Abstract
The honey bee queen produces pheromones that function in both releaser and primer roles such as attracting a retinue of workers around her, attracting drones on mating flights, preventing workers from reproducing at the individual (worker egg-laying) and colony (swarming) level, and regulating several other aspects of colony functioning. The queen mandibular pheromone (QMP), consisting of five synergistic components, is the only pheromone chemically identified in the honey bee (Apis mellifera L.) queen, but this pheromone does not fully duplicate the pheromonal activity of a full queen extract. To identify the remaining unknown compounds for retinue attraction, honey bee colonies were selectively bred to have low response to synthetic QMP and high response to a queen extract in a laboratory retinue bioassay. Workers from these colonies were then used in the bioassay to guide the isolation and identification of the remaining active components. Four new compounds were identified from several glandular sources that account for the majority of the difference in retinue attraction between synthetic QMP and queen extract: methyl (Z)-octadec-9-enoate (methyl oleate), (E)-3-(4-hydroxy-3-methoxyphenyl)-prop-2-en-1-ol (coniferyl alcohol), hexadecan-1-ol, and (Z9,Z12,Z15)-octadeca-9,12,15-trienoic acid (linolenic acid). These compounds were inactive alone or in combination, and they only elicited attraction in the presence of QMP. There was still unidentified activity remaining in the queen extract. The queen therefore produces a synergistic, multiglandular pheromone blend of at least nine compounds for retinue attraction, the most complex pheromone blend known for inducing a single behavior in any organism.
The semiochemicals released by a honey bee queen have many effects within the colony (1, 2). Most obvious is the retinue attractant, which encourages workers to feed and groom the queen and acquire and distribute her pheromone messages to other workers throughout the colony. These messages, which may or may not involve the same chemical components, inhibit reproduction by workers, control swarming and the production of sexuals, act as nestmate and queen recognition cues, and regulate worker tasks critical to colony growth and survival. They are also important outside of the colony during mating flights and swarming (2).
The queen's mandibular glands were recognized long ago as a source of pheromonal activity, including retinue attraction. The first component of the queen mandibular pheromone, (E)-9-oxodec-2-enoic acid (9-ODA), and shortly thereafter, (E)-9-hydroxydec-2-enoic acid (9-HDA), were identified >40 years ago (1). However, these compounds did not match the pheromonal activity of the mandibular glands for retinue attraction. Almost 30 years passed before the chemical identity of the queen mandibular pheromone (QMP) was more fully described (3). In addition to 9-ODA and both enantiomers of 9-HDA, methyl p-hydroxybenzoate (HOB) and 4-hydroxy-3-methoxyphenylethanol (HVA) act synergistically to elicit retinue attraction, making it one of the most complex pheromone blends known. Individually, the activity of each component is not significantly different from a solvent control. The mandibular glands of a mated, laying queen contain ≈200 μg of 9-ODA, 80 μg of 9-HDA [85% (R)-(−)], 20 μg of HOB, and 2 μg of HVA, defined as a queen equivalent (Qeq) (4). A queen secretes ≈1 Qeq every 24 h, which is translocated onto other parts of her body by self-grooming and worker grooming such that ≈10−3 Qeq of QMP is present on her body surface at any time (5). Virgin queens produce significantly less 9-HDA, HOB, and undetectable HVA and are less attractive to workers than mated queens.
Soon after the identification of QMP, variation between colonies in the magnitude of response to QMP in the retinue bioassay was found to be independent of the queen's QMP production, the workers' age, the workers' fostering colony, and the dose used in the bioassay (6). Instead, it seemed to have a genetic component. Two-way selection for high and low QMP response in the retinue bioassay quickly yielded colonies significantly different in their response to QMP, with the high strains at least nine times more responsive to QMP than low strains (7). The impact of low and high QMP-responding workers on colony function has only begun to be tested (8, 9). Although the retinue attraction of workers to QMP in the bioassay can be very low, the workers attend their queen normally within the colony (10), suggesting the presence of additional semiochemicals in the queen to maintain the retinue attraction. Bees that respond poorly to QMP, yet have a strong retinue response to queen extract, could be powerful tools to guide the isolation and identification of new queen pheromone components.
The tergite glands, found on the dorsal surface of the abdomen, have been considered as the source of the remaining pheromonal activity in the queen (1, 11–13). After several years of unsuccessfully seeking the identity of compounds in the tergite glands responsible for retinue activity, Slessor et al. (5) reexamined the hypothesis that the tergite glands possess this remaining pheromonal activity. Using low QMP-responding workers, they compared the retinue attraction of several body washes and extracts to the matching QMP found in each treatment. They found that an abdomen extract was no more attractive than the small but significant quantity of QMP translocated onto the abdomen. Instead, a head extract was significantly more attractive than the matching QMP. In addition, the mandibular glands were no more attractive than the QMP they contained. These results suggested that the unidentified pheromone component(s) are located in the head but are not in the mandibular glands. The remaining activity was thought to be polar and labile, losing activity on fractionation and storage.
This paper describes the bioassay-guided isolation and identification of four new components of the queen retinue pheromone by using low QMP-responding worker bees.
Methods
Source of Bees.
All honey bees (similar to Apis mellifera ligustica L.) used in this project originated from queen stock imported from New Zealand, Australia, or Hawaii and were maintained at Simon Fraser University under normal beekeeping practices (14).
Preparation of Extract.
Mated wild-type queens were removed from their colonies and immediately immobilized on dry ice. Whole queens were homogenized and extracted repeatedly in distilled diethyl ether to give a combined total extract of 1,000 μl per queen. Queen heads, thoraces, and abdomens were extracted similarly in 100, 500, and 500 μl, respectively, per queen.
Bioassay.
The pseudoqueen laboratory retinue bioassay using 150 × 25-mm petri dishes was used (15). Pseudoqueen bioassay lures were fashioned from Pasteur pipettes to be approximately the size of a queen and had a dimple to hold the test treatment. Fifteen unnarcotized nurse (worker) bees from comb that contained uncapped brood were placed into each dish. Unless specified otherwise, lures were spotted with 10−2 Qeq of treatment and bioassayed under ambient laboratory fluorescent lighting at room temperature. The bioassay began, after solvent had evaporated, with the insertion of the lure into the bioassay dish. The number of bees contacting the lure 30 s later, and every 30 s thereafter for 5 min, was recorded. Only contacts of the wide portion of the lure with the bees' mouthparts or antenna were recorded. These 10 counts were treated as independent measurements, and the sum provided the score for each dish. Each lure and dish of bees were used only once. jmp software (SAS Institute, Cary, NC) was used for statistical analysis.
Colony Selection.
A selective mating program was established to produce colonies whose workers responded with both low QMP and high queen extract response. Colonies from Simon Fraser University's apiaries were screened to find those colonies whose workers responded strongly to a queen extract and poorly to synthetic QMP (PheroTech, Delta, BC, Canada) in the bioassay. One- to 2-day-old female larvae from one of these colonies were grafted into queen cells and reared into new queens by using normal queen-rearing practices (14). The resulting virgin queens were established in small mating colonies in an area otherwise free from honey bees. Other colonies that also were found to have low QMP and high queen extract responses were placed nearby and served as drone sources to mate with the virgin queens. After 2 weeks, those queens that had mated were removed from their colonies and used to requeen normal-sized colonies. These colonies were not used for the bioassay for at least 3 months to ensure that all nurse bees would be descendents of the selected queen. To increase the power of the bioassay, a colony to supply workers for the bioassay was chosen from the group of selectively bred colonies to maximize the difference in retinue response between queen extract and the known pheromone components. Initially, a colony was chosen based on the greatest difference between QMP and queen extract retinue responses. As each additional component was identified, another colony was chosen from the selectively bred colonies that had the greatest difference in retinue response between QMP with the new component(s) and queen extract.
GC and GC–MS Analysis.
GC was performed on a Hewlett–Packard 5890 gas chromatograph with a split/splitless injector with helium as the carrier gas, a flame ionization detector, and DB-1, -23, and -210 fused silica columns (J & W Scientific, Folsom, CA). GC–MS was performed on a Varian Saturn ion trap mass spectrometer coupled to a Varian 3400 GC with a splitless temperature programmable injector and a DB-5ms fused silica column (J & W Scientific). Extracts were reacted with N,O-bis(trimethylsilyl)trifluoroacetamide (Sigma) before GC analysis to form trimethylsilyl (TMS) derivatives to improve chromatographic characteristics. Retention indices (RIs) were used to compare the elution characteristics of different compounds relative to n-alkanes (16). Double bond positions were located by GC–MS analysis of dimethyl disulfide (17) or 2-alkenyl-4,4-dimethyloxazoline derivatives (18).
Quantitative Analysis.
The quantity of a particular compound in an extract or fraction was quantified by GC–MS as described (19). For the final quantitative analysis of QMP and the new pheromone components, 10 wild-type mated queens that were at least 1 year old were individually extracted in diethyl ether with internal standard. Standard solutions containing QMP and the new pheromone components were used to calibrate the GC–MS with respect to the internal standard over a 1,000-fold range in concentration.
Liquid Chromatography.
HPLC was performed on a Waters 625 LC system with a Rheodyne 9125 injector, 50- or 250-μl sample loop, and a Waters 486 UV detector. RP chromatography was conducted on a 4.6-mm i.d. × 250 mm C18 column with a 4.6-mm i.d. × 30-mm guard column (Phenomenex Columbus C18). Normal phase chromatography was conducted on a 4.6-mm i.d. × 250-mm silica column (Phenomenex Luna Silica-2). Flow rates were 1 ml/min on both columns. Helium-sparged HPLC grade methanol, acetonitrile, hexane (redistilled), diethyl ether (redistilled), and deionized water (Alpha-Q, Millipore) were used for mobile phases.
Location of Pheromone Components.
Following previous research that showed that the remaining retinue activity was in the head, isolation of new pheromone components began with the head extract. As each new pheromone component was identified, the body section that contained the greatest remaining retinue activity was determined and used to isolate additional components. To determine the glandular source of each new component once identified, workers and virgin and mated queens were dissected into body sections, extracted, and analyzed by GC–MS. Sections containing the majority of each component were dissected further under distilled water to identify the gland that held the majority of each new component. In addition, hemolymph was collected from 10 virgin queens, and the nonpolar fraction was extracted following Francis et al. (20) and analyzed by GC–MS.
Dose–Response.
To assess the activity of QMP with the newly identified compounds over a range of doses, four colonies (two wild-type and two selectively mated) were used to bioassay over five orders of magnitude of dose. Synthetic treatments were matched to the quantities in the whole multiple queen extracts.
Results
Selective Mating.
On average, the queen extract response was more than three times greater than the QMP response for over 20 colonies (21). The large difference in response between QMP and queen extract (>10-fold difference for some colonies) and the consistency of response over the bioassay season made the isolation and identification of additional pheromone components possible.
Isolation and Identification of New Components.
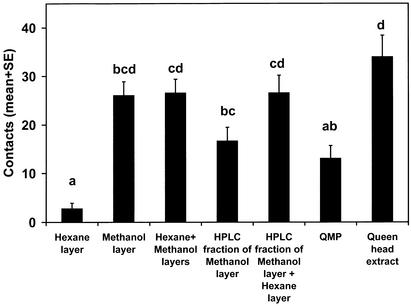
The polar portion of a queen head extract was investigated first by partitioning it between hexane and methanol. The methanol layer was removed, and the hexane layer further extracted with methanol. The combined methanol layers were injected onto the RP-HPLC column with methanol as the mobile phase, and a 1- to 11-min fraction was collected and bioassayed (Fig. 1). Although the hexane layer was not active alone and the methanol layer was as active as the combination, the activity of the methanol layer decreased when passed through the RP-HPLC column. However, addition of the hexane layer to the 1- to 11-min RP-HPLC fraction returned activity to the level of the methanol layer before HPLC. The hexane layer seemed to contain a pheromone component that was inactive alone. The methanol layer also must have contained this component, but it must not have eluted within 11 min from the RP-HPLC column. Thus, it seemed that there were at least two new components to be identified, one polar and one relatively nonpolar, and both seemed to be inactive alone.
Figure 1.
RP-HPLC of partitioned queen head extract. Workers in the bioassay were from a wild-type (not selectively bred) colony with a low QMP response. One-way ANOVA: F = 11.1, P < 0.0001, n = 10. In Figs. 1–5, treatments with the same letter are not significantly different (P < 0.05, by Tukey–Kramer test).
Using RP-HPLC to fractionate the methanol layer of the queen head extract, the column was eluted with a mobile phase of methanol:water in various mixtures and gradients. All fractions were bioassayed in combination with a corresponding dose of the hexane layer. After many bioassays, an active fraction could not be isolated that did not also contain a QMP component (21). Instead, several fractions, each containing a QMP component, needed to be combined together, with the hexane layer, for activity. These results suggested that QMP was synergistic with an unknown component in the hexane layer.
Two nonpolar compounds in a queen head extract, methyl oleate (MO) and ethyl oleate, were previously identified as active to worker antenna by GC–electroantennographic detection but were inactive when bioassayed individually or in combination (unpublished results). These esters were present in the nonpolar fraction and were investigated further. Retention indices and mass spectra of the synthetics matched the corresponding peaks in the extract (RIDB-1 = 2,082, RIDB-5ms = 2,100, and RIDB-210 = 2,380 for MO). Lesser amounts of methyl palmitate, ethyl palmitate, methyl palmitoleate, ethyl palmitoleate, and ethyl stearate also were identified.
Double bond position and stereochemistry of MO was confirmed to be (Z)-9 based on the retention indices of a series of synthetic methyl (Z)-octadecenoates on both DB-5ms and DB-23 columns, GC–MS analysis of the dimethyl disulfide derivative, and the unmatched retention index of methyl (E)-octadec-9-enoate. Dimethyl disulfide derivatization of synthetic methyl (Z)-octadec-9-enoate (Sigma) yielded an identical mass spectrum and retention index as the derivatized unknown. In a similar manner, the double bond was confirmed to be in the 9 position for the methyl palmitoleate, ethyl palmitoleate, and ethyl oleate present in the extract. GC–MS analysis of the esters in the body sections of the queen indicated that they were not specific to the head but rather distributed throughout the queen. Initial bioassays suggested that, of these esters, only MO was synergistic with QMP (21), and this was later confirmed (Fig. 2). Bioassay of QMP with all of the other esters was not significantly different from QMP alone (n = 40, t test, P = 0.13). However, QMP+MO accounted for only a portion of the retinue attraction of the whole queen extract.
Figure 2.
QMP with MO and other esters. EO, ethyl oleate; MP, methyl palmitate; EP, ethyl palmitate; MPL, methyl palmitoleate; EPL, ethyl palmitoleate; ES, ethyl stearate. One-way ANOVA: F = 59.3, P < 0.0001, n = 60.
Unexpected decreases in activity were observed while bioassaying fractions in methanol, suggesting that the methanol had a detrimental effect on the activity. Bioassays of queen head extract in different solvents and in the presence or absence of light indicated that activity significantly decreased when a protic solvent was used in ambient fluorescent room light during spotting and storing of the lures (e.g., in methanol, 30.4 ± 2.4 versus 44.0 ± 2.5, n = 40, t test, P = 0.0002). Subsequently, the bioassay procedure was changed so that all treatments were kept in the dark as much as possible. The lures were spotted under a 7.5-W red, incandescent, photographic darkroom light and then kept in the dark until needed. The bioassay itself was conducted in dim incandescent light.
Comparison by GC–MS of N,O-bis(trimethylsilyl)trifluoroacetamide-derivatized samples of queen head extract in methanol that had been kept on a bioassay lure either in the dark or exposed to fluorescent room light for 1 h identified a peak that significantly decreased in intensity after exposure to light. The retention indices [RIDB-1(TMS) = 1,931 and RIDB-5ms(TMS) = 1,933] and mass spectrum of this peak were identical to the TMS derivative of (E)-3-(4-hydroxy-3-methoxyphenyl)-prop-2-en-1-ol [(E)-coniferyl alcohol, CA, Sigma], a compound known to degrade photochemically in protic solvents (22). Subsequently, other related phenylpropanoids were identified in the queen head extract and confirmed by comparison of mass spectra and retention indices with synthetics including (E)-ferulic acid, dihydroferulic acid, and dihydroconiferyl alcohol (21), which did not degrade significantly when exposed to light in methanol. The addition of these aromatics to QMP+MO significantly increased retinue attraction in the dark (11.7 ± 2.1 versus 22.6 ± 2.4, n = 30, t test, P = 0.0012), and this retinue activity seemed light sensitive (not shown). However, bioassay of QMP with all of the identified esters and phenolics was not significantly different from QMP+MO+CA alone (n = 40, t test, P = 0.60), indicating that these additional compounds were not involved in retinue attraction. CA alone was later confirmed to significantly increase activity (n = 40, t test, P = 0.02).
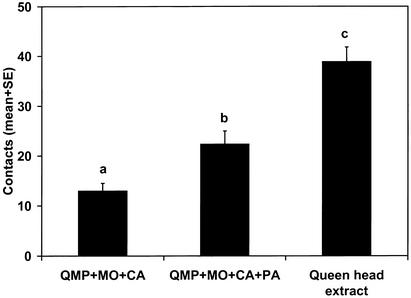
Fractions of high, low, and intermediate polarity were prepared by dissolving the queen head extract in methanol, extracting with hexane, and then back-extracting the hexane layer with methanol. In combination with QMP+MO+CA, the remaining activity appeared only in the fraction of intermediate polarity. The queen head extract was evaporated, redissolved in methanol, injected onto the RP-HPLC column, and eluted with methanol. The full activity of the queen head extract was reproduced by QMP+MO+CA in combination with the 6- to 11-min section. Activity was further isolated to the 7- to 8-min fraction, but this fraction did not fully match the remaining activity of the queen head extract. GC–MS analysis of this fraction derivatized with N,O-bis(trimethylsilyl)trifluoroacetamide identified a peak unique to this fraction with a retention index (RIDB-5ms = 1,956) and mass spectrum identical to the TMS derivative of hexadecan-1-ol. Bioassay of synthetic hexadecan-1-ol (PA, Kodak) in combination with QMP+MO+CA with all components matched to the quantities in the extract showed a significant improvement in activity consistent with the activity in the 7- to 8-min fraction but did not equal the queen extract (Fig. 3). The three new components elicited little retinue activity themselves; only when combined with QMP did they induce retinue activity (Fig. 4), but they still did not equal the retinue attraction of a queen extract.
Figure 3.
Retinue activity of PA. One-way ANOVA: F = 30.2, P < 0.0001, n = 40.
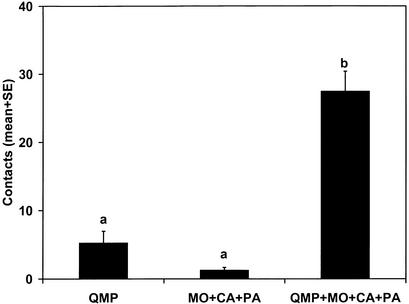
Figure 4.
Synergy of QMP with the three new components. One-way ANOVA F = 51.6, P < 0.0001, n = 20.
A portion of the remaining activity was still of intermediate polarity. A queen abdomen extract dissolved in hexane was added to a silica Sep-Pak Light cartridge (Waters) conditioned with hexane. The cartridge was eluted with increasingly more polar solvents from 100% hexane to 100% diethyl ether, and five 1-ml fractions were collected. Bioassay of these fractions in combination with QMP+MO+CA+PA indicated that the remaining activity was located in the second fraction (75:25 hexane:ether).
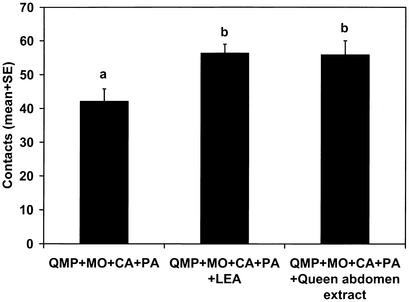
The second fraction was injected onto the normal phase HPLC column and eluted with the following gradient: 100:0 to 50:50 hexane:ether linearly over 50 min. Fractions were collected by monitoring absorbance at 210 nm. Bioassay of fractions in combination with QMP+MO+CA+PA indicated that the strongest UV-absorbing peak, at 20.9–22.1 min, also held activity. This fraction was evaporated, redissolved in acetonitrile, and then injected onto the RP-HPLC column and eluted with acetonitrile. Fractions were collected by monitoring absorbance at 210 nm. The largest UV-absorbing peak, at 6.4–8.4 min, also gave the strongest bioassay response when combined with QMP+MO+CA+PA. GC–MS analysis of this fraction as its TMS derivative suggested an octadecatrienoic acid. Unsaturation was located at positions 9, 12, and 15 by GC–MS analysis of its 2-alkenyl-4,4-dimethyloxazoline derivative. The retention index and mass spectrum for synthetic linolenic acid [(9Z,12Z,15Z)-octadec-9,12,15-trienoic acid, LEA, Sigma] were identical to that in the fraction [RIDB-5ms(TMS) = 2,213, RIDB-23(TMS) = 2,620, and RIDB-210(TMS) = 2,450]. The mass spectrum and retention index of the 2-alkenyl-4,4-dimethyloxazoline derivative were identical. Bioassay of synthetic LEA in combination with QMP+MO+CA+PA confirmed its activity (Fig. 5).
Figure 5.
Retinue activity of LEA. One-way ANOVA: F = 5.4, P = 0.006, n = 30.
Location of Pheromone Components.
On average, there was 48% more MO in mated queens than in virgin queens, and it was distributed evenly between body sections in both queens. The hemolymph contained ≈300 pg/μl MO. Workers contained no detectable MO. CA was concentrated in the head extracts of mated queens. Analysis of the glands in the head established CA as originating from the mandibular glands. CA could not be detected in worker or young virgin queen head extracts. The PA in virgin queens was concentrated in the abdomen, whereas mated queens had more PA in the head than in the abdomen. PA was distributed as follows: 6 ± 1%, 4 ± 1%, and 90 ± 2% and 59 ± 5%, 6 ± 1%, and 35 ± 4% in the head, thorax, and abdomen of virgin and mated queens, respectively (mean ± SE, n = 7). The overall amounts were similar between virgin and mated queens. PA in mated queens was concentrated in the Dufour's gland of the abdomen and the cephalic labial gland of the head. LEA was concentrated in the thorax and abdomen of both virgin and mated queens, but no glandular source was found. The thorax of a mated queen had ≈40 times more LEA than a thorax of a nurse bee.
Quantitative Analysis.
The quantities of the QMP components and the new pheromone components in mated queens were as follows: 9-ODA, 240 ± 30; 9-HDA, 110 ± 10; HOB, 36 ± 10; HVA, 3.6 ± 1.0; MO, 3.8 ± 0.8; CA, 0.15 ± 0.04; PA, 1.1 ± 0.1; and LEA, 22 ± 6 μg/Qeq (mean ± SE, n = 10). Quantitative analysis of the aliphatic esters not involved in retinue attraction was described elsewhere (21).
Dose–Response.
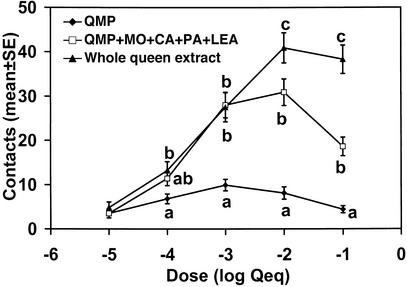
The four new pheromone components significantly increased the attraction of QMP over most doses, but the whole queen extract was still significantly more attractive than the nine-component blend at the higher doses (Fig. 6). The synthetic blend matched the whole queen extract at 10−3 Qeq per lure, a dose similar to that expected on the surface of a queen (5).
Figure 6.
Dose–response. The bioassay results from two wild-type and two selectively mated colonies were combined (n = 48; n = 12 per colony). Treatments at each dose with the same letter are not significantly different (P < 0.05, by Tukey–Kramer test).
Discussion
Unlike previous research that selected for either low or high QMP retinue response (7), the isolated mating program successfully selected for colonies with two traits, low QMP and high queen extract response. These selected colonies were essential for the bioassay-guided isolation and identification of the new pheromone components. These four components were not attractive alone or together but instead synergized with QMP to elicit increased retinue attraction. The nine compounds now identified still do not fully match the retinue activity of a whole queen extract at higher doses; other components may still require identification. Bioassays suggest that the remaining activity is of intermediate polarity and that hydrocarbons and the known phenols and esters are not involved.
The selected colonies noticeably varied in their responses to the new components. For example, some colonies could be classified as low or high MO responders. This variation was used to advantage to maximize the power of the bioassay when identifying the later components. Most wild-type colonies screened had a lower retinue response for QMP than queen extract (not shown). A high QMP-responding colony is apparently low responding to the other components. This variability in response to the different pheromone components is expected in such a complex system and may be correlated with the natural genetic variation of other honey bee phenotypes.
The pheromone blend contains contributions from several glands, and, like QMP (4), we found a distinct ontogeny to the new components. Both mated and virgin queens contained MO throughout their bodies including their hemolymph, although mated queens had more than virgins. Like pheromones of some other insects, MO might be biosynthesized in oenocytes located in the integument or hemocoel, rather than in a specific gland, and then transported through the hemolymph to the site of secretion by lipid carrier proteins (23). CA was located in the mandibular glands of mated but not virgin queens. Its sensitivity to light in methanol precluded its earlier discovery. PA was located in the Dufour's gland and the cephalic labial gland of mated queens but predominantly in the abdomen of virgin queens. LEA was found in all three body sections but chiefly in the thorax and abdomen.
Multicomponent pheromones are the norm, but a pheromone for one distinct behavior originating from several glandular sources is rare. Some closely related species of ants use a common pheromone arising from one gland but use secretions from another gland to convey species specificity to the signal (24). Ants may also use multiple glandular sources to modulate the different aspects of trail following and recruitment. Because the honey bee pheromone components originate from several sources within the queen, the retinue response may be modulated by the metabolic processes of each gland. For example, like the mandibular gland components (4), other glandular components may change significantly when a mated queen begins to fail and starts laying unfertilized eggs. Because the retinue pheromone is used to facilitate the dispersion of other queen signals throughout the colony, changes to the retinue attraction may influence such things as queen rearing, swarming, and worker reproduction. Further study of each gland involved is needed to understand its role in the queen.
Queens contained several aliphatic esters in addition to methyl oleate, and most have previously been identified on honey bee brood. Le Conte and coworkers (25) have reported the many pheromonal effects these brood esters have on colony function. Many of the effects shown with these esters in the brood, including primer effects, are likely to occur with the esters in the queen. Thus, the queen seems to act in concert with her brood to modulate the behavior and physiology of worker bees. The retinue pheromone, which elicits the workers to lick, antennate, and groom their queen, entices workers to pick up these other control sociochemicals from their queen.
Queens in all Apis species analyzed produce 9-ODA and 9-HDA in their mandibular glands (26). Queens of Apis cerana and Apis nigrocincta, the species most closely related to Apis mellifera, produce HOB but neither HVA nor CA (19). However, they do produce many other aromatic compounds including some containing the 4-hydroxy-3-methoxyphenyl group. The queens of the other species of honey bees do not produce HOB or HVA (26), and it is not known whether MO, PA, LEA, or any aromatic compounds are produced. Further research is needed to establish the importance of these compounds in the other honey bees.
During this study, no colony was ever found that had a retinue response significantly higher for QMP than the queen extract, suggesting that queen recognition cues may only increase, and not inhibit, retinue response. The likelihood of encountering the effects of queen recognition increased as the retinue activity of the synthetic blend approached that of the queen extract. Cuticular hydrocarbons often are suggested as the source of nest-mate cues (27). However, we found no evidence that worker bees use hydrocarbons as queen recognition cues; workers in the retinue bioassay seemed indifferent to fractions rich in queen-derived hydrocarbons. Nevertheless, the variation seen in retinue response for different colonies with the same queen extract and the same colony with different queen extracts suggests that there may be other chemical cues involved in queen recognition.
Some of the new components have previously been identified in honey bee queens, but no pheromonal activity had been demonstrated or suggested. PA, MO, ethyl oleate, and ethyl palmitate are found in the mandibular gland extracts of honey bee queens (Apis mellifera carnica) (28). MO is found in both cuticular and tergal gland extracts of virgin and mated queens of A. m. scutellata and A. m. capensis (29). MO has not been reported in workers except for A. m. capensis pseudoqueens (A. m. capensis workers treated like queens by A. m. scutellata workers).
Although the components of QMP are thought to be queen specific, workers of most Apis species contain detectable amounts of 9-ODA and 9-HDA (26). Worker, drone, and queen larvae all produce esters, including MO (30). CA was not detected in drones or workers (21), but PA can be detected in the cocoon and pupa of both drones and workers but not in larvae or adults (31). The quantity of LEA detected in a worker's thorax was significantly less than in a queen's. Thus, all of the new pheromone components except CA can be detected in either drones or workers at some life stage. However, their synergy with QMP and the queen specificity of HOB, HVA, and CA confer a queen specificity to the retinue response.
Although presence does not indicate function, queens of other Hymenoptera also contain some of the new components. MO is found in the queen leg extracts of the social wasps Vespa crabro, Vespa orientalis, and Polistes dominulus (32) and the Dufour's and cephalic labial glands of the queen bumble bee Bombus terrestris (33). PA is found in the tarsal gland extracts of the queen bumble bee B. terrestris (33). CA has not previously been identified in Hymenoptera, and LEA is not mentioned in most analyses of Hymenoptera. Pollen is known to contain a unique fatty acid, (2E,9Z,12Z)-octadeca-2,9,12-trienoic acid, attractive to foraging honey bees (34), but there was no evidence of this structurally related compound in the queen.
Because this work focused on the isolation and identification process, many questions about each new component remain unanswered. The location of biosynthesis and secretion of both MO and LEA need to be determined. More detailed studies of the ontogeny and phylogeny of all components are necessary. Further studies are needed to establish whether the new pheromone components synergize with QMP as a whole or with certain components of QMP. Studies to examine other pheromonal effects of the new components have begun (unpublished work).
Our study has increased the total number of components known to be involved in retinue attraction to nine, originating from several glandular sources. This is the most complex pheromone system known for a single behavior in any organism and provides an example of the complexity to be expected for pheromone communication in other social insects. Not all of the components involved need to be unique to the queen because synergy and the contextual presentation are critical to elicit a response. This emphasizes the necessity of a bioassay-guided approach to pheromone isolation and identification. QMP should no longer be considered a distinct retinue pheromone but rather a portion of a more complex queen retinue pheromone. In addition to QMP and the four new components, the queen also possesses compounds that, although not essential for retinue attraction, may be crucial for the control she exerts on her colony.
Acknowledgments
We thank P. Balcar, Z. Chan, R. Cheng, R. Falk, P. Hick, A. Lee, H. T. Ngo, and T. Smith for assistance with the bioassays, and the Simon Fraser University Swarm Team for apicultural support. We acknowledge the early work of E. Plettner and R. Gries with GC–electroantennography and the former for manuscript review. C.I.K. was supported by graduate fellowships from Simon Fraser University, the Marshall Noble Memorial Graduate Bursary in Chemical Ecology, and the Frank A. Linville Graduate Scholarship in Olfaction. This work was supported by Natural Sciences and Engineering Research Council of Canada grants (to K.N.S. and M.L.W.) and a Killam fellowship (to M.L.W.).
Abbreviations
- CA
(E)-coniferyl alcohol
- 9-HDA
(E)-9-hydroxydec-2-enoic acid
- HOB
methyl 4-hydroxybenzoate
- HVA
4-hydroxy-3-methoxyphenylethanol
- LEA
linolenic acid
- MO
methyl oleate
- 9-ODA
(E)-9-oxodec-2-enoic acid
- PA
hexadecan-1-ol
- Qeq
queen equivalent
- QMP
queen mandibular pheromone
- RI
retention index
- TMS
trimethylsilyl
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Free J B. Pheromones of Social Bees. London: Chapman & Hall; 1987. [Google Scholar]
- 2.Winston M L, Slessor K N. Apidologie. 1998;29:81–95. [Google Scholar]
- 3.Slessor K N, Kaminski L-A, King G G S, Borden J H, Winston M L. Nature. 1988;332:354–356. [Google Scholar]
- 4.Pankiw T, Winston M L, Plettner E, Slessor K N, Pettis J S, Taylor O R., Jr J Chem Ecol. 1996;22:605–615. doi: 10.1007/BF02033573. [DOI] [PubMed] [Google Scholar]
- 5.Slessor K N, Foster L J, Winston M L. In: Pheromone Communication in Social Insects: Ants, Wasps, Bees, and Termites. Vander Meer R K, Breed M D, Espelie K E, Winston M L, editors. Boulder, CO: Westview Press; 1998. pp. 331–343. [Google Scholar]
- 6.Pankiw T, Winston M L, Slessor K N. J Insect Behav. 1994;7:1–15. [Google Scholar]
- 7.Pankiw T, Winston M L, Fondrk K M, Slessor K N. Naturwissenschaften. 2000;87:487–490. doi: 10.1007/s001140050764. [DOI] [PubMed] [Google Scholar]
- 8.Pankiw T, Huang Z-Y, Winston M L, Robinson G E. J Insect Physiol. 1998;44:685–692. doi: 10.1016/s0022-1910(98)00040-7. [DOI] [PubMed] [Google Scholar]
- 9.Pankiw T. Can Entomol. 1997;129:679–690. [Google Scholar]
- 10.Pankiw T, Winston M L, Slessor K N. Insectes Soc. 1995;42:371–378. [Google Scholar]
- 11.De Hazan M, Lensky Y, Cassier P. Comp Biochem Physiol A. 1989;93:777–784. [Google Scholar]
- 12.Wossler T C, Crewe R M. Apidologie. 1999;30:311–320. [Google Scholar]
- 13.Wossler T C, Crewe R M. J Insect Behav. 1999;12:343–351. [Google Scholar]
- 14.Graham J M, editor. The Hive and the Honey Bee. Hamilton, IL: Dadant & Sons; 1992. [Google Scholar]
- 15.Kaminski L-A, Slessor K N, Winston M L, Hay N W, Borden J H. J Chem Ecol. 1990;16:841–850. doi: 10.1007/BF01016494. [DOI] [PubMed] [Google Scholar]
- 16.van den Dool H, Kratz P D. J Chromatogr. 1963;11:463–471. doi: 10.1016/s0021-9673(01)80947-x. [DOI] [PubMed] [Google Scholar]
- 17.Buser H-R, Arn H, Guerin P, Rauscher S. Anal Chem. 1983;55:818–822. [Google Scholar]
- 18.Garrido J L, Medina I. J Chromatogr. 1994;673:101–105. [Google Scholar]
- 19.Keeling C I, Otis G W, Hadisoesilo S, Slessor K N. Apidologie. 2001;32:243–252. [Google Scholar]
- 20.Francis B R, Blanton W E, Littlefield J L, Nunamaker R A. Ann Entomol Soc Am. 1989;82:486–494. [Google Scholar]
- 21.Keeling C I. Ph.D. thesis. Burnaby, BC, Canada: Simon Fraser Univ.; 2001. [Google Scholar]
- 22.Radotic K, Zakrzewska J, Sladic D, Jeremic M. Photochem Photobiol. 1997;65:284–291. [Google Scholar]
- 23.Gu X, Quilici D, Juarez P, Blomquist G J, Schal C. J Insect Physiol. 1995;41:257–267. [Google Scholar]
- 24.Hölldobler B. Proc Natl Acad Sci USA. 1995;92:19–22. doi: 10.1073/pnas.92.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohammedi A, Paris A, Crauser D, Le Conte Y. Naturwissenschaften. 1998;85:455–458. [Google Scholar]
- 26.Plettner E, Otis G W, Wimalaratne P D C, Winston M L, Slessor K N, Pankiw T, Punchihewa P W K. J Chem Ecol. 1997;23:363–377. [Google Scholar]
- 27.Breed M D. In: Pheromone Communication in Social Insects: Ants, Wasps, Bees, and Termites. Vander Meer R K, Breed M D, Espelie K E, Winston M L, editors. Boulder, CO: Westview Press; 1998. pp. 57–78. [Google Scholar]
- 28.Engels W, Rosenkranz P, Adler A, Taghizadeh T, Lübke G, Francke W. J Insect Physiol. 1997;43:307–313. doi: 10.1016/s0022-1910(96)00110-2. [DOI] [PubMed] [Google Scholar]
- 29.Wossler T C, Crewe R M. J Apic Res. 1999;38:137–148. [Google Scholar]
- 30. Le Conte, Y., Sreng, L., Sacher, N., Trouiller, J., Dusticier, G. & Poitout, S. H. (1994/1995) Chemoecology5/6, 6–12.
- 31.Donzé G, Schnyder-Candrian S, Bogdanov S, Diehl P-A, Guerin P M, Kilchenman V, Monachon F. Arch Insect Biochem Physiol. 1998;37:129–145. [Google Scholar]
- 32.Moneti G, Dani F R, Pieraccini G, Turillazzi S. Rapid Commun Mass Spectrom. 1997;11:857–862. [Google Scholar]
- 33.Hefetz A, Taghizadeh T, Francke W. Z Naturforsch. 1996;51:409–422. [Google Scholar]
- 34.Hopkins C Y, Jevans A W, Boch R. Can J Biochem. 1969;47:433–436. doi: 10.1139/o69-067. [DOI] [PubMed] [Google Scholar]