Abstract
Hepatitis C virus (HCV) infects nearly 3% of the population of the world and is a major cause of liver disease. However, the mechanism whereby the virus targets the liver for infection remains unknown, because none of the putative cellular receptors for HCV are both expressed specifically in the liver and capable of binding HCV envelope glycoproteins. Liver/lymph node-specific intercellular adhesion molecule-3-grabbing integrin (L-SIGN) is a calcium-dependent lectin expressed on endothelial cells of liver and lymph nodes. Dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin (DC-SIGN), a homologous molecule expressed on dendritic cells, binds HIV and promotes infection. By using a virus-binding assay, we demonstrate that L-SIGN and DC-SIGN specifically bind naturally occurring HCV present in the sera of infected individuals. Further studies demonstrate that binding is mediated by the HCV envelope glycoprotein E2 and is blocked by specific inhibitors, including mannan, calcium chelators, and Abs to the lectin domain of the SIGN molecules. Thus, L-SIGN represents a liver-specific receptor for HCV, and L-SIGN and DC-SIGN may play important roles in HCV infection and immunity.
An estimated 170 million individuals worldwide are infected with hepatitis C virus (HCV), an enveloped RNA virus that often establishes chronic infection leading to liver cirrhosis and hepatocellular carcinoma (1). The HCV genome encodes a single polyprotein that is processed by viral and cellular proteases into structural and nonstructural proteins (1). The HCV structural proteins include the envelope glycoproteins E1 and E2, which mediate viral binding and entry into host cells. E1 and E2 form heterodimers and undergo extensive posttranslational modification by N-linked glycans (1).
A fundamental riddle of HCV is how the virus targets the liver for infection. The tissue and cellular tropisms of viruses are often regulated by one or more host receptors that mediate distinct functions such as viral attachment, internalization, fusion, and trafficking (2, 3). CD81 and the low-density lipoprotein receptor (LDL-R) have been identified as putative attachment and entry receptors for HCV (4, 5). CD81 binds the HCV envelope glycoprotein E2 in vitro but its wide tissue distribution fails to explain the tropism of HCV. Although the expression pattern of LDL-R is consistent with HCV tropism, direct binding of the envelope glycoproteins has not been demonstrated. Rather, LDL-R binds and internalizes virus-like particles in complex with LDL. Neither CD81 nor LDL-R has been shown to mediate viral fusion and entry. Recently, a broadly expressed lipoprotein binding receptor, human scavenger receptor class B type I, was shown to bind to E2 protein in vitro, but binding to virus was not demonstrated (6).
Dendritic cell (DC)-specific intercellular adhesion molecule-3 (ICAM-3)-grabbing nonintegrin (DC-SIGN; CD209) is a 44-kDa type II integral membrane protein with a short amino-terminal cytoplasmic domain and a carboxyl-terminal C-type (calcium-dependent) lectin domain. A membrane-proximal extracellular heptad-repeat region promotes oligomerization of DC-SIGN via a novel coiled-coil motif. DC-SIGN is expressed at high levels on myeloid-lineage DCs in tissues where it interacts with ICAM-3 on T cells as part of the immunological synapse of T cell activation (7). DC-SIGN functions as a specific attachment factor for HIV and other lentiviruses, and binding is mediated by the viral surface glycoprotein gp120 (7). DC-SIGN does not mediate HIV entry directly but rather enhances infection of susceptible target cells in trans (8). DCs may bind HIV via DC-SIGN at a site of mucosal exposure and carry the virus to target cells within the draining lymph node, thereby facilitating establishment of infection (8). DC-SIGN also binds other pathogens, including Ebola virus, cytomegalovirus, and Leishmania (9–11).
Liver/lymph node-specific ICAM-3-grabbing integrin (L-SIGN) or DC-SIGN-related (CD209L) shares 77% amino acid sequence identity with DC-SIGN and has functional similarity in its interactions with ICAM-3 and HIV-1 (12, 13). L-SIGN is abundantly expressed on endothelial cells of liver and lymph nodes but not on DCs (12, 13).
Here we report that HCV particles bind specifically to L-SIGN and DC-SIGN and that this interaction is mediated by the viral glycoprotein HCV-E2, the functional equivalent of HIV gp120 that similarly contains abundant high-mannose-type oligosaccharides. Thus, L-SIGN and DC-SIGN function as capture receptors for HCV and may play critical roles in viral pathogenesis and tissue tropism.
Methods
Abs and Recombinant Proteins.
Conformation-specific anti-E2 mAbs H31, H33, H44, H48, H50, H52, H53, H54, H60, and H61 were generously provided by J. Dubuisson (Institut Pasteur de Lille, Lille, France) (14, 15). mAb 4F6/2 (Austral Biological) reacts with a linear epitope in E2. mAbs to the lectin-binding domain of DC-SIGN (mAb 507D) and L-SIGN (mAb 604L) or both lectins (mAb 612X) were obtained from R & D Systems. mAbs DC6 and DC28 (AIDS Research and Reference Reagent Program, Rockville, MD) recognize the repeat regions of DC-SIGN (DC6) or DC-SIGN and L-SIGN (DC28). Control isotype-matched murine IgG (mIgG; Caltag, South San Francisco, CA) was used to establish background levels of binding. Recombinant E2 protein (Accurate Chemicals) was expressed in secreted form in Chinese hamster ovary cells and encompassed amino acids 384–665 of the HCV polyprotein. Recombinant ICAM-2 and ICAM-3 were used as soluble Fc fusion proteins (R & D Systems).
Plasmids and Cell Lines.
Plasmids pcDNA3-DC-SIGN and pcDNA3-DC-SIGN-related (AIDS Research and Reference Reagent Program) were transfected into HeLa cells by using Effectene (Qiagen, Valencia, CA). Cells were treated with standard growth media comprising DMEM with 10% heat-inactivated FBS (HyClone), penicillin/streptomycin (Life Technologies, Carlsbad, CA), and l-glutamine (Life Technologies) supplemented with 600 μg/ml Geneticin (Life Technologies). After 2 weeks, surviving colonies were selected, expanded, and screened for expression by flow cytometry. High expressors were cloned and passaged in growth media with Geneticin (600 μg/ml).
Fluorescence-Activated Cell Sorter (FACS) Analysis of DC-SIGN and L-SIGN Expression.
Cell-surface expression of DC-SIGN and L-SIGN on transfected cell lines was measured by using flow cytometry. Cells were harvested with cell dissociation solution (Sigma), and FACS analysis was performed as described (16).
HCV-E2 Binding Assay.
The assay is a modification of that described for gp120 binding (8). Briefly, HeLa cells were removed from culture as described previously and washed three times in adherence buffer [AB; 20 mM Tris⋅HCl (pH 8.0)/150 mM NaCl/1 mM CaCl2/2 mM MgCl2/0.5% BSA]. Cells (5 × 105) were preincubated with mannan (20 μg/ml; Sigma), Abs (20 μg/ml), EDTA (5 mM), or EGTA (5 mM) for 10 min at room temperature. HCV-E2-coated fluorescent beads were prepared with the indicated anti-E2 capture mAb and added to cells (20 beads per cell) for 30 min at 37°C. Binding was determined by flow cytometry.
Virus Binding Assay.
Virus-cell binding.
HeLa cell lines were cultured overnight in DMEM containing 10% FBS in a 96-well plate at 1 × 104 cells per well. Cells were blocked with AB containing 10% heat-inactivated goat serum for 20 min at 37°C. Cells were washed once with AB, and mannan (20 μg/ml) was added for 15 min in AB at room temperature. After washing, sera (10–20 μl) from HCV RNA+ (virus-positive) or HCV RNA− serum (virus-negative) donors (all HIV sero-negative) were diluted in AB and allowed to bind to cells for 1 h at 37°C with gentle agitation every 15 min, after which cells were washed five times with AB.
RNA extraction.
Viral RNA was extracted from cells by using a QIAmp Viral RNA Mini Spin kit (Qiagen) with modifications. Briefly, RNA was extracted with lysis buffer followed by binding to spin columns, and DNA was removed by treatment with RNase-free DNase (Qiagen). RNA was washed and eluted in elution buffer.
Southern blot.
HCV RNA was amplified by RT-PCR as described (17) with modifications. Primer KY78 (5′-CTCGCAAGCACCCTATCAGGCAGT-3′, 0.5 nmol; nucleotides 276–299) was combined with 0.5 μl of extracted RNA in a final volume of 6 μl and preheated followed by addition of cDNA synthesis buffer, 10 mM DTT, 5 mM dNTPs, and 7.5 units of ThermoScript (Invitrogen), incubated at 58°C for 50 min then 85°C for 5 min before cooling to 4°C. From this RT reaction, 5 μl was used as the template for PCR in a 50-μl reaction containing 1× High-Fidelity PCR buffer, 2 mM MgSO4, 2 mM dNTPs, 50-pmol primers KY80 (5′-GCAGAAAGCGTCTAGCCATGGCGT-3′; nucleotides 56–79) and KY78, and 1.25 units Platinum Taq high-fidelity DNA polymerase (Invitrogen). The amplification product was resolved on a 1% agarose gel and blotted onto a nitrocellulose membrane (Bio-Rad).
For detection, the blot was incubated for 4 h at 63°C in prehybridization solution {5× Denhardt's [0.2% (wt/vol) fatty-acid-free BSA (JRH Biosciences, Lenexa, KS)/0.2% (wt/vol) polyvinylpyrrolidonpolyvinyl (Sigma)/0.2% (wt/vol) Ficoll-400 (Sigma)]; 6× SSC (0.9 M NaCl/90 mM sodium citrate, pH 7.4), 0.5% (wt/vol) SDS (Promega), and 0.1 mg/ml herring sperm DNA (Invitrogen)}. After incubation, 1 pmol/ml primer RJD-6 (5′ biotin-GGAGAGCCATAGTGGTCTGCGGAA C-3′; nucleotides 120–144) or KY88 (5′ biotin-GTTGGGTCGCGAAAGGCCTTGTGGT-3′; nucleotides 251–275) was added to the prehybridization solution and incubated overnight at 63°C. After extensive high-stringency washing, the blot was incubated with streptavidin-horseradish peroxidase (Pierce), washed, and developed by using Western Lightening Plus (NEN/Perkin–Elmer). An HCV RNA+ signal is exemplified by a specific band of 243 bp.
Real-time PCR.
HCV Quantasure Plus assay was used at LabCorp (Research Triangle Park, NC) and has been demonstrated to be sensitive, specific to HCV, and has a linear dynamic range of 10–100,000,000 units/ml in comparative studies to 228 Roche COBAS Amplicor assay.§ Briefly, a 4-μl aliquot of extracted RNA was added to a one-step RT-PCR reaction mixture containing sense and antisense primers specific for HCV and a TaqMan probe (proprietary sequences; LabCorp). The cycle at which the amplification plot crossed the threshold was defined as the threshold cycle (CT) and was predictive of the number of HCV RNA copies in the sample. A standard curve was calculated for quantification by using serial 10-fold dilutions of a reference HCV (RNA+) sample.
Results
HCV-E2 Binding to L-SIGN and DC-SIGN.
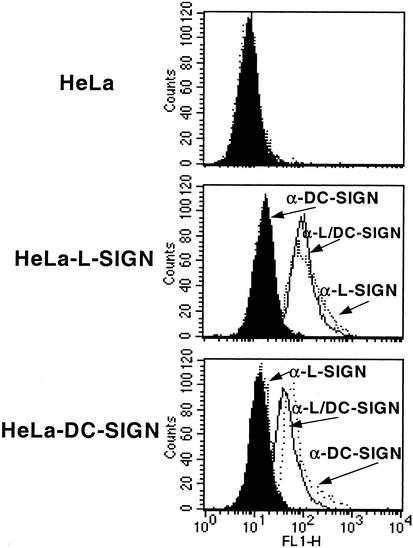
We first generated HeLa cells that stably express L-SIGN and DC-SIGN at the cell surface, as determined by flow cytometry using a panel of mAbs (Fig. 1). Because these mAbs recognize conformational epitopes (18, 19), their binding indicates that L-SIGN and DC-SIGN are expressed in native form on the HeLa transfectants.
Figure 1.
Cell-surface expression of L-SIGN and DC-SIGN in stably transfected HeLa cell lines. Flow cytometry was performed by using the cross-reactive mAb 612X (line), which binds to both transfected cell lines, the L-SIGN-specific mAb 614L (dotted line), and DC-SIGN-specific mAb 507D (dashed line), which bind only to the HeLa-L-SIGN or HeLa-DC-SIGN transfectants, respectively. Binding of an isotype-matched control mAb is indicated by filled histogram, and one representative result out of three independent analyses is shown.
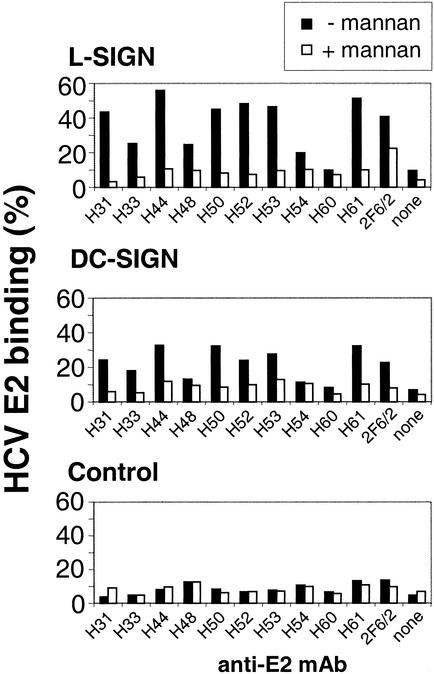
To investigate the binding of HCV-E2 to L-SIGN and DC-SIGN, we used a flow cytometric adhesion assay similar to that described for HIV-1 gp120 (8). Purified HCV-E2 protein was captured on fluorescent beads by using a panel of anti-E2 and control mAbs. The E2-coated beads bound efficiently to HeLa transfectants but not to parental HeLa (Fig. 2). Background levels of binding were observed for E2 and an isotype-matched mAb of irrelevant specificity and for anti-E2 mAb in the absence of E2 (data not shown). Binding levels depended on the anti-E2 mAb used for coating the beads (Fig. 2), a finding that could reflect differences in E2 capture efficiency of the mAbs or in their masking/presentation of the epitopes recognized by L-SIGN and DC-SIGN. L-SIGN and DC-SIGN showed similar patterns of reactivity for the various E2–mAb combinations.
Figure 2.
L-SIGN and DC-SIGN transfectants bind HCV-E2. L-SIGN (Top), DC-SIGN (Middle), and control HeLa cells (Bottom) were incubated with HCV-E2-coated beads prepared by using a panel of anti-E2 mAbs. Adhesion was quantified by fluorescence-activated cell sorter analysis in the presence of adherence buffer (filled bars) and was blocked by mannan (open bars). Different anti-E2 mAbs are indicated on the x axis, and the y axis represents the percentage of cells that have bound beads. One representative experiment out of three is shown.
E2 binding to both SIGN molecules was efficiently inhibited by mannan (Fig. 2), which binds to the lectin domain of L-SIGN and DC-SIGN. Binding was also abrogated by EDTA and EGTA (data not shown), which are chelators for the calcium ions required for the structural integrity and carbohydrate-binding properties of the C-type lectins.
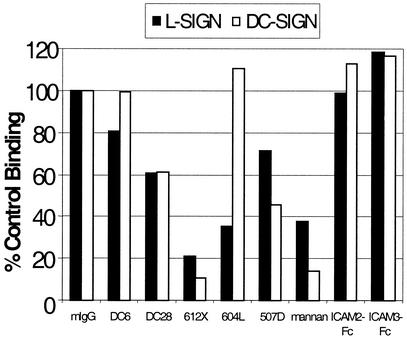
We also tested a panel of anti-L-SIGN and anti-DC-SIGN mAbs for their effect on E2 binding (Fig. 3). mAbs to the lectin domain of the SIGN molecules mediated similar levels of inhibition compared with mannan, whereas mAbs to the membrane-proximal heptad-repeat region were less effective. The patterns of inhibition of E2 binding by these mAbs largely parallel those observed for inhibition of HIV gp120 binding (16).
Figure 3.
Effect of mAbs or soluble ICAMs on adhesion of HCV-E2 to L-SIGN or DC-SIGN. HeLa cells expressing L-SIGN (filled bars) or DC-SIGN (open bars) were incubated with individual mAbs that bind the repeat region (DC6 and DC28) or the lectin-binding domain (612X, 604L, and 507D) or soluble ICAM-Fc conjugates as described. Binding of E2 beads was quantified by fluorescence with a FACScan machine, and results were normalized to isotype control (mIgG) levels. One representative data set from three experiments is shown.
Soluble ICAM-2 and ICAM-3 Fc fusion proteins had little effect on E2 binding to either SIGN molecule (Fig. 3). Studies have suggested differences in the recognition of gp120 and ICAM-3 by DC-SIGN (19, 20), and a similar situation may apply to HCV. Similarly, anti-E2 mAbs did not inhibit binding of E2 beads to either SIGN molecule (data not shown). This finding is consistent with lectin recognition of glycans distributed over the surface of E2, and the attendant difficulty of blocking such interactions with monospecific agents. Similarly, gp120 binding to DC-SIGN is not blocked either by anti-gp120 mAbs or by mutation of individual N-linked glycosylation sites on gp120 (21).
HCV Virus Binding to L-SIGN and DC-SIGN.
We next compared the SIGN molecules for their abilities to bind HCV virions. In the absence of a method for culturing HCV in vitro, we developed an assay to measure the interaction of the SIGN molecules with HCV virions present in the sera of infected individuals. There are no prior reports on the binding of naturally occurring viruses to either L-SIGN or DC-SIGN.
In our virus-binding assay, HCV-RNA positive or -negative sera were combined for 1 h with HeLa cells expressing L-SIGN, DC-SIGN, or neither receptor. After removal of unbound virus and RNA extraction, HCV genomes were detected by real-time PCR (TaqMan) or by RT-PCR followed by qualitative Southern blot.
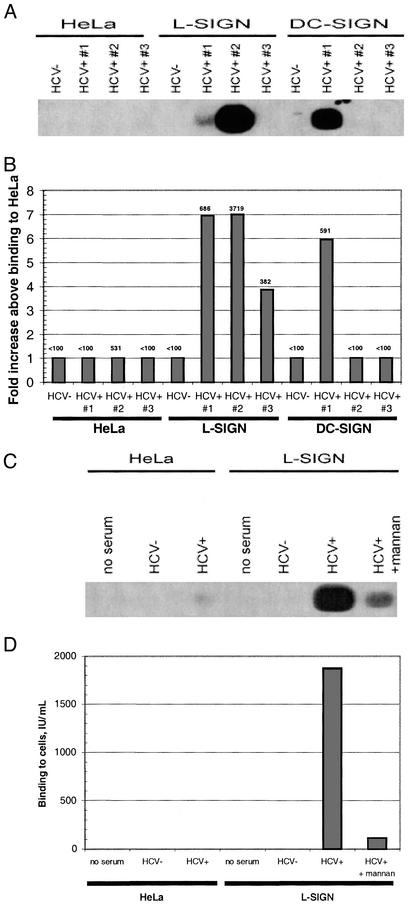
L-SIGN transfectants specifically bound three of three HCV-positive sera as determined by TaqMan analysis, whereas DC-SIGN mediated specific binding for one of three sera. The levels of virus binding to L-SIGN ranged from 4- to 7-fold greater than the background levels observed for parental HeLa (Fig. 4 A and B). HCV binding to L-SIGN was abrogated by >90% after mannan treatment (Fig. 4 C and D). Overall, there was a good concordance in results obtained in the hybridization and TaqMan assays, although the latter was more quantitative and sensitive to detect the low levels of binding (Fig. 4).
Figure 4.
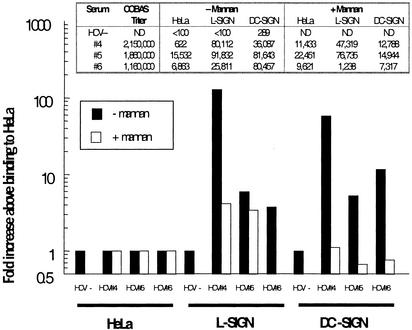
L-SIGN and DC-SIGN bind to HCV virions from infected patients. HeLa transfectants or control cells were incubated with sera from three HCV RNA+ patients. HCV sera RNA titers (copies per ml) were no. 1, 850,000; no. 2, 242,000; and no. 3, 161,000 by COBAS MONITOR assay (Roche Molecular Systems). After washing, cells were lysed and RNA was extracted. HCV RNA was measured by a qualitative RT-PCR and Southern blot assay (A) or by a quantitative real-time PCR assay (B). Data are presented as fold increase above HeLa control cell binding for each matched sera, and absolute values (units/ml) are depicted for each sample. Binding to L-SIGN was inhibited by mannan. The cells were preincubated with mannan before addition of serum 2 as described previously. Bound HCV RNA was extracted and analyzed either by Southern blot (C) or quantitative real-time PCR (D).
During natural infection, HCV serum titers show significant interpatient variation (1). To investigate further the breadth and robustness of HCV binding to L-SIGN and DC-SIGN, we repeated the assay by using sera from patients with higher viral loads. For these, both L- and DC-SIGN mediated specific, mannan-inhibitable binding for three of three sera (Fig. 5), with the highest-titered sera exhibiting 129- and 58-fold enhanced binding to L-SIGN and DC-SIGN cells, respectively. Only background signals were observed for HCV-negative sera. In toto, L-SIGN specifically bound HCV for six of six donor sera independent of viral load, whereas DC-SIGN mediated binding in four of six cases with a bias toward high-titered sera.
Figure 5.
Inhibition of HCV virion binding to L-SIGN and DC-SIGN by mannan. Cells were incubated with sera from three HCV RNA+ patients as described in Fig. 4. HCV sera RNA titers (copies per ml) were no. 4, 2.15 million; no. 5, 1.86 million; and no. 6, 1.16 million. Each serum was incubated with cells that had been pretreated with adherence buffer (filled bars) or mannan (open bars), and bound RNA was analyzed by real-time PCR as described. Fold increases above HeLa cell binding are indicated in the graph, and absolute levels (units/ml) are depicted (Inset).
In addition, mAbs to the lectin domains of the SIGN molecules inhibited virus binding to L-SIGN and DC-SIGN by 78% and 91%, respectively. In contrast, mAbs to E2 had no effect on virus binding (data not shown). The effects of mannan, anti-SIGN mAbs, and anti-E2 mAbs on native virion binding to SIGNs were concordant with those observed for purified E2 protein, and thus support the notion that E2 plays a major role in mediating HCV binding to the SIGN molecules.
Discussion
Our findings demonstrate that HCV interacts specifically with L-SIGN and DC-SIGN, and that this interaction is mediated at least in part by E2. Binding was blocked by relevant inhibitors, including mannan, calcium chelators, and mAbs to L-SIGN and DC-SIGN. Intriguingly, L-SIGN was somewhat more efficient than DC-SIGN at capturing virions at low viral loads.
The interactions of HCV with L-SIGN and DC-SIGN in liver and lymph nodes may have important consequences for the virus life cycle. L-SIGN is expressed on liver sinusoidal endothelial cells (LSECs), which form the vessels that separate hepatic blood flow from hepatocytes. LSECs provide a diffusional barrier to viruses but also mediate transcytosis in a receptor-driven process (22). In addition to regulating transport, LSECs are nonmyeloid professional antigen-presenting cells that may contribute to hepatic immune surveillance in association with resident macrophages that adhere to the LSEC (23). Although LSECs and DCs use similar mechanisms for cross-presentation of antigens to CD8+ T cells, LSEC presentation results in tolerance rather than immunity (24).
Studies with another hepatotropic virus, hepatitis B virus (HBV), indicate that the surveillance functions of LSEC can be subverted by pathogens. Receptor-mediated transcytosis of HBV results in efficient delivery of virus from the circulation to the target cell population (23, 25) but the identity of the transcytosing receptor has not been determined. If operative in the setting of natural infection, this process could facilitate the establishment of initial as well as chronic infection. By analogy with the ability of DC-SIGN to facilitate HIV-1 infection in trans, it also is possible that L-SIGN promotes infection of adjacent hepatocytes after transcytosis of virus.
The patterns of HCV binding and inhibition suggest that the interaction is mediated by high-mannose glycans on HCV and E2. That is, binding was competitively inhibited by mannan and by mAbs to the lectin domain of the SIGN molecules. E1 and other molecules on the native virion envelope may contribute to binding but the absence of specific reagents precluded analysis in this study. Binding also was abrogated by chelators of the calcium ions that are required by these C-type lectins. However, binding was not inhibited by mAbs to other regions of the SIGN molecules or by anti-E2 mAbs, at least when used individually. Similar patterns of inhibition have been observed for HIV-1 gp120, whose binding to DC-SIGN has recently been shown to be mediated by high-mannose sugars (21). There are potential conformational differences between the purified glycoproteins (E2 or gp120) and the virion-associated oligomers to consider; however, the glycan-dependence of HIV and HCV binding to L-SIGN and DC-SIGN may cause the interactions to be less dependent on protein conformation.
Although HCV and HIV-1 interact with the SIGN molecules in qualitatively similar manners, important differences in the viral life cycles might subtly influence these interactions. Whereas HCV envelope proteins and budding virus are generally thought to localize to the ER and/or cis-Golgi compartments (26), HIV-1 particles assemble and bud at the cell surface (27). Thus, unlike HIV-1 gp120, virion-associated E2 is unlikely to encounter the full N-linked protein glycosylation processing pathway, which initiates with the transfer of presynthesized Glu-3Man9GlcNAc2 from dolichylidphosphate to the canonical N-linked glycosylation sites of the newly synthesized peptide. The terminal glucose residues are removed in the endoplasmic reticulum before transfer of the protein to the cis-Golgi, where the high-mannose residues are trimmed by α-mannosidases. Further trimming of mannose residues and the addition of complex glycans occurs in the medial- and trans-Golgi (28). The net result would be a decreased percentage of high-mannose structures on HIV-1 compared with HCV. This differential processing of the viral envelope glycans may influence their interactions with L-SIGN and/or DC-SIGN, whose selective recognition of model oligosaccharides has been mapped to specific amino acid differences within their lectin domains (29, 30).
It will be important to determine the molecular events that ensue after HCV binding to L-SIGN and DC-SIGN. Although model ligands are routed to late endosomes/lysosomes for degradation after DC-SIGN binding (31), DC-SIGN-bound HIV retains infectivity for prolonged periods (8). Remarkably, internalization is crucial for enhancement of infectivity, whereas simple tethering of virus is insufficient (32). The ability of DC-SIGN to chaperone endocytosed virus past the lysosomal pathway may be central to its function as a capture receptor for HIV, and similar principles may apply to HCV. Clearly, a scavenging receptor that targets virus for degradation would promote clearance rather than infection. Such considerations may cause receptors of similar binding specificities to have divergent effects on viral infectivity.
Immune system disorders such as cryoglobulinemia are the chief extrahepatic complications of HCV infection (33), and the interaction of E2 with CD81 on B cells has been posited to be a contributing factor (34). HCV transcripts have been observed at low levels in DCs and other lymphoid cells (35, 36) but these do not seem to represent significant reservoirs of HCV. However, L-SIGN and DC-SIGN interactions may contribute to immune dysregulation, including the impaired DC function observed in chronic HCV infection (37–39). Migratory DCs may also mediate trafficking of HCV to liver and other sites, and virus binding to DC-SIGN or L-SIGN may modulate HCV immunity to promote maintenance of chronic infection.
The events and receptors involved in HCV entry remain obscure, and progress has been hampered by the lack of a reliable means of culturing HCV in vitro. Although DC-SIGN is not a requisite receptor for HIV fusion, DC-SIGN expression increases the susceptibility of cells to HIV by reducing the threshold level of fusion receptor expression, thereby rendering otherwise impermissive cells susceptible to HIV infection (40). Similarly, DC-SIGN and L-SIGN confer permissivity to cells that are otherwise refractory to infection by Ebola and cytomegalovirus pseudoviruses (9, 11).
L-SIGN is unlikely to be the sole determinant of HCV liver tropism, because the receptor is also expressed in lymph nodes and is not expressed on hepatocytes. Thus, additional liver-specific factors are likely to play a critical role in HCV infection. Alternatively, HCV bound to L-SIGN may undergo differential processing in liver and lymph nodes. Coexpression of L-SIGN and DC-SIGN in term placenta may also contribute to vertical transmission of HCV (1).
HCV is one of a growing number of human pathogens shown to interact with L-SIGN and/or DC-SIGN. However, the specificity of these interactions is underscored by the observation that several viruses have envelopes that are glycosylated yet show little or no avidity for DC-SIGN and L-SIGN (11, 21). The findings in this report indicate that HCV has coopted host glycosylation pathways in ways that promote binding to L-SIGN and DC-SIGN.
In conclusion, we have demonstrated that HCV interacts specifically with L-SIGN and DC-SIGN, and this interaction is mediated, at least in part, by the viral envelope glycoprotein E2. L-SIGN thus represents a liver-specific receptor that avidly binds HCV envelope. These findings raise the hypothesis that interaction of HCV with these molecules in liver and lymph nodes may have important consequences for HCV infection and immunity.
Acknowledgments
We thank R. Klein, J. Dubuisson, R. Doms, M. Cohen, and the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program for reagents and technical assistance. This work was supported by Progenics Pharmaceuticals, Inc., and in part by NIH Grant AI051134 (to J.P.G.) and the Speaker's Fund for Biomedical Research (to T.D.).
Abbreviations
- LDL
low-density lipoprotein
- ICAM
intercellular adhesion molecule
- DC-SIGN
dendritic cell-specific ICAM-3-grabbing nonintegrin
- HCV
hepatitis C virus
- LSEC
liver sinusoidal endothelial cell
- L-SIGN
liver/lymph node-specific ICAM-3-grabbing nonintegrin
Note Added in Proof.
While this article was in press, two groups confirmed the interaction between the SIGN molecules and E2 in studies limited to recombinant forms of the HCV glycoproteins (41, 42).
Footnotes
Turnmire, C., 18th Annual Clinical Virology Symposium, April 30, 2002, Clearwater Beach, FL.
References
- 1.Lauer G M, Walker B D. N Engl J Med. 2001;345:41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 2.Doms R W, Moore J P. J Cell Biol. 2000;151:F9–F14. doi: 10.1083/jcb.151.2.f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sieczkarski S B, Whittaker G R. J Gen Virol. 2002;83:1535–1545. doi: 10.1099/0022-1317-83-7-1535. [DOI] [PubMed] [Google Scholar]
- 4.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner A J, Houghton M, Rosa D, Grandi G, et al. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 5.Agnello V, Abel G, Elfahal M, Knight G B, Zhang Q X. Proc Natl Acad Sci USA. 1999;96:12766–12771. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scarselli E, Ansuini H, Cerino R, Roccasecca R M, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. EMBO J. 2002;21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geijtenbeek T B, Torensma R, van Vliet S J, van Duijnhoven G C, Adema G J, van Kooyk Y, Figdor C G. Cell. 2000;100:575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- 8.Geijtenbeek T B, Kwon D S, Torensma R, van Vliet S J, van Duijnhoven G C, Middel J, Cornelissen I L, Nottet H S, KewalRamani V N, Littman D R, et al. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 9.Alvarez C P, Lasala F, Carrillo J, Muniz O, Corbi A L, Delgado R. J Virol. 2002;76:6841–6844. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colmenares M, Puig-Kroger A, Muniz P O, Corbi A L, Rivas L. J Biol Chem. 2002;277:36766–36769. doi: 10.1074/jbc.M205270200. [DOI] [PubMed] [Google Scholar]
- 11.Halary F, Amara A, Lortat-Jacob H, Messerle M, Delaunay T, Houles C, Fieschi F, Arenzana-Seisdedos F, Moreau J F, Dechanet-Merville J. Immunity. 2002;17:653–664. doi: 10.1016/s1074-7613(02)00447-8. [DOI] [PubMed] [Google Scholar]
- 12.Pohlmann S, Soilleux E J, Baribaud F, Leslie G J, Morris L S, Trowsdale J, Lee B, Coleman N, Doms R W. Proc Natl Acad Sci USA. 2001;98:2670–2675. doi: 10.1073/pnas.051631398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bashirova A A, Geijtenbeek T B, van Duijnhoven G C, van Vliet S J, Eilering J B, Martin M P, Wu L, Martin T D, Viebig N, Knolle P A, et al. J Exp Med. 2001;193:671–678. doi: 10.1084/jem.193.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deleersnyder V, Pillez A, Wychowski C, Blight K, Xu J, Hahn Y S, Rice C M, Dubuisson J. J Virol. 1997;71:697–704. doi: 10.1128/jvi.71.1.697-704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flint M, Thomas J M, Maidens C M, Shotton C, Levy S, Barclay W S, McKeating J A. J Virol. 1999;73:6782–6790. doi: 10.1128/jvi.73.8.6782-6790.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baribaud F, Pohlmann S, Leslie G, Mortari F, Doms R W. J Virol. 2002;76:9135–9142. doi: 10.1128/JVI.76.18.9135-9142.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young K K, Resnick R M, Myers T W. J Clin Microbiol. 1993;31:882–886. doi: 10.1128/jcm.31.4.882-886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jameson B, Baribaud F, Pohlmann S, Ghavimi D, Mortari F, Doms R W, Iwasaki A. J Virol. 2002;76:1866–1875. doi: 10.1128/JVI.76.4.1866-1875.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu L, Martin T D, Vazeux R, Unutmaz D, KewalRamani V N. J Virol. 2002;76:5905–5914. doi: 10.1128/JVI.76.12.5905-5914.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geijtenbeek T B, van Duijnhoven G C, van Vliet S J, Krieger E, Vriend G, Figdor C G, van Kooyk Y. J Biol Chem. 2002;277:11314–11320. doi: 10.1074/jbc.M111532200. [DOI] [PubMed] [Google Scholar]
- 21.Hong P W, Flummerfelt K B, De Parseval A, Gurney K, Elder J H, Lee B. J Virol. 2002;76:12855–12865. doi: 10.1128/JVI.76.24.12855-12865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tavassoli M, Kishimoto T, Kataoka M. J Cell Biol. 1986;102:1298–1303. doi: 10.1083/jcb.102.4.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knolle P A, Gerken G. Immunol Rev. 2000;174:21–34. doi: 10.1034/j.1600-0528.2002.017408.x. [DOI] [PubMed] [Google Scholar]
- 24.Limmer A, Ohl J, Kurts C, Ljunggren H G, Reiss Y, Groettrup M, Momburg F, Arnold B. Nat Med. 2000;6:1348–1354. doi: 10.1038/82161. [DOI] [PubMed] [Google Scholar]
- 25.Breiner K M, Schaller H, Knolle P A. Hepatology. 2001;34:803–808. doi: 10.1053/jhep.2001.27810. [DOI] [PubMed] [Google Scholar]
- 26.Dubuisson J. Curr Top Microbiol Immunol. 2000;242:135–148. doi: 10.1007/978-3-642-59605-6_7. [DOI] [PubMed] [Google Scholar]
- 27.Frankel S S, Steinman R M, Michael N L, Kim S R, Bhardwaj N, Pope M, Louder M K, Ehrenberg P K, Parren P W, Burton D R, et al. J Virol. 1998;72:9788–9794. doi: 10.1128/jvi.72.12.9788-9794.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright A, Morrison S L. Trends Biotechnol. 1997;15:26–32. doi: 10.1016/S0167-7799(96)10062-7. [DOI] [PubMed] [Google Scholar]
- 29.Feinberg H, Mitchell D A, Drickamer K, Weis W I. Science. 2001;294:2163–2166. doi: 10.1126/science.1066371. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell D A, Fadden A J, Drickamer K. J Biol Chem. 2001;276:28939–28945. doi: 10.1074/jbc.M104565200. [DOI] [PubMed] [Google Scholar]
- 31.Engering A, Geijtenbeek T B, van Vliet S J, Wijers M, van Liempt E, Demaurex N, Lanzavecchia A, Fransen J, Figdor C G, Piguet V, et al. J Immunol. 2002;168:2118–2126. doi: 10.4049/jimmunol.168.5.2118. [DOI] [PubMed] [Google Scholar]
- 32.Kwon D S, Gregorio G, Bitton N, Hendrickson W A, Littman D R. Immunity. 2002;16:135–144. doi: 10.1016/s1074-7613(02)00259-5. [DOI] [PubMed] [Google Scholar]
- 33.Dammacco F, Sansonno D, Piccoli C, Racanelli V, D'Amore F P, Lauletta G. Semin Liver Dis. 2000;20:143–157. doi: 10.1055/s-2000-9613. [DOI] [PubMed] [Google Scholar]
- 34.Flint M, McKeating J A. Rev Med Virol. 2000;10:101–117. doi: 10.1002/(sici)1099-1654(200003/04)10:2<101::aid-rmv268>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 35.Navas M C, Fuchs A, Schvoerer E, Bohbot A, Aubertin A M, Stoll-Keller F. J Med Virol. 2002;67:152–161. doi: 10.1002/jmv.2204. [DOI] [PubMed] [Google Scholar]
- 36.Mellor J, Haydon G, Blair C, Livingstone W, Simmonds P. J Gen Virol. 1998;79:705–714. doi: 10.1099/0022-1317-79-4-705. [DOI] [PubMed] [Google Scholar]
- 37.Kanto T, Hayashi N, Takehara T, Tatsumi T, Kuzushita N, Ito A, Sasaki Y, Kasahara A, Hori M. J Immunol. 1999;162:5584–5591. [PubMed] [Google Scholar]
- 38.Bain C, Fatmi A, Zoulim F, Zarski J P, Trepo C, Inchauspe G. Gastroenterology. 2001;120:512–524. doi: 10.1053/gast.2001.21212. [DOI] [PubMed] [Google Scholar]
- 39.Auffermann-Gretzinger S, Keeffe E B, Levy S. Blood. 2001;97:3171–3176. doi: 10.1182/blood.v97.10.3171. [DOI] [PubMed] [Google Scholar]
- 40.Lee B, Leslie G, Soilleux E, O'Doherty U, Baik S, Levroney E, Flummerfelt K, Swiggard W, Coleman N, Malim M, et al. J Virol. 2001;75:12028–12038. doi: 10.1128/JVI.75.24.12028-12038.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lozach, P.-Y., Lortat-Jacob, H., de Lacroix de Lavalette, A., Staropoli, I., Foung, S., Amara, A., Houlès, C., Fieschi, F., Schwartz, O., Virelizier, J.-L., et al. (2003) J. Biol. Chem., 10.1074/jbc.M301284200. [DOI] [PubMed]
- 42.Pohlmann S, Zhang J, Baribaud F, Chen Z, Leslie G J, Lin G, Granelli-Piperno A, Doms R W, Rice C M, McKeating J A. J Virol. 2003;77:4070–4080. doi: 10.1128/JVI.77.7.4070-4080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]