Abstract
We report studies testing the importance of Watson–Crick hydrogen bonding, base-pair geometry, and steric effects during DNA replication in living bacterial cells. Nonpolar DNA base shape mimics of thymine and adenine (abbreviated F and Q, respectively) were introduced into Escherichia coli by insertion into a phage genome followed by transfection of the vector into bacteria. Genetic assays showed that these two base mimics were bypassed with moderate to high efficiency in the cells and with very high efficiency under damage-response (SOS induction) conditions. Under both sets of conditions, the T-shape mimic (F) encoded genetic information in the bacteria as if it were thymine, directing incorporation of adenine opposite it with high fidelity. Similarly, the A mimic (Q) directed incorporation of thymine opposite itself with high fidelity. The data establish that Watson–Crick hydrogen bonding is not necessary for high-fidelity replication of a base pair in vivo. The results suggest that recognition of DNA base shape alone serves as the most powerful determinant of fidelity during transfer of genetic information in a living organism.
Keywords: replication bypass‖isostere‖difluorotoluene‖mutagenesis
It is essential that genetic information be transferred with high fidelity during replication. This fidelity has been historically attributed to the specificity of Watson–Crick hydrogen bonding in each new base pair. More recently, however, the importance of base-pair geometry and steric effects has also been considered (1–5). To date, experiments addressing these issues have been performed only in vitro with purified polymerases. Significantly, this has not been tested inside cells, where the environment is much more complex and several polymerases and repair enzymes are present. Indeed, it has been shown that the mutagenicity and replication-blocking power of a DNA lesion in vitro may not necessarily correlate well with intracellular experiments (6).
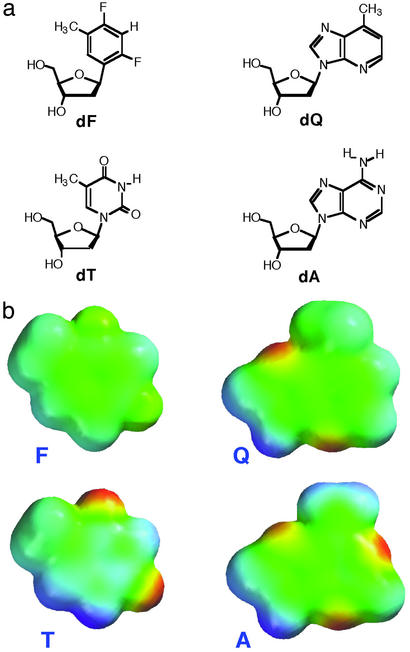
Nonpolar nucleoside isosteres such as those in this study act as shape mimics of the natural nucleosides (Fig. 1). For example, the dT mimic, difluorotoluene deoxynucleoside (dF), contains difluorotoluene as a replacement for the natural base thymine, and it is extremely close in size and shape to the natural molecule (7, 8). However, difluorotoluene is quite nonpolar and shows no evidence of hydrogen bonding even in low-polarity solvents (9). Similarly, the isostere of deoxyadenosine 9-(1-aza-4-methyl-benzimidazolyl)-1′-β-2′-deoxyriboside (dQ) (10), is also nonpolar and forms essentially no hydrogen bonds in water along its Watson–Crick edge. Although the analog dQ is a somewhat imperfect shape mimic for dA because of the presence of the proton that is missing at N1 of dA, the dF analog is a near-perfect mimic for dT. Both dF and dQ display very similar pairing abilities in DNA in the absence of a polymerase enzyme. Consistent with their nonpolar character, dF and dQ exhibit no selective pairing affinity for dA and dT (respectively) over the other natural bases when situated in a synthetic DNA duplex (9–11). This observation supports the idea that in DNA alone (in the absence of a replicating enzyme), Watson–Crick hydrogen bonding is a strong determinant of pairing selectivity, and shape differences have relatively little influence on this selectivity.
Figure 1.
(a) Structures of nucleoside shape mimics dF and dQ, which lack Watson–Crick hydrogen-bonding ability but possess shapes very similar to dT and dA (shown for comparison), respectively. (b) Space-filling models of nucleobase analogs F and Q with comparison to T and A, showing shapes with electrostatic potentials mapped on the surfaces (red, negative potential; blue, positive).
In marked contrast to this, studies with some purified DNA polymerases have shown that such analogues can have activity remarkably similar to their natural congeners. The analogues dF and dQ, for example, can encode genetic information with efficiency and fidelity approaching those of natural DNA bases by using purified enzymes such as the Klenow fragment of Escherichia coli DNA polymerase I or T7 DNA polymerase (5, 10, 11). This suggests that at least some enzymes are readily able to process base pairs lacking hydrogen bonds in vitro. Importantly, however, some polymerases [such as Pol β (an X-family enzyme) and Pol η (Y family)] appear not to process such molecules as efficiently (M. T. Washington, S.A.H., E.T.K., L. Prakash, and S. Prakash, unpublished data). The present studies were therefore aimed at testing hydrogen bonding and shape requirements in replication within the whole cellular context, with multiple polymerases and repair mechanisms operating.
Materials and Methods
Nucleoside analogs dF and dQ were synthesized and incorporated into oligonucleotides by using phosphoramidite solid-phase methods as described (10, 13). The oligonucleotides were ligated into an M13mp7(L2) single-stranded viral genome by using reported methods (14–16).
Genome construction and lesion-bypass experiments in wild-type AB1157 E. coli were performed by using oligodeoxynucleotides (5′-GCGAAGACCGXAGCGTCCG-3′, X is the base analog or the guanine control) that were ligated into a single-stranded bacteriophage M13mp7(L2) genome as described (16). Briefly, cells were grown in 100 ml of 2× yeast tryptone (YT) medium to an OD600 of 0.3, after which they were pelleted, resuspended in 50 ml of 0.1 M MgSO4, and split. To induce bypass polymerases (the SOS response), half of the cells were irradiated with 254-nm light at 45 Joules/m2 (UV Stratalinker 2400, Stratagene). Both the irradiated and uninduced cells were diluted 2-fold with 2× YT, grown with agitation at 37°C for 20 min (which allowed for the expression of SOS proteins for the UV-treated cells before genome transfection), and made chemically competent and plated by using 5 ng of DNA and ≈108 cells as described (16). The number of plaques formed for each lesion relative to that of the guanine control genome is indicated as “phage survival” (Fig. 2), with the guanine reference averaging ≈240 plaques. Mutational analysis was performed by using the restriction endonuclease and postlabeling (REAP) assay (15, 16).
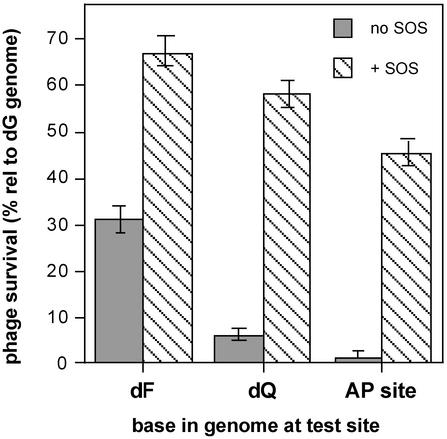
Figure 2.
Replication-bypass efficiencies in E. coli for phage templates containing the F and Q isosteres, with comparison to a chemically stable tetrahydrofuran abasic analog (AP) site. The solid bars show data for normal replication, and crosshatched bars show results under UV light-induced SOS damage-response conditions from individual genomes constructed in triplicate.
Results and Discussion
In an effort to explore the importance of nucleobase shape and hydrogen bonding within a cellular context, we tested a nonpolar analog from each of the pyrimidine and purine families of bases [the analogs F and Q (Fig. 1)]. In the current cellular study, initial experiments were directed to the question of translesion-bypass efficiency, which measures the ability of the replicative polymerase, in addition to the SOS polymerases, to incorporate a nucleotide encoded by the (natural or modified) base of interest. Also necessary is that the polymerase can synthesize additional nucleotides beyond the probe nucleotide, passing it through the active site where additional contacts are made. Finally, bypass efficiency also depends on the rate of addition of a nucleotide (opposite the probe base) relative to the rate at which it is removed by the 3′ proofreading function of the enzyme (17).
We measured the bypass efficiency and mutagenic potential of the F and Q isosteres by incorporating them into oligonucleotides using phosphoramidite solid-phase synthesis (10, 13), and the oligonucleotides then were ligated into an M13mp7(L2) single-stranded viral genome (14–16). These genomes then were passaged through E. coli as described (16) to score for the biological responses to the F and Q isosteres. A feature of this system is that there is no complementary strand opposite the probe bases, thus ensuring that signals for replication blocking and mutagenesis are derived solely from the initial translesion bypass of the adducts, which is also important in that a lesion-free strand of a DNA duplex genome may be preferentially replicated (18). Note that in these experiments one does not expect literal double-stranded replication of the isosteres, replacing (for example) dF by dF again, because the free dF 5′-triphosphate was not present in the cells. Thus, after synthesis of a strand complementary to the phage insert, a second round of synthesis would contain only natural nucleotides.
Translesion DNA Polymerase-Bypass Efficiency.
The ability of the F and Q isosteres to block DNA replication in vivo was addressed by using a common phage-survival assay (19–23). Briefly, genomes were constructed either bearing or lacking a probe isostere (lesion), and an equal amount of each genome was transfected into E. coli. The ability of an isostere to block DNA replication was scored as a percentage of the number of plaques formed from the isostere-bearing genome with respect to the genome containing an unmodified DNA insert. Presumably, if an isostere were a strong enough block to DNA replication, a region of the single-stranded genome that had not yet been converted to the duplex form by DNA polymerase(s) would persist longer than that of a lesion-free genome and thus be more susceptible to single-stranded endonucleases within the cell. This outcome would result in the formation of fewer phage plaques, because the plaque forming ability of linear single-stranded genomes is at least 5 orders of magnitude smaller than that of the analogous single-stranded circular DNA (24). In the present study we constructed a genome containing guanine as a control for both the lesion-bypass and mutagenesis studies, because it is unlike the two isosteres studied, allowing for a measure of the background of the assays used.
In E. coli that have not been induced for the SOS response to translesion DNA replication, we find that the pyrimidine isostere analog of thymidine, dF, was bypassed relatively efficiently (31% with respect to a guanine control genome; Fig. 2). The isosteric analog of deoxyadenosine, dQ was not as well bypassed (6% with respect to the unmodified control genome). However, this isostere exhibited better translesion synthesis than a tetrahydrofuran abasic site analog (lacking both steric fit as well as hydrogen-bonding features for a DNA helix), which gave only a 1% bypass efficiency in SOS-uninduced E. coli. This chemically stable abasic site was used as a positive control known to be a strong block to DNA replication in vitro (25) and in vivo (22).
A portion of E. coli was treated with UV light and grown for a brief period to express the SOS response to DNA damage before the introduction of the lesion-containing genomes. The bypass efficiencies of polymerases past F, Q, and the abasic site in these SOS-induced E. coli were high and of similar magnitude (respectively 67%, 58%, and 45% compared with the G control; Fig. 2). The fact that the isosteres and abasic site exhibited significant increases in lesion-bypass efficiency when compared with non-SOS-induced E. coli implies that SOS factors were active during translesion synthesis for each molecule (see below).
Fidelity of Information Transfer in Vivo.
Next we determined the identity of nucleotides inserted opposite base analogs F or Q by the replicative enzymes for both uninduced and SOS-induced E. coli. The recently developed REAP assay was used for determination of mutation frequency and for quantitative identification of the nucleotide(s) inserted opposite a given template base (15). Briefly, the method involves cutting a duplex form of DNA obtained from the progeny of genomes with a type IIs restriction endonuclease. This enzyme cleaves a fixed number of bases away from its recognition sequence, regardless of the intervening sequence. Consequently, the lesion to be studied is programmed such that the lesion site becomes the 5′ base after cleavage. The newly exposed site is then dephosphorylated, radiolabeled, size-fractionated, and ultimately digested to 5′ dNMPs, which are resolved and quantified by using TLC and PhosphorImager (Molecular Dynamics) analysis. Thus, the base composition at the lesion site (and hence the mutation frequency and specificity) is obtained.
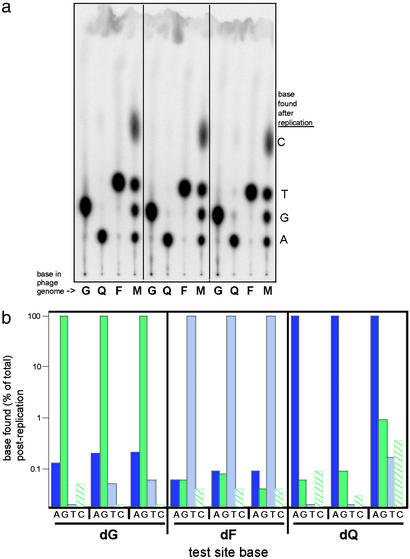
Analysis of the TLC chromatogram for the Q and F base isosteres in uninduced E. coli (Fig. 3a) revealed, remarkably, that the thymidine isostere (dF), as well as the deoxyadenosine isostere (dQ), each directed the insertion of the “complementary” deoxynucleoside triphosphate with very high fidelity. The lesion site that was dF before biological processing became dT via the quantitative, specific incorporation of dATP during translesion synthesis. Likewise, the site hosting the dQ analog was converted to dA, implying high-fidelity insertion of dTTP during translesion synthesis (Fig. 3b). Surprisingly, the average fidelities (over three experiments) for dQ and dF were similar to that of the natural nucleobase, guanine. Although no studies have been carried out in vitro with DNA Pol III, this in vivo finding correlates well with our earlier in vitro studies, which showed that DNA Pol I Klenow fragment (exo−) efficiently incorporates A opposite F (≈25% of the efficiency of A opposite T) with a fidelity similar to or slightly higher than the incorporation of A opposite a natural T in the template (4). Also, studies with analog Q in the template showed that the efficiency of dTTP insertion was significantly lower than that opposite A, but the selectivity for dTTP (over the other three nucleotides) was also relatively high (10).
Figure 3.
Mutagenesis data for nonpolar DNA base isosteres in E. coli under normal (SOS-uninduced) conditions. (a) Chromatogram from the REAP assay using genomes that were constructed in triplicate with oligonucleotides containing G, Q, or F at the type IIs (BbsI) cleavage site. These genomes were passaged through E. coli that were not induced for the SOS response. The lanes designated “M” contain markers generated from an oligonucleotide with a degenerate 5′ end that was carried through the assay at the 32P-labeling step. (b) Plots of mutagenesis data (from a) showing high fidelity of replication. A log10 plot was used to make visible all insertion events.
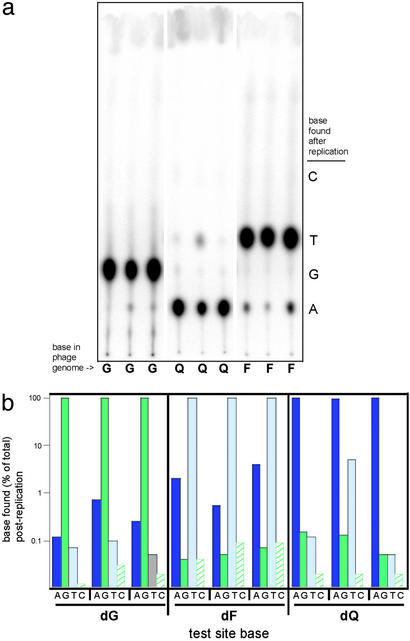
Mutagenesis data for the isosteres in SOS-induced E. coli are shown in Fig. 4a and graphically in Fig. 4b. As was the case for the uninduced cells, the isosteric base analogs appeared to be highly selective in their coding properties. dF instructed the insertion of primarily dATP, whereas the dQ isostere directed incorporation of dT. We do note, however, that 5% of dATP was inserted opposite the dQ isostere for one replicate, whereas 2% and 4% of dTTP was inserted opposite dF for two replicates. This lowered fidelity is consistent with the high error rates observed for eubacterial lesion-bypass polymerases (26).
Figure 4.
DNA base isostere mutagenesis data in SOS-induced E. coli. (a) Chromatogram from the REAP assay using G-, Q-, and F-containing genomes constructed in triplicate and passaged through E. coli that were induced for the SOS response with UV light. (b) Graphical representation of mutagenesis data from G, Q, and F genomes passaged through E. coli that were induced for the SOS response with UV light (from a). The log10 plot shows that the isosteres provide for direct coding when SOS replication proteins are expressed. The graph illustrates the near lack of ambiguous pairing of the isosteres with an incipient dNTP during DNA replication in vivo even with low-fidelity lesion-bypass polymerases.
We observed that although bypass efficiencies of nonpolar analogues dQ and dF were relatively efficient compared with the abasic control, they are significantly lower under normal replication conditions than the unmodified insert control. Analog dF shows a bypass efficiency of 31% that of guanine, and dQ shows a bypass efficiency of 6% that of guanine (Fig. 2). To account for this lowered efficiency, we hypothesize that bypass of dF may require an unfavorable minor groove interaction after the A–F pair is synthesized, an effect observed in vitro with E. coli Pol I Klenow fragment (27). Although this unfavorable interaction is not likely an issue for dQ (which has a minor groove H-bond acceptor), the added size of dQ relative to dA may cause a pause on insertion of dT or thereafter (28). Additional experiments with other analogs could test some of these possibilities.
Significantly, we note that with SOS induction, bypass of dF and dQ isosteres, as well as the tetrahydrofuran abasic site analog, becomes more efficient, and approaches that of the unmodified control (at 67%, 58%, and 45% of dG, respectively; Fig. 2). This result suggests that the SOS polymerases that aid in translesion DNA synthesis were actively replicating the isosteres in UV-irradiated E. coli. The SOS cellular response to DNA damage is a complex system involving many polymerases and accessory proteins, the mechanisms of which are not all well understood at present. E. coli possesses the SOS polymerases Pol II (synonymous with PolB and DinA), Pol IV (DinB), and Pol V (UmuD C), in addition to RecA, single-strand binding protein, and the β-sliding clamp and clamp-loading γ-complex from Pol III used during translesion synthesis. Expression of these Lex A-regulated polymerases is increased after cellular exposure of DNA-damaging agents such as UV light (29). Because the cells used in the present study were proficient in all of the above proteins, the polymerase(s) responsible for translesion bypass past the isosteres cannot be determined at present. Pol II allows for replication restart from damaged DNA almost immediately (30, 31) and may at first be an attractive candidate responsible for the SOS translesion bypass in our study; however, the cells in the present study were harvested 20 min after UV treatment. Cells lacking Pol II experience synthesis 50 min later, coincidental with maximal expression of UmuD′ (30–32). Consequently, the UmuD′-dependent Pol V complex should not be ruled out as the lesion-bypass polymerase for several reasons. In vitro gel-fidelity assays show that Pol V may be able to bypass some lesions dramatically better than Pol III or Pol IV (33). Furthermore, a round of DNA replication pausing at the lesion in the current study would leave a single-stranded region ≈500-nt long between the RNA-primed origin of replication and the lesion site; however, the optimal gap size for Pol II has been estimated to be <100 nt (34). The issue of finding the polymerase responsible may be moot, because it has also been shown that a direct competition by the bypass polymerases for a lesion may result in different mutagenic outcomes (35, 36). This may explain the small but measurable amount of transversion mutagenesis for independent SOS-induced isostere mutagenesis experiments. The use of E. coli deficient in combinations of the SOS-bypass polymerases may in the future aid in identification of the polymerase/complex responsible for the bypass of these analogs.
C), in addition to RecA, single-strand binding protein, and the β-sliding clamp and clamp-loading γ-complex from Pol III used during translesion synthesis. Expression of these Lex A-regulated polymerases is increased after cellular exposure of DNA-damaging agents such as UV light (29). Because the cells used in the present study were proficient in all of the above proteins, the polymerase(s) responsible for translesion bypass past the isosteres cannot be determined at present. Pol II allows for replication restart from damaged DNA almost immediately (30, 31) and may at first be an attractive candidate responsible for the SOS translesion bypass in our study; however, the cells in the present study were harvested 20 min after UV treatment. Cells lacking Pol II experience synthesis 50 min later, coincidental with maximal expression of UmuD′ (30–32). Consequently, the UmuD′-dependent Pol V complex should not be ruled out as the lesion-bypass polymerase for several reasons. In vitro gel-fidelity assays show that Pol V may be able to bypass some lesions dramatically better than Pol III or Pol IV (33). Furthermore, a round of DNA replication pausing at the lesion in the current study would leave a single-stranded region ≈500-nt long between the RNA-primed origin of replication and the lesion site; however, the optimal gap size for Pol II has been estimated to be <100 nt (34). The issue of finding the polymerase responsible may be moot, because it has also been shown that a direct competition by the bypass polymerases for a lesion may result in different mutagenic outcomes (35, 36). This may explain the small but measurable amount of transversion mutagenesis for independent SOS-induced isostere mutagenesis experiments. The use of E. coli deficient in combinations of the SOS-bypass polymerases may in the future aid in identification of the polymerase/complex responsible for the bypass of these analogs.
Possible Influence of Proofreading and Repair on Mutational Outcome in Vivo.
There is an outside chance that our observed mutations may not have been a result of the primary insertion event by the DNA polymerase, because an equal insertion of all dNTPs followed by selective DNA polymerase proofreading of three of the pairings might conceivably result in the same mutational outcome. However, we consider this to be unlikely, because we have shown previously that the Vmax/Km specificity constants for the efficiency of Klenow exonuclease proofreading of G, A, T, or C opposite a template F are identical (37).
Also worth considering is the influence of mismatch repair (MMR) mechanisms on this outcome. Because our cells were proficient in MMR, there is a possibility that what we observed as a mutation outcome may not have been from the direct insertion of the (preferred) base by the polymerase but rather from the selective repair of all but one (the observed) pairing by the MMR machinery. However, we have demonstrated previously that the binding affinities of MutS (the MMR recognition protein from E. coli) to DNA containing F opposite each of the four natural bases were essentially the same (38), suggesting a lack of selectivity in MMR recognition. Although these data strongly suggest that the selectivity we observed in vivo was due to the replicative enzymes, they do not prove so conclusively, because there are a number of other repair pathways in the cell (such as nucleotide excision repair, base excision repair, and yet-undiscovered repair systems) for which we do not have isostere repair data at present.
Origins of Fidelity in Bacterial Cells.
The REAP assay results establish that in E. coli, accurate DNA replication can occur without Watson–Crick hydrogen bonds. This is in marked contrast to the pairing selectivities of these DNA base mimics alone (in the absence of enzyme), which are virtually nil. It therefore is clear that, in vivo, the replicative enzyme places strong constraints on a base pair that the base otherwise would not have. We and others have proposed that this constraint is steric and geometric in nature (1–5).
The current data suggest that shape complementarity (3, 12) in the active site of the replicative polymerase may be the most important determinant of replication fidelity in vivo. During formation of a new phosphodiester bond, the active-site pocket for a replicative enzyme tightly surrounds the incipient base pair. The shape of the template base being addressed determines the shape of the complementary void that is to be filled by the incoming nucleotide partner. This tightly defined void can reject most incorrectly shaped nucleotides by steric clashing (2, 12). Conversely, the lesion-bypass polymerases are more error-prone and may function in part by providing a looser fit with the DNA (12), which consequently may explain the measurable “F-to-A transversion mutagenesis” seen in SOS-induced cells (Fig. 4) that was not observed in the absence of the SOS response (Fig. 3). The F–T pair may well be the smallest mismatch that could be made, consistent with a bypass polymerase dNTP selection process governed by steric constraints.
This simple mechanism alone cannot explain why small nucleotides (e.g., thymidine) are excluded from being paired with other small nucleobases such as T or C. We have hypothesized that this type of undesired mispairing is prevented also by steric clashing, not by the bases directly but by waters of solvation on their polar Watson–Crick pairing edges (3). This is supported further by the observation that the small F nucleotide, which lacks these tightly bound waters, is efficiently inserted opposite another template F in in vitro experiments with the Klenow fragment of E. coli Pol I (5). To test this hypothesis further it would be of interest in future experiments to examine such nonpolar–nonpolar pairing in vivo.
Acknowledgments
We thank Jacob Lai (Stanford University) for assistance with molecular modeling. This work was supported by National Institutes of Health Grants GM52956, CA80024, CA26731, and ES07020.
Abbreviations
- dF
difluorotoluene deoxynucleoside
- dQ
9-(1-aza-4-methyl-benzimidazolyl)-1′-β-2′-deoxyriboside
- Pol
DNA polymerase
- REAP
restriction endonuclease and postlabeling
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Goodman M F. Proc Natl Acad Sci USA. 1997;94:10493–10495. doi: 10.1073/pnas.94.20.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kunkel T A, Bebenek K. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 3.Kool E T. Biopolymers. 1998;48:3–17. doi: 10.1002/(SICI)1097-0282(1998)48:1<3::AID-BIP2>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 4.Moran S, Ren R X-F, Rumney S, IV, Kool E T. J Am Chem Soc. 1997;119:2056–2057. doi: 10.1021/ja963718g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moran S, Ren R X-F, Kool E T. Proc Natl Acad Sci USA. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McNees A G, O'Donnell M, Horton P H, Kim H Y, Kim S J, Harris C M, Harris T M, Lloyd R S. J Biol Chem. 1997;272:33211–33219. doi: 10.1074/jbc.272.52.33211. [DOI] [PubMed] [Google Scholar]
- 7.Schweitzer B A, Kool E T. J Org Chem. 1994;59:7238–7242. doi: 10.1021/jo00103a013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guckian K M, Kool E T. Angew Chem Int Ed Engl. 1998;36:2825–2828. doi: 10.1002/anie.199728251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schweitzer B A, Kool E T. J Am Chem Soc. 1995;117:1863–1872. doi: 10.1021/ja00112a001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morales J C, Kool E T. J Am Chem Soc. 1999;121:2323–2324. doi: 10.1021/ja983502+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kool E T, Morales J C, Guckian K M. Angew Chem Int Ed Engl. 2000;39:990–1009. doi: 10.1002/(sici)1521-3773(20000317)39:6<990::aid-anie990>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 12.Kool E T. Annu Rev Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 13.Ren R X-F, Chaudhuri N C, Paris P L, Rumney S, IV, Kool E T. J Am Chem Soc. 1996;118:7671–7678. doi: 10.1021/ja9612763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horsfall M J, Borden A, Lawrence C W. J Bacteriol. 1997;179:2835–2839. doi: 10.1128/jb.179.9.2835-2839.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delaney J C, Essigmann J M. Chem Biol. 1999;6:743–753. doi: 10.1016/s1074-5521(00)80021-6. [DOI] [PubMed] [Google Scholar]
- 16.Henderson P T, Delaney J C, Gu F, Tannenbaum S R, Essigmann J M. Biochemistry. 2002;41:914–921. doi: 10.1021/bi0156355. [DOI] [PubMed] [Google Scholar]
- 17.Goodman M F, Creighton S, Bloom L B, Petruska J. Crit Rev Biochem Mol Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- 18.Fink S P, Reddy G R, Marnett L J. Proc Natl Acad Sci USA. 1997;94:8652–8657. doi: 10.1073/pnas.94.16.8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawrence C W, Borden A, Banerjee S K, LeClerc J E. Nucleic Acids Res. 1990;18:2153–2157. doi: 10.1093/nar/18.8.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Latham G J, Zhou L, Harris C M, Harris T M, Lloyd R S. J Biol Chem. 1993;268:23427–23434. [PubMed] [Google Scholar]
- 21.Hanrahan C J, Bacolod M D, Vyas R R, Liu T, Geacintov N E, Loechler E L, Basu A K. Chem Res Toxicol. 1997;10:369–377. doi: 10.1021/tx9601925. [DOI] [PubMed] [Google Scholar]
- 22.Reuven N B, Tomer G, Livneh Z. Mol Cell. 1998;2:191–199. doi: 10.1016/s1097-2765(00)80129-x. [DOI] [PubMed] [Google Scholar]
- 23.Ramos L A, Sayer J M, Yagi H, Shah J H, Dipple A, Jerina D M. Chem Res Toxicol. 2001;14:1082–1089. doi: 10.1021/tx010076o. [DOI] [PubMed] [Google Scholar]
- 24.Yarema K J, Lippard S J, Essigmann J M. Nucleic Acids Res. 1995;23:4066–4072. doi: 10.1093/nar/23.20.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeshita M, Chang C N, Johnson F, Will S, Grollman A P. J Biol Chem. 1987;262:10171–10179. [PubMed] [Google Scholar]
- 26.Goodman M F. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 27.Morales J C, Kool E T. Biochemistry. 2000;39:12979–12988. doi: 10.1021/bi001578o. [DOI] [PubMed] [Google Scholar]
- 28.Morales J C, Kool E T. J Am Chem Soc. 2000;122:1001–1007. doi: 10.1021/ja993464+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 30.Pham P, Rangarajan S, Woodgate R, Goodman M F. Proc Natl Acad Sci USA. 2001;98:8350–8354. doi: 10.1073/pnas.111007198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rangarajan S, Woodgate R, Goodman M F. Proc Natl Acad Sci USA. 1999;96:9224–9229. doi: 10.1073/pnas.96.16.9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Opperman T, Murli S, Smith B T, Walker G C. Proc Natl Acad Sci USA. 1999;96:9218–9223. doi: 10.1073/pnas.96.16.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang M, Pham P, Shen X, Taylor J S, O'Donnell M, Woodgate R, Goodman M F. Nature. 2000;404:1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- 34.Kornberg A, Baker T A. DNA Replication. New York: Freeman; 1992. [Google Scholar]
- 35.Becherel O J, Fuchs R P. Proc Natl Acad Sci USA. 2001;98:8566–8571. doi: 10.1073/pnas.141113398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Napolitano R, Janel-Bintz R, Wagner J, Fuchs R P. EMBO J. 2000;19:6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morales J C, Kool E T. Biochemistry. 2000;39:2626–2632. doi: 10.1021/bi992173a. [DOI] [PubMed] [Google Scholar]
- 38.Schofield M J, Brownewell F E, Nayak S, Du C, Kool E T, Hsieh P. J Biol Chem. 2001;276:45505–45508. doi: 10.1074/jbc.C100449200. [DOI] [PubMed] [Google Scholar]