Google Scholar]
- 10.Scott J D. Pharmacol Ther. 1991;50:123–145. doi: 10.1016/0163-7258(91)90075-w. [DOI] [PubMed] [Google Scholar]
- 11.Barsony J, Marks S J. Proc Natl Acad Sci USA. 1990;87:1188–1192. doi: 10.1073/pnas.87.3.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Ma Y, Taylor S S, Tsien R Y. Proc Natl Acad Sci USA. 2001;98:14997–15002. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaccolo M, Pozzan T. Science. 2002;295:1711–1715. doi: 10.1126/science.1069982. [DOI] [PubMed] [Google Scholar]
- 14.Colledge M, Scott J D. Trends Cell Biol. 1999;9:216–221. doi: 10.1016/s0962-8924(99)01558-5. [DOI] [PubMed] [Google Scholar]
- 15.Lohmann S M, DeCamili P, Enig I, Walter U. Proc Natl Acad Sci USA. 1984;81:6723–6727. doi: 10.1073/pnas.81.21.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bregman D B, Bhattacharyya N, Rubin C S. J Biol Chem. 1989;264:4648–4656. [PubMed] [Google Scholar]
- 17.Carr D W, Stofko-Hahn R E, Fraser I D C, Cone R D, Scott J D. J Biol Chem. 1992;267:16816–16823. [PubMed] [Google Scholar]
- 18.Carr D W, Hausken Z E, Fraser I D, Stofko-Hahn R E, Scott J D. J Biol Chem. 1992;267:13376–13382. [PubMed] [Google Scholar]
- 19.Carr D W, DeManno D A, Atwood A, Hunzicker-Dunn M, Scott J D. J Biol Chem. 1993;268:20729–20732. [PubMed] [Google Scholar]
- 20.Huang L J, Durick K, Weiner J A, Chun J, Taylor S S. J Biol Chem. 1997;272:8057–8064. doi: 10.1074/jbc.272.12.8057. [DOI] [PubMed] [Google Scholar]
- 21.Huang L J, Durick K, Weiner J A, Chun J, Taylor S S. Proc Natl Acad Sci USA. 1997;94:11184–11189. doi: 10.1073/pnas.94.21.11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reinton N, Collas P, Haugen T B, Skalhegg B S, Hansson V, Jahnsen T, Tasken K. Dev Biol. 2000;223:194–204. doi: 10.1006/dbio.2000.9725. [DOI] [PubMed] [Google Scholar]
- 23.Angelo R, Rubin C S. J Biol Chem. 1998;273:14633–14643. doi: 10.1074/jbc.273.23.14633. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Degenhardt B, Tobin D, Yao Z X, Tasken K, Papadopoulos V. Mol Endocrinol. 2001;15:2211–2228. doi: 10.1210/mend.15.12.0736. [DOI] [PubMed] [Google Scholar]
- 25.Kussel-Andermann P, El-Amraoui A, Safieddine S, Hardelin J P, Nouaille S, Camonis J, Petit C. J Biol Chem. 2000;275:29654–29659. doi: 10.1074/jbc.M004393200. [DOI] [PubMed] [Google Scholar]
- 26.Carr D W, Stofko-Hahn R E, Fraser I D C, Bishop S M, Acott T S, Brennan R G, Scott J D. J Biol Chem. 1991;266:14188–14192. [PubMed] [Google Scholar]
- 27.Hausken Z E, Dell'Acqua M L, Coghlan V M, Scott J D. J Biol Chem. 1996;271:29016–29022. doi: 10.1074/jbc.271.46.29016. [DOI] [PubMed] [Google Scholar]
- 28.Glantz S B, Li Y, Rubin C S. J Biol Chem. 1993;268:12796–12804. [PubMed] [Google Scholar]
- 29.Rosenmund C, Carr D W, Bergeson S E, Nilaver G, Scott J D, Westbrook G L. Nature. 1994;368:853–856. doi: 10.1038/368853a0. [DOI] [PubMed] [Google Scholar]
- 30.Lester L B, Langeberg L K, Scott J D. Proc Natl Acad Sci USA. 1997;94:14942–14947. doi: 10.1073/pnas.94.26.14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vijayaraghavan S, Goueli S A, Davey M P, Carr D W. J Biol Chem. 1997;272:4747–4752. doi: 10.1074/jbc.272.8.4747. [DOI] [PubMed] [Google Scholar]
- 32.Westphal R S, Tavalin S J, Lin J W, Alto N M, Fraser I D, Langeberg L K, Sheng M, Scott J D. Science. 1999;285:93–96. doi: 10.1126/science.285.5424.93. [DOI] [PubMed] [Google Scholar]
- 33.Klussmann E, Maric K, Wiesner B, Beyermann M, Rosenthal W. J Biol Chem. 1999;274:4934–4938. doi: 10.1074/jbc.274.8.4934. [DOI] [PubMed] [Google Scholar]
- 34.Fink M A, Zakhary D R, Mackey J A, Desnoyer R W, Apperson-Hansen C, Damron D S, Bond M. Circ Res. 2001;88:291–297. doi: 10.1161/01.res.88.3.291. [DOI] [PubMed] [Google Scholar]
- 35.Moita M A, Lamprecht R, Nader K, LeDoux J E. Nat Neurosci. 2002;5:837–838. doi: 10.1038/nn901. [DOI] [PubMed] [Google Scholar]
- 36.Newlon M G, Roy M, Morikis D, Hausken Z E, Coghlan V, Scott J D, Jennings P A. Nat Struct Biol. 1999;6:222–227. doi: 10.1038/6663. [DOI] [PubMed] [Google Scholar]
- 37.Newlon M G, Roy M, Morikis D, Carr D W, Westphal R, Scott J D, Jennings P A. EMBO J. 2001;20:1651–1662. doi: 10.1093/emboj/20.7.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nauert J B, Klauck T M, Langeberg L K, Scott J D. Curr Biol. 1997;7:52–62. doi: 10.1016/s0960-9822(06)00027-3. [DOI] [PubMed] [Google Scholar]
- 39.Feliciello A, Cardone L, Garbi C, Ginsberg M D, Varrone S, Rubin C S, Avvedimento E V, Gottesman M E. FEBS Lett. 1999;464:174–178. doi: 10.1016/s0014-5793(99)01585-9. [DOI] [PubMed] [Google Scholar]
- 40.Herberg F W, Maleszka A, Eide T, Vossebein L, Tasken K. J Mol Biol. 2000;298:329–339. doi: 10.1006/jmbi.2000.3662. [DOI] [PubMed] [Google Scholar]
- 41.Frank R. Tetrahedron. 1992;48:123–132. [Google Scholar]
- 42.Hausken Z E, Coghlan V M, Scott J D. In: Protein Targeting Protocols. Clegg R A, editor. Vol. 88. Totowa, NJ: Humana; 1998. pp. 47–64. [Google Scholar]
- 43.Grundy W N, Bailey T L, Elkan C P, Baker M E. Comput Appl Biosci. 1997;13:397–406. doi: 10.1093/bioinformatics/13.4.397. [DOI] [PubMed] [Google Scholar]
- 44.Fisinger S, Serrano L, Lacroix E. Protein Sci. 2001;10:809–818. doi: 10.1110/ps.34901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corbin J D, Reimann E M. Methods Enzymol. 1974;38:287–294. doi: 10.1016/0076-6879(74)38044-5. [DOI] [PubMed] [Google Scholar]
- 46.Scott J D, Glaccum M B, Fischer E H, Krebs E G. Proc Natl Acad Sci USA. 1986;83:1613–1616. doi: 10.1073/pnas.83.6.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Westphal R S, Soderling S H, Alto N M, Langeberg L K, Scott J D. EMBO J. 2000;19:4589–4600. doi: 10.1093/emboj/19.17.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alto N M, Soderling J, Scott J D. J Cell Biol. 2002;158:659–668. doi: 10.1083/jcb.200204081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eisenberg D, Marcotte E M, Xenarios I, Yeates T O. Nature. 2000;405:823–826. doi: 10.1038/35015694. [DOI] [PubMed] [Google Scholar]
- 50.Diviani D, Soderling J, Scott J D. J Biol Chem. 2001;276:44247–44257. doi: 10.1074/jbc.M106629200. [DOI] [PubMed] [Google Scholar]
- 51.Miki K, Eddy E M. J Biol Chem. 1998;273:34384–34390. doi: 10.1074/jbc.273.51.34384. [DOI] [PubMed] [Google Scholar]
- 52.Miki K, Eddy E M. J Biol Chem. 1999;274:29057–29062. doi: 10.1074/jbc.274.41.29057. [DOI] [PubMed] [Google Scholar]
- 53.Burton K A, Johnson B D, Hausken Z E, Westenbroek R E, Idzerda R L, Scheuer T, Scott J D, Catterall W A, McKnight G S. Proc Natl Acad Sci USA. 1997;94:11067–11072. doi: 10.1073/pnas.94.20.11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kammerer S, Burns-Hamuro L L, Ma Y, Hamon S C, Cànaves J M, Shi M M, Nelson M R, Sing C F, Cantor C R, Taylor S S, Braun A. Proc Natl Acad Sci USA. 2003;100:4066–4071. doi: 10.1073/pnas.2628028100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burns-Hamuro L L, Ma Y, Kammerer S, Reineke U, Self C, Cook C, Olson G L, Cantor C R, Braun A, Taylor S S. Proc Natl Acad Sci USA. 2003;100:4072–4077. doi: 10.1073/pnas.2628038100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.