Abstract
In previous studies, we demonstrated the sufficiency of short nucleotide sequences to facilitate internal initiation of translation in mammalian cells. By using a selection methodology, we have now identified comparable sequences in Saccharomyces cerevisiae. For these studies, a library of constructs expressing dicistronic mRNAs with the HIS3 gene as the second cistron and 18 random nucleotides in the intercistronic region was introduced into a yeast strain in which the endogenous HIS3 gene was deleted. Untransformed cells or those containing the parent construct failed to grow on medium lacking histidine. Intercistronic sequences recovered from cells that did grow were evaluated by using various criteria. Fifty-six of the 18-nt sequences (≈1/400,000) functioned as synthetic internal ribosome entry sites (IRESes). The 14 most active sequences allowed growth in the presence of 0.1–0.6 mM 3-amino-1,2,4-triazole, a competitive inhibitor of the HIS3 gene product. In addition, eight sequences were identified that were not IRESes, but that enhanced HIS3 expression by an alternative mechanism that depended on the 5′ end of the mRNA and appeared to involve either shunting or reinitiation. Comparisons among the 56 selected IRESes identified eight significant sequence matches containing up to 10 nucleotides. Many of the selected sequences also contained extensive complementary matches to yeast 18S rRNA, some at overlapping sites. The identification of cis sequences that facilitate translation initiation in yeast enables detailed biochemical and genetic analyses of underlying mechanisms and may have practical applications for bioengineering.
The translation of some eukaryotic mRNAs is initiated in a cap-independent manner at internal ribosome entry sites (IRESes) contained within those mRNAs. IRESes were first discovered in uncapped RNAs from poliovirus (1) and encephalomyocaritis virus (2), and have subsequently been identified in other viral as well as cellular mRNAs from mammals, insects, and yeast (3, 4). IRESes appear to be used by some mRNAs to facilitate translation when initiation by the cap-dependent mechanism is less efficient or blocked, for example, during poliovirus infection (5), during the G2/M phase of the cell cycle (6, 7), and in dendrites (8). Internal initiation also appears to facilitate the translation of mRNAs with 5′ leaders that are encumbered by numerous upstream AUGs or RNA secondary structures (see refs. 9 and 10).
Naturally occurring and synthetic IRESes comprise a heterogeneous group of sequences that range in length from <10 to several hundred nucleotides and that appear to vary in their requirements for primary, secondary, and tertiary structures of RNA. For example, although secondary or tertiary structures are important for the activities of some viral IRESes (e.g., refs. 11 and 12), there is little evidence to suggest that comparable RNA structures occur within cellular IRESes or are required for their activities (see ref. 10). Rather, the data suggest that the activities of some cellular IRESes are determined by combined effects of numerous shorter elements of which they are comprised. For example, in various studies, discrete 5′ and 3′ boundaries could not be defined for particular IRESes, and, for some IRESes, activity was distributed between two or more nonoverlapping fragments (13–17). In an earlier study, we investigated the modular composition of a cellular IRES contained within the 5′ leader of the Gtx homeodomain mRNA (18), and more recently, of an IRES contained within the cold-stress-induced Rbm3 mRNA (unpublished data). For both 5′ leaders, several nonoverlapping fragments were found to function as IRESes when tested in isolation. Other evidence supporting the notion that short cis sequences can contribute to IRES activity came from a selection study in mammalian cells in which we identified two sequences of 9 and 15 nucleotides having IRES activity from libraries containing random nucleotide sequences (19).
Although most IRESes have been identified and characterized in mammalian cells or cell-free lysates, yeast appears to be ideally suited for the detailed analyses of IRES cis sequences. For example, it has been found that starving yeast cells die very rapidly if all translation is inhibited by using cycloheximide, but they survive for long periods of time if only cap-dependent translation is blocked (20). These results suggest that cells can survive by using a cap-independent mechanism of initiating translation. Other studies have identified IRESes within the yeast TFIID and HAP4 mRNAs, which were shown to function in yeast cell-free lysates (21), and in the YAP1 and p150 mRNAs, which we showed can function in vegetatively growing cells (22). The ability of yeast cells to initiate translation internally is also supported by the observations that RNA sequences from various organisms and sources, including the cricket paralysis virus, can function as IRESes in yeast (23–25).
The identification and analysis of cis sequences within IRESes is critical to understanding the mechanisms by which these sequences facilitate translation. However, their identification on an mRNA-by-mRNA basis is labor-intensive. In the present study, we developed a selection method to identify IRES elements more efficiently. We used a library of constructs expressing dicistronic mRNAs with the HIS3 gene as the second cistron and 18 random nucleotides (N18) in the intercistronic region. This library of constructs was introduced into a yeast strain in which the endogenous HIS3 gene was deleted, and the transformed cells were selected on medium lacking histidine. The constructs used for these studies were engineered to eliminate possible sources of false positives, and the selected sequences were subjected to a series of stringent tests to identify any that enhanced HIS3 expression by other mechanisms. Based on various criteria, 56 selected sequences were determined to function as IRESes. In addition, eight other sequences were shown to enhance HIS3 expression by a different mechanism that appeared to involve the 5′ end of the mRNA.
Methods
Construction of Dicistronic Libraries.
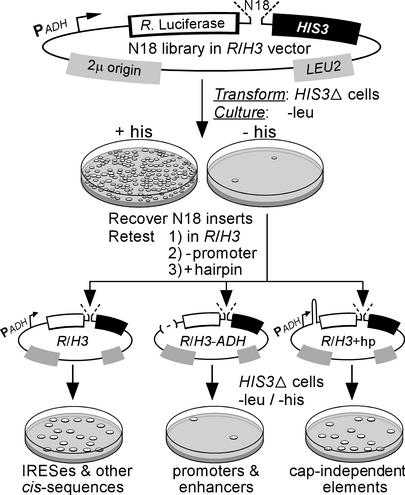
A library of constructs was generated in the R/H3 vector, which expresses a dicistronic mRNA with the coding regions of the Renilla luciferase and HIS3 genes as the first and second cistrons, respectively (see Fig. 1). An oligonucleotide containing 18 random nucleotides (N18) was cloned in the intercistronic region of these constructs. Dicistronic mRNA transcription was driven by the ADH promoter. The vector also contained the LEU2 gene to allow selection on medium lacking leucine, and the 2μ origin of replication to maintain the construct as a high copy episome. The ADH promoter was removed from the R/H3 vector by using the restriction endonucleases SphI and SacI to generate the R/H3-ADH vector. A hairpin structure with a predicted stability of −50 kcal/mol (14, 22) was introduced into the 5′ UTR of the dicistronic mRNA to generate the R/H3+hp vector.
Figure 1.
IRES selection strategy in yeast. A library of random 18-nt sequences was generated in the R/H3 parent vector. These constructs were used to transform HIS3Δ cells, which were cultured on leu−/his− medium. N18 inserts were recovered from selected cells and recloned into the parent R/H3 vector, the promoterless R/H3-ADH vector, or the hairpin-containing R/H3+hp vector (see text). These constructs were transformed into HIS3Δ cells and cultured on leu−/his− medium.
Transformation and Selection of Cells.
Constructs containing 18 random nucleotides in the intercistronic region were transformed into histidine auxotrophic strain no. 4741 (American Type Culture Collection) in which the endogenous HIS3 gene was deleted. We refer to this strain as HIS3Δ. Transformed cells were plated on medium containing glucose, but lacking leucine and histidine (Fig. 1). Renilla luciferase activities were determined as previously described from 0.3 OD600 units of exponentially growing cells (22).
Recovery and Analysis of Selected Sequences.
Cells selected on his− medium were recovered and the intercistronic sequences were isolated by PCR, cloned into the R/H3 vector, and retested for IRES activity. To identify non-IRES sequences that enhanced HIS3 expression by promoting the transcription of monocistronic HIS3 transcripts or by mechanisms that depend on the 5′ end of the mRNA, the recovered sequences were also cloned into the R/H3-ADH vector and into the R/H3+hp vector (Fig. 1). All of these constructs were individually transformed into the HIS3Δ strain and cultured on leu−/his− medium by using glucose as a carbon source. Total RNA was prepared from transformed cells and used for both Northern blot analyses (22) and RNase protection assays (using RPAIII kit; Ambion, Austin, TX).
Sequence analyses were performed by using the fasta program (ref. 26; Genetics Computer Group, version 10.2) to compare selected sequences to each other with the following parameters: −WOR = 1; −GAP = 30; −LEN = 6. Two of the adenosine residues that flank the N18 sequences were included for these comparisons. The significance of the sequence matches was evaluated by using the BESTFIT program (Genetics Computer Group, version 10.2). The individual sequences were compared with a sequence file containing the sequence match of interest embedded in 1,000 random nucleotides. The resulting BESTFIT quality scores were compared with those obtained after randomly shuffling the 1,000-nt sequence file 100-times. Calculation of significance matches did not include the flanking adenosines.
Results
To identify IRES elements, we developed the described selection method which uses constructs based on a Renilla/HIS3 (R/H3) parent vector that encode dicistronic mRNAs with Renilla luciferase and HIS3 as the first and second cistrons, respectively (Fig. 1). A library of constructs was generated by inserting oligonucleotides containing 18 random nucleotides into the intercistronic region of the R/H3 parent vector. Because the HIS3 cistron encodes IGP dehydratase, an enzyme required for histidine biosynthesis, oligonucleotide sequences that permit the translation of this cistron should enable a histidine auxotrophic yeast strain (HIS3Δ) to grow on medium lacking histidine. Although this selection method will identify N18 sequences with IRES activity, cells might also be selected because they grow by other mechanisms. For example, cell survival might result from mutations in the vector or in host cells that are independent of the intercistronic sequences. Alternatively, cell survival might result from intercistronic sequences that facilitate leaky scanning, reinitiation, shunting, or the production of monocistronic HIS3 transcripts via promoter or splicing activities. It was therefore of particular importance to take these possibilities into consideration in the design of the constructs and of the selection method.
IRES Selection Vector Designed to Minimize False Positives.
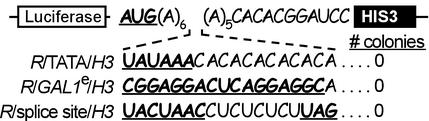
In yeast, transcription begins 40–100 nt downstream of a TATA box or equivalent promoter sequence (27). To minimize selection for transcription start sites, we placed the entire N18 sequence less than 40 nucleotides upstream of the HIS3 initiation codon. In this configuration, N18-sequences with promoter activity should generate truncated HIS3 transcripts that lack the HIS3 initiation codon and fail to produce functional IGP dehydratase enzyme. To test this aspect of the vector design, we introduced a TATA box promoter sequence (TATAAA) at the extreme 5′ end of the N18 sequence (Fig. 2, construct R/TATA/H3), and then transformed HIS3Δ cells with this construct. None of the transformed cells survived on medium lacking histidine, suggesting that transcripts produced by intercistronic promoter sequences would not be translated into a functional gene product.
Figure 2.
Control experiments to test N18 intercistronic sequences with potential transcriptional or splicing activities. Schematic representations of dicistronic mRNAs containing promoter (TATA), enhancer, (GAL1e), and splice site sequences are indicated. These constructs were transformed into HIS3Δ cells and cultured on leu−/his− medium. As mentioned in the text, cells containing these constructs failed to form colonies.
Monocistronic HIS3 transcripts might also be generated by N18 sequences that enhance the activity of cryptic transcriptional promoters located 5′ of the N18 sequence. To block the translation of these monocistronic HIS3 transcripts, the nucleotide sequence AUG was introduced upstream of the N18 sequence (Fig. 2) to function as a decoy initiation codon. Ribosomes scanning such a monocistronic HIS3 mRNA should initiate translation at the upstream AUG and not at the downstream HIS3 cistron (28). Any ORF resulting from the upstream AUG would either overlap and be out-of-frame with the HIS3 cistron or will terminate within the N18 sequence. We tested this aspect of the vector design by using an 18-nt sequence containing a known enhancer sequence from the GAL1 promoter (Fig. 2, construct R/GAL1e/H3). A construct containing this enhancer was transformed into HIS3Δ cells, which were cultured on medium lacking histidine but containing galactose. None of the transformed cells survived under these culture conditions, suggesting that intercistronic transcriptional enhancers are not likely to be selected in this assay.
A third mechanism by which monocistronic HIS3 mRNAs might be generated is by a splicing mechanism that removes the Renilla luciferase sequence from the dicistronic mRNA. In the R/H3 vector, all potential 3′ splice donor sites (YAG) in the intercistronic region were removed. However, splicing might still be mediated by splice acceptor sites contained within N18 sequences if the appropriate splice donor and branch point sequences were present upstream. This possibility appears to be unlikely, as 5′ splice site/branch point sequences are highly conserved in S. cerevisiae (29, 30) and sequence analysis of the Renilla/HIS3 dicistronic mRNA failed to identify any potential combinations (31) upstream of the intercistronic region. Experimental evidence was also inconsistent with the possibility of splicing in these constructs. For example, the introduction of a perfect branch point/3′ splice site into the intercistronic region of the R/H3 mRNA (Fig. 2, construct R/splice site/H3) did not enable cells containing this construct to grow on medium lacking histidine. In addition, cells containing other dicistronic constructs, for example, with the β-globin 5′ UTR in the intercistronic region did not grow on medium lacking histidine despite the presence of potential 3′ splice acceptor sites (YAG).
Identification of 56 IRES Elements and Eight Sequences That Enhance Translation by an Alternative Mechanism.
To identify short IRES elements, the R/N18/H3 library was transformed into a HIS3Δ cell line and cells were selected on leu−/his− medium for up to 4 weeks. Under these same growth conditions, cells transformed with the parent R/H3 construct failed to survive; however, ≈0.004% of the cells transformed with the library constructs survived selection (Table 1). Intercistronic sequences were recovered from selected colonies by PCR. Four-hundred and twenty-seven PCR products were recovered from 1,017 colonies: 122 contained a single N18 sequence, whereas the other 305 contained multiple N18 sequences that were ligated to each other in the original cloning. The 122 single N18 sequences were recloned into the R/H3 vector and retested. Seventy-five of the 122 sequences survived the second round of selection, suggesting that the other 47 sequences were originally contained within cells that survived selection on the basis of genomic or vector mutations.
Table 1.
Summary of screening data
| Construct* | Growth condition | Colonies per μg of construct DNA | No. of single N18 inserts | Histidine prototrophic† | |
|---|---|---|---|---|---|
| Parent dicistronic | R/H3 | leu− | 5 × 105 | ||
| R/H3 | his−/leu− | 0 | |||
| First round of selection | |||||
| N18 library | R/N18/H3 | leu− | 4 × 105 | ||
| R/N18/H3 | his−/leu− | 15 (0.00375%) | 122/1,017 colonies‡ | ||
| Second round of selection and controls | |||||
| Selected N18-mers§ | R/N181–122/H3 | his−/leu− | 122 | 75 | |
| -ADH promoter | R/N181–75/H3-ADH | his−/leu− | 75 | 11 | |
| in hairpin vector | R/N181–64/H3 + hp | his−/leu− | 64 | 56 | |
All constructs were transformed into HIS3Δ strain #4741 (ATCC).
Number of N18 inserts that rendered HIS3Δ cells histidine prototrophic.
Number of single N18 inserts recovered by PCR; the total number of inserts recovered was 427.
Second round of screening using single N18 sequences recovered from selected cells and recloned into the R/H3 vector.
Although the R/H3 vector was specifically designed to minimize the number of false positives resulting from promoter or enhancer activity, the 75 selected sequences with single N18 sequences were further evaluated with these possibilities in mind. Selected sequences were tested in a vector in which the ADH promoter upstream of the R/H3 dicistronic mRNA was deleted (R/H3-ADH, Fig. 1). Cells transformed with the parent promoterless vector (−ADH promoter) did not express the R/H3 dicistronic mRNA or have any detectable Renilla luciferase activity (data not shown). Eleven of the 75 selected sequences drove expression of the HIS3 cistron in these constructs (Table 1), suggesting that these sequences might have facilitated the production of monocistronic HIS3 transcripts by transcriptional mechanisms.
The remaining 64 sequences recovered from selected cells were further tested to assess whether any of them enhanced HIS3 expression by mechanisms that were dependent on the 5′ end of the mRNA for recruitment of the translation machinery. Selected sequences were tested in a vector containing a stable stem-loop or hairpin (hp) structure in the 5′ UTR of the mRNA (Fig. 1, construct R/H3+hp). The stem-loop structure appeared to function as a physical obstruction that reduced translation of the Renilla luciferase cistron by >99%, presumably by blocking scanning ribosomes. For 56 of the 64 constructs, the presence of the stem-loop structure did not reduce HIS3 expression based on cell growth on his− medium (Table 1). Indeed, cell growth was enhanced slightly in all cases (data not shown). These results indicated that the activities of these 56 selected sequences did not require translation of the first cistron. However, for the other eight constructs translation of the HIS3 cistron depended on the 5′ end of the mRNA for ribosome recruitment (Table 1), as the presence of a stem-loop structure 5′ of the Renilla luciferase cistron blocked the translation of both cistrons. We will refer to these eight selected sequences as “cap-dependent” selected sequences. A list of these cap-dependent sequences can be found in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org.
The 14 most active selected sequences, based on cell growth on his− medium, were subjected to Northern analyses using RNA prepared from cells transformed with the parent R/H3 or experimental constructs. All showed multiple bands; similar results were obtained by using constructs containing the GAL1 promoter. These bands are possibly the consequence of premature termination events occurring within the HIS3 cistron; all bands hybridized to both Renilla luciferase and HIS3 cistron probes (data not shown). The possibility of premature termination events occurring within the second cistron is also consistent with the results obtained with a similar vector (pMyr-RP) that differed from the R/H3 vector only in the second cistron (Photinus luciferase vs. HIS3), but that produced single band on a Northern blot (22).
Quantification of HIS3 Gene Product.
Based on the criteria used in this study, 56 of the selected sequences (Table 1) appeared to function as IRESes. A complete list of the selected sequences can be found in Table 3, which is published as supporting information on the PNAS web site. Quantitatively, the activities of the synthetic IRES sequences appeared to vary based on the time required for transformed HIS3Δ host cells to form colonies under selective conditions. Colonies containing the most active constructs were formed within 5–7 days. Twenty-one constructs resulted in colony formation within the second week, 14 constructs resulted in colony formation within the third week, and seven constructs resulted in colony formation within the fourth week. The time required for transformed cells to form colonies when the selected individual intercistronic sequences were retested was approximately the same as it was in the first round of selection.
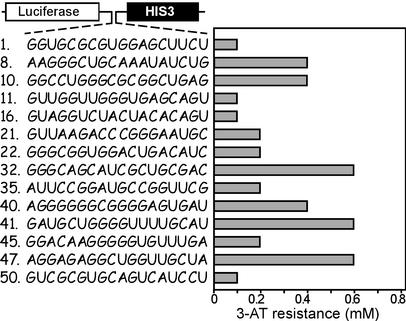
To obtain a relative measure of HIS3 gene expression, cells transformed with the 14 most active IRES elements were cultured on his− medium in the presence of different concentrations of 3-amino-1,2,4-triazole (3-AT), a competitive inhibitor that specifically blocks the activity of the HIS3 gene product without affecting the activity of other factors in yeast (see ref. 32). The results showed that cells transformed with the 14 most active constructs enhanced IGP dehydratase production to different extents that ranged in resistance from 0.1 to 0.6 mM 3-AT (Fig. 3). The relative activities of these IRES elements were determined by normalizing the 3-AT resistance to the activity of the first cistron (Renilla luciferase), which should reflect mRNA levels. With one exception, the Renilla luciferase activities obtained for all of the constructs varied <2-fold, and the normalized IRES activities were very similar to the 3-AT resistance (Fig. 6, which is published as supporting information on the PNAS web site).
Figure 3.
Quantification of IRES activity of the 14 most active N18 IRES sequences by using 3-AT to inhibit the activity of the HIS3 gene product. A schematic representation of the dicistronic mRNAs used in this analysis is indicated. The numbering of the N18 sequences is based on the order in which they were identified. Constructs were transformed into HIS3Δ cells and cultured for up to 2 weeks on leu−/his− medium in the presence of up to 0.8 mM 3-AT.
Sequence Analysis of Selected IRES Elements Identifies Potential Motifs.
Sequence comparisons among the selected N18 IRES sequences did not identify any sequence that occurred frequently, and that might therefore represent a potential IRES-motif. However, eight significant sequence matches of up to 10 nucleotides were identified in two or three different selected sequences (Fig. 4). In contrast, no significant sequence matches were identified when the cap-dependent N18 sequences were compared with each other.
Figure 4.
Significant sequence matches contained within N18 IRES sequences. The numbers to the left of the sequence matches indicate the N18 sequences in which the matches are contained. Stretches of sequence similarity are indicated by the shaded nucleotides. Although calculations of significance did not consider matches to adenosine nucleotides that flank the N18 sequences, these flanking nucleotides (indicated in italics) are indicated in cases where they generate a more extensive sequence match. The probability of similar matches occurring by chance is <0.05 (*) or <0.01 (**) as indicated.
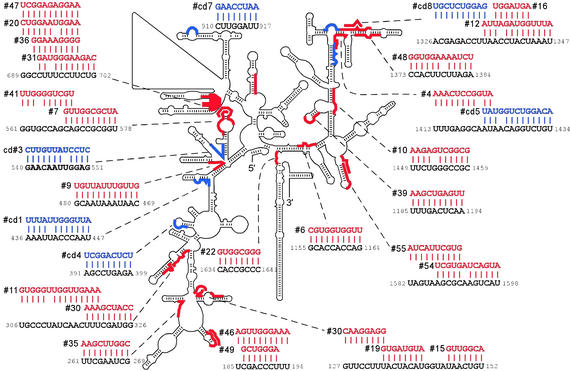
We have hypothesized (33) that some IRESes recruit the translation machinery by directly binding to 40S ribosomal subunits and that base pairing to 18S rRNA is one mechanism by which this may occur. We therefore compared the selected N18 sequences to those contained within yeast 18S rRNA and identified several extensive complementary matches of up to 12 consecutive nucleotides (Fig. 5). For both the IRESes and cap-dependent sequences, many of the complementary matches are to relatively unstructured regions of the 18S rRNA. Moreover, several of the complementary matches are in close proximity to each other and appear to be clustered at particular sites of the 18S rRNA.
Figure 5.
Complementarity of selected IRES and cap-dependent sequences to 18S rRNA. Both the schematic secondary structure of the S. cerevisiae 18S rRNA (www.rna.icmb.utexas.edu) and the individual 18S rRNA sequences (U53879) are indicated in black. Sites of complementarity with selected IRES and cap-dependent (cd) sequences are indicated in red and blue, respectively. The numbers flanking the 18S rRNA sequences indicate their location within the full-length rRNA. Potential base pairing interactions are indicated by the longer vertical bars; GU base pairs are indicated by the shorter vertical bars.
Discussion
Numerous studies indicate that particular cellular IRESes are composed of shorter elements that can function independently when tested in isolation. An increased understanding of how IRESes facilitate translation initiation requires elucidation of these cis sequences. To expedite the identification of IRES elements in yeast, we developed a selection method. These studies used dicistronic mRNAs with a selectable marker (HIS3) as the second cistron and an 18-nt random sequence in the intercistronic region. Extensive control experiments were performed to minimize the isolation of non-IRES elements, and indicated that the identification of such sequences was unlikely. Moreover, our selection method enabled us to identify non-IRES elements. Although, as based on various criteria, 56 of the selected sequences were confirmed to function as IRESes, eight others enhanced translation by a mechanism that depended on the 5′ end of the mRNA. Sequence analysis of the selected IRES sequences identified several significant sequence matches among them. In addition, several of the IRES and cap-dependent sequences contained extensive complementary matches to yeast 18S rRNA.
In a previous study, we screened mammalian cells containing libraries of random nucleotide sequences to identify two synthetic IRES elements, of 9 and 15 nucleotides (19). In addition, Dasgupta et al. (34) identified two other (50-nt) sequences by using a method that differed primarily in how the constructs were introduced into cells. For both screens, cells were transfected with dicistronic constructs encoding two different fluorescent proteins. The dicistronic vectors were integrated into the cellular genomes and cells were sorted based on their fluorescence. The yeast method used here differs from those used in mammalian cells in the use of a selectable marker. In addition, dicistronic constructs were maintained in an episomal state in the yeast method, avoiding variations that might arise after their integration into different genomic contexts.
Thus far, the identification of common IRES motifs within naturally occurring mRNAs has been difficult, possibly reflecting the heterogeneity of the component cis sequences. Computer analysis of the 56 selected IRESes identified here revealed only eight significant matches; these may represent IRES motifs. A larger set of selected IRESes should increase the likelihood of identifying common elements. The acquisition and analysis of these sequences is the focus of ongoing studies. Additional studies will be required to identify the functional nucleotides within the selected IRES elements. If these functional nucleotides can be identified, any mRNAs containing them can be recovered and tested to determine whether the presence of such sequences are predictive of IRES activity.
The rate at which IRESes were recovered in this study was ≈1/480,000, which in this study might be affected by the specifics of our selection vector. For instance, the spacing between the IRES element and the HIS3 initiation codon might be critical for activity. Alternatively, particular IRES elements might be sterically masked, for example, by forming stem-loop structures with other RNA sequences contained within the N18 sequence or within the dicistronic mRNA.
One plausible explanation for the diversity of IRES sequences identified is that internal initiation of translation occurs by multiple mechanisms. The notion of mechanistic diversity is supported by various findings; for example, different IRESes vary greatly in their requirements for initiation factors or other trans-factors (see refs. 4 and 35). The extreme examples appear to be the hepatitis A virus IRES, which requires all of the same initiation factors as cap-dependent initiation, and the cricket paralysis virus IRES, which requires only 40S ribosomal subunits. In previous studies, we noted that both cellular and synthetic IRESes contain numerous short segments with complementarity to different regions of the 18S rRNA (9, 19, 22). In the case of the Gtx IRES, a fragment of the 5′ leader containing the 9-nt IRES module was able to base pair to its rRNA complement within 40S ribosomal subunits (36).
It is possible that internal initiation of translation is enhanced by any mechanism that increases the local concentration of 40S ribosomal subunits in the vicinity of the mRNA (33). We have previously suggested that direct binding between mRNA sequences and 40S ribosomal subunits might be the basis for such a mechanism and that complementary sequence matches to 18S rRNA appear to be a defining feature of many cellular IRESes (33, 37). The three IRES-modules identified in our earlier studies all contained stretches of 8–11 nt with complementarity to different regions of 18S rRNA (18, 19). In all three cases, the complementary nucleotides were shown to function as IRESes when tested in isolation. In the present study, a number of complementary matches to 18S rRNA were also identified. It may be significant that most of these matches were localized to relatively unstructured regions of the rRNA (see www.rna.icmb.utexas.edu). In addition, several of the matches were to overlapping segments of the 18S rRNA, or to segments in close physical proximity to each other, suggesting that these sites may represent accessible regions on the surface of the 40S ribosomal subunit. The notion that base pairing between mRNA cis sequences and 18S rRNA mediates IRES-activity or shunting can be directly tested by altering complementary sequences within either the mRNA or the yeast rRNA, and is the focus of ongoing studies.
In addition to IRESes, the yeast screen identified eight cap-dependent sequences that facilitated HIS3 gene expression in a manner that depended on the translation of the first cistron. This was shown by the finding that the presence of a hairpin structure in the 5′ UTR of the dicistronic mRNA blocked HIS3 expression. These results are consistent with the notion that the ribosomes that translated the HIS3 cistron were recruited by the 5′ cap structure and reached the HIS3 cistron by a leaky scanning, reinitiation, or shunting mechanism (see ref. 38). Leaky scanning is thought to occur when scanning ribosomes fail to recognize an initiation codon and initiate translation at a downstream AUG. Reinitiation occurs when ribosomes that have terminated translation remain associated with the mRNA and reinitiate translation at a downstream cistron. Shunting is a mechanism that enables scanning or translating ribosomes to bypass segments of the mRNA. The presence of 32 ORFs and the distance between the initiation codons of the Renilla luciferase and HIS3 cistrons do not appear to be consistent with leaky scanning or reinitiation as possible mechanisms. Shunting remains plausible, however. Shunting has been described for cauliflower mosaic virus and adenovirus RNAs (39, 40). In the case of adenovirus, segments with complementarity to 18S rRNA have been implicated in the shunting process (40). We suggest that the cap-dependent N18 sequences may interact with and facilitate the shunting of ribosomal subunits that are recruited at the cap. Shunting to the intercistronic region might also occur from within the 5′ UTR, or the first cistron.
The identification in yeast of large numbers of RNA cis sequences that facilitate internal initiation of translation or shunting provides reagents that will facilitate both biochemical and genetic analyses of underlying mechanisms of translation.
Supplementary Material
Acknowledgments
We thank Brian Head and Luke Burman for excellent technical assistance and Drs. Joseph Gally and Kathryn Crossin for critical reading of the manuscript. Funding was generously provided by the G. Harold and Leila Y. Mathers Charitable Foundation, National Institutes of Health Grant GM61725, National Science Foundation Grant MCB9982574 (to V.P.M.), and U.S. Public Health Service Grant NS39837 (to G.M.E.).
Abbreviations
- IRES
internal ribosome entry site
- 3-AT
3-amino-1,2,4-triazole
- hp
hairpin
References
- 1.Pelletier J, Sonenberg N. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 2.Jang S K, Krausslich H G, Nicklin M J, Duke G M, Palmenberg A C, Wimmer E. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hellen C U, Sarnow P. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 4.Bonnal S, Boutonnet C, Prado-Lourenco L, Vagner S. Nucleic Acids Res. 2003;31:427–428. doi: 10.1093/nar/gkg003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johannes G, Sarnow P. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pyronnet S, Pradayrol L, Sonenberg N. Mol Cell. 2000;5:607–616. doi: 10.1016/s1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- 7.Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S, Beyaert R. Mol Cell. 2000;5:597–605. doi: 10.1016/s1097-2765(00)80239-7. [DOI] [PubMed] [Google Scholar]
- 8.Pinkstaff J K, Chappell S A, Mauro V P, Edelman G M, Krushel L A. Proc Natl Acad Sci USA. 2001;98:2770–2775. doi: 10.1073/pnas.051623398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chappell S A, Owens G C, Mauro V P. J Biol Chem. 2001;276:36917–36922. doi: 10.1074/jbc.M106008200. [DOI] [PubMed] [Google Scholar]
- 10.Le Quesne J P, Stoneley M, Fraser G A, Willis A E. J Mol Biol. 2001;310:111–126. doi: 10.1006/jmbi.2001.4745. [DOI] [PubMed] [Google Scholar]
- 11.Fletcher S P, Jackson R J. J Virol. 2002;76:5024–5033. doi: 10.1128/JVI.76.10.5024-5033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kieft J S, Zhou K, Grech A, Jubin R, Doudna J A. Nat Struct Biol. 2002;9:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- 13.Yang Q, Sarnow P. Nucleic Acids Res. 1997;25:2800–2807. doi: 10.1093/nar/25.14.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoneley M, Paulin F E M, Le Quesne J P C, Chappell S A, Willis A E. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 15.Huez I, Creancier L, Audigier S, Gensac M-C, Prats A-C, Prats H. Mol Cell Biol. 1998;18:6178–6190. doi: 10.1128/mcb.18.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gan W, La Celle M, Rhoads R E. J Biol Chem. 1998;273:5006–5012. doi: 10.1074/jbc.273.9.5006. [DOI] [PubMed] [Google Scholar]
- 17.Bernstein J, Sella O, Le S-Y, Elroy-Stein O. J Biol Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- 18.Chappell S A, Edelman G M, Mauro V P. Proc Natl Acad Sci USA. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Owens G C, Chappell S A, Mauro V P, Edelman G M. Proc Natl Acad Sci USA. 2001;98:1471–1476. doi: 10.1073/pnas.98.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paz I, Choder M. J Bacteriol. 2001;183:4477–4483. doi: 10.1128/JB.183.15.4477-4483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iizuka N, Najita L, Franzusoff A, Sarnow P. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou W, Edelman G M, Mauro V P. Proc Natl Acad Sci USA. 2001;98:1531–1536. doi: 10.1073/pnas.98.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson S R, Gulyas K D, Sarnow P. Proc Natl Acad Sci USA. 2001;30:12972–12977. doi: 10.1073/pnas.241286698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paz I, Abramovitz L, Choder M. J Biol Chem. 1999;274:21741–21745. doi: 10.1074/jbc.274.31.21741. [DOI] [PubMed] [Google Scholar]
- 25.Dorokhov Y L, Skulachev M V, Ivanov P A, Zvereva S D, Tjulkina L G, Merits A, Gleba Y Y, Hohn T, Atabekov J G. Proc Natl Acad Sci USA. 2002;99:5301–5306. doi: 10.1073/pnas.082107599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearson W R, Lipman D J. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santangelo G M, Tornow J, McLaughlin C S, Moldave K. Mol Cell Biol. 1988;8:4217–4224. doi: 10.1128/mcb.8.10.4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pestova T V, Hellen C U. Trends Biochem Sci. 1999;24:85–87. doi: 10.1016/s0968-0004(99)01356-0. [DOI] [PubMed] [Google Scholar]
- 29.Lopez P J, Seraphin B. Nucleic Acids Res. 2000;28:85–86. doi: 10.1093/nar/28.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis C A, Grate L, Spingola M, Ares M., Jr Nucleic Acids Res. 2000;28:1700–1706. doi: 10.1093/nar/28.8.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spingola M, Grate L, Haussler D, Ares M., Jr RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horecka J, Sprague G F., Jr Methods Enzymol. 2000;326:107–119. doi: 10.1016/s0076-6879(00)26049-7. [DOI] [PubMed] [Google Scholar]
- 33.Mauro V P, Edelman G M. Proc Natl Acad Sci USA. 2002;99:12031–12036. doi: 10.1073/pnas.192442499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venkatesan A, Dasgupta A. Mol Cell Biol. 2001;21:2826–2837. doi: 10.1128/MCB.21.8.2826-2837.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pestova T V, Kolupaeva V G, Lomakin I B, Pilipenko E V, Shatsky I N, Agol V I, Hellen C U. Proc Natl Acad Sci USA. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu M C-Y, Tranque P, Edelman G M, Mauro V P. Proc Natl Acad Sci USA. 1999;96:1339–1344. doi: 10.1073/pnas.96.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mauro V P, Edelman G M. Proc Natl Acad Sci USA. 1997;94:422–427. doi: 10.1073/pnas.94.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson R J. In: Translational Control of Gene Expression. Sonenberg N, Hershey J W B, Mathews M B, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 2000. pp. 127–183. [Google Scholar]
- 39.Ryabova L A, Pooggin M M, Hohn T. Prog Nucleic Acid Res Mol Biol. 2002;72:1–39. doi: 10.1016/S0079-6603(02)72066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yueh A, Schneider R J. Genes Dev. 2000;14:414–421. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.