Abstract
Recent research in the field of nanometer-scale electronics has focused on the operating principles of small-scale devices and schemes to realize useful circuits. In contrast to established “top-down” fabrication techniques, molecular self-assembly is emerging as a “bottom-up” approach for fabricating nanostructured materials. Biological macromolecules, especially proteins, provide many valuable properties, but poor physical stability and poor electrical characteristics have prevented their direct use in electrical circuits. Here we describe the use of self-assembling amyloid protein fibers to construct nanowire elements. Self-assembly of a prion determinant from Saccharomyces cerevisiae, the N-terminal and middle region (NM) of Sup35p, produced 10-nm-wide protein fibers that were stable under a wide variety of harsh physical conditions. Their lengths could be roughly controlled by assembly conditions in the range of 60 nm to several hundred micrometers. A genetically modified NM variant that presents reactive, surface-accessible cysteine residues was used to covalently link NM fibers to colloidal gold particles. These fibers were placed across gold electrodes, and additional metal was deposited by highly specific chemical enhancement of the colloidal gold by reductive deposition of metallic silver and gold from salts. The resulting silver and gold wires were ≈100 nm wide. These biotemplated metal wires demonstrated the conductive properties of a solid metal wire, such as low resistance and ohmic behavior. With such materials it should be possible to harness the extraordinary diversity and specificity of protein functions to nanoscale electrical circuitry.
Nanometer-scale structures are of great interest as potential building blocks for future electronic devices. One significant challenge is the construction of nanowires to enable the electrical connection of such structures. Biomolecules may provide a solution to the difficulty of manufacturing wires at this scale because they naturally exist in the nanometer size range (1–13). In addition, biomolecules can exhibit self-assembly, which would remove the need to individually pattern them into structures, and greatly aid the mass production of nanostructures.
The intrinsic properties of biomolecules are generally unsuitable for conducting electrical currents; therefore they are usually combined with an inorganic compound that acts as a conductor. This conductivity is achieved through a hierarchical assembly process where the first step is to form a regular scaffold by using biological molecules followed by a second step where the inorganic components are guided to aggregate selectively along the scaffold (2, 8, 14–17).
The first biomolecular templates used for microstructures were phospholipid tubules (18), and since then other self-assembling rod-like structures have been assessed for their strengths and weaknesses as nanostructural templates, including DNA, bacteriophages, and microtubules (2, 3, 5, 9, 13, 14, 19–24). These materials have many positive characteristics as nanostructure materials. DNA has good recognition capabilities, mechanical rigidity, and amenability to high-precision processing. Recent studies using DNA as a template for gold plating produced wires with ohmic conductivity [resistance, R = 86 Ω and a linear current–voltage (I–V) curve] (23); however, DNA is unstable under conditions (pH 10–12 and temperatures >60°C) necessary for industrial metallization. Bacteriophages are expected to have similar chemical and thermal constraints and they do not readily polymerize to form continuous fibers.
Proteins are an attractive alternative material for the construction of nanostructures. Their physical size is appropriate and they are capable of many types of highly specific interactions; indeed, as many as 93,000 different protein–protein interactions have been predicted in yeast (25–27). Moreover, proteins provide an extraordinary array of functionalities that could potentially be coupled to electronic circuitry in the building of nanoscale devices. Protein tubules have the advantage of a high degree of stiffness and greater stability than DNA. In addition they exhibit good adsorption to technical substrates like glass, silicon oxide, or gold. Various protein tubules such as microtubules and rhapidosomes (3, 14, 22) have been assessed, but all have important limitations such as relatively high resistance once metallized (of the order of 200 kΩ) (3), morphology that cannot withstand metallization under industrial conditions, or they aggregate once they are metallized (14). Therefore, there is a need to explore alternative biomaterials.
A good candidate protein for circumventing these problems is the N-terminal and middle region (NM) of yeast Saccharomyces cerevisiae Sup35p. NM forms self-assembling β-sheet-rich amyloid fibers that are suitably sized and shaped for nanocircuitry with diameters of 9–11 nm (28). The highly flexible structure of soluble NM rapidly converts to form amyloid fibers when it associates with preformed fibers that act as seeds for fiber formation (29–31). The fibers grow by extension from either end (32), and this bidirectional formation is likely to be useful for forming varied fiber patterns: a crucial property for the production of circuitry.
NM has several properties that make it amenable to the manufacturing process. Once the fibers are formed it has a higher than average chemical stability as demonstrated by its resistance to proteases and protein denaturants (29). Indeed, PrP, the mammalian prion counterpart of Sup35p, is infamous for its extraordinary resistance to destruction. (However, neither Sup35p nor NM are infectious to humans and therefore can be handled safely.) The stability of NM suggests that it might even withstand diverse metallization procedures necessary for creating electric circuits in industrial settings. In addition, NM fibers do not form aggregates as readily as other amyloids. Furthermore, under some circumstances such as different surface treatments, methods of fiber deposition, and solutions in which they are suspended, NM fibers tend not to aggregate with each other (T.S. and S.L.L., unpublished observation). The solubility of NM in physiological buffers greatly facilitates handling before and during fiber formation (32). These characteristics of NM led us to investigate the use of NM fibers as templates for nanocircuitry.
In this study, the NM of the yeast prion determinant Sup35p was successfully used as a template for constructing wires with nanometer dimensions. The nanowires were able to conduct electricity with low resistance and produced linear current–voltage curves by using a protein template. These results demonstrate that self-assembling NM molecules have great potential for the construction of nanoscale devices.
Materials and Methods
Protein Expression and Purification.
NM/NMK184C was recombinantly expressed in Escherichia coli BL21 [DE3] as described (32) and purified by chromatography with Q-Sepharose (Amersham Pharmacia), hydroxyapatite (Bio-Rad), and Poros HQ (Roche Molecular Biochemicals) as a final step. All purification steps were performed in the presence of 8 M urea.
Fiber Assembly.
Solutions with protein (NM or NMK184C) concentrations >25 μM were rotated at high speed (60 rpm) to increase turbulence and surface area. At this protein concentration many seeding events initiate simultaneous fiber assembly, which results in many short fibers (average fiber length from 60 to 200 nm). These short fibers were then used to seed further soluble NM. Fibers of different average length were generated by changing the ratios of seed to soluble NM (keeping the soluble NM concentration constantly at 5 μM).
Analysis of Fiber Structure.
After fiber assembly, three techniques were used to examine the fibrous state of NM: far-UV CD, Congo red (CR) binding, and atomic force microscopy (AFM). CD spectra were obtained by using a Jasco (Easton, MD) 715 spectropolarimeter equipped with a temperature control unit. All spectra were taken with a 0.1-cm pathlength quartz cuvette (Hellma, Forest Hills, NY) in 5 mM potassium phosphate (pH 7.4)/150 mM NaCl (standard buffer). The settings for wavelength scans were 5-nm bandwidth; 0.25-sec response time; speed, 20 nm/min; and four accumulations.
CR-binding was carried out as described (28). Proteins were diluted to a final concentration of 1 μM into standard buffer plus 10 μM CR and incubated for 1 min at 25°C before measuring the absorbance at 540 and 477 nm.
Samples for AFM analysis were placed on freshly cleaved mica attached to 15-mm AFM sample disks (Ted Pella, Redding, CA). After 3 min of adsorption at 25°C, disks were rinsed once with buffer and twice with Millipore filtered distilled H2O. The samples were then allowed to air dry. Contact and tapping-mode imaging were performed on a Digital Instruments (Santa Barbara, CA) multimode scanning probe microscope (Veeco, Santa Barbara, CA) by using long, thin-leg standard silicon nitride (Si3N4) probes for contact mode and standard etched silicon probes for tapping mode.
Analysis of Fiber Stability.
To investigate fiber stability at elevated temperatures, NM fibers were incubated in standard buffer for 90 min at 98°C, before assessment by CD, CR binding, and AFM. The stability of the fibers was also tested under other temperatures for varying lengths of time, i.e., several months at 25°C and after freezing at −20°C and −80°C. Chemical stability was tested by the addition of high concentrations of salt (2.5 M NaCl) or denaturants [8 M urea or 2 M guanidiniumchloride (Gdm⋅Cl)] to the standard buffer (5 mM sodium phosphate, pH 6.8) and assessed by CD, CR binding, and AFM. NM fiber stability in strong alkaline or acidic solutions and in organic solvents was tested by immobilizing the fibers on mica, air-drying them, and treating them with NaOH (pH 10), HCl (pH 2), or 100% ethanol for several hours. These conditions were not compatible with CD and CR-binding assessment, therefore only AFM was used.
Electrode Assembly and Visualization.
Electrodes were prepared on Si3N4 membrane substrates as described (33). Transmission electron microscopy (TEM) images of electrodes in the absence and presence of protein fibers were obtained with a CM120 transmission electron microscope (Phillips, FEI, Hillsboro, OR) with a LaB6 filament, operating at 120 kV in low-dose mode at a magnification of ×45,000, and recorded on Kodak SO163 film. Alternatively, samples were imaged by AFM in contact mode. Conductivity measurements were performed as described (33).
Gold Toning.
Monomaleimido Nanogold (Nanoprobes, Yaphank, NY) with a particle diameter of 1.4 nm was covalently cross-linked to NMK184C fibers as described (32). The 1.4-nm Nanogold particles were then subjected to “gold toning.” There, the Nanogold particles act first as promoters for reducing silver ions from a solution. After subjecting the Nanogold-labeled fibers to silver enhancement with LI Silver (Nanoprobes; enhancement was performed according to the manufacturer's protocol; solutions A and B were mixed in a 1:1 ratio and incubated at 25°C), the resulting silver-coated fiber-bound Nanogold particles were gold-enhanced with GoldEnhance LM (Nanoprobes; enhancement was performed according to the manufacturer's protocol; solutions A–D were mixed in a 1:1:1:1 ratio and incubated at 25°C). Exposure times varied from 3 min of silver enhancement and 3 min of gold enhancement to 25 min of silver enhancement and 25 min of gold enhancement.
Results
NM Fibers Are Highly Stable.
To investigate the feasibility of using NM fibers in building nanoscale devices we first tested their stability under extreme conditions such as those that might be encountered in industrial manufacturing processes. NM fibers assembled at physiological pH and room temperature were assayed for stability by three established techniques that differentiate between NM in its soluble and amyloid state. Far-UV CD distinguishes the β-sheet-rich secondary structure of NM fibers from the random coil-rich structure of soluble NM. CR exhibits a spectral shift when it intercalates into the cross-pleated β-strands of NM fibers, which is not observed with soluble NM. AFM and EM were used to monitor the maintenance of fiber morphology.
NM fibers were incubated in standard buffer at high and low temperatures, in the absence or presence of high salt (2.5 M NaCl), and in denaturants (8 M urea or 2 M guanidiniumchloride, Gdm⋅Cl). By all three techniques (data not shown), fibers were stable in standard buffer after incubation for 90 min at 98°C, for several months at 25°C, and after freezing at −20° and −80°C. (Some shearing of long fibers occurred with repeated cycles of freeze-thawing.) Fibers were completely stable to prolonged incubation in the absence of salt and at 2.5 M salt. They dissociated in <2 h at concentrations of Gdm⋅Cl >4 M but remained intact in the presence of 2 M Gdm⋅Cl and 8 M urea.
To test whether NM fibers can withstand strong alkaline or acidic solutions and incubation in organic solvents, which are incompatible with CD and CR-binding assays, we immobilized NM fibers on mica, imaged them by AFM, incubated them with test solutions [NaOH (pH 10), HCl (pH 2), or 100% ethanol], and reimaged the same fibers. No morphological changes were apparent after any of these treatments (data not shown). Therefore, NM fibers show unusually high chemical and thermal stability for a biological material.
NM Fibers of Variable Lengths Can Be Produced Repeatedly.
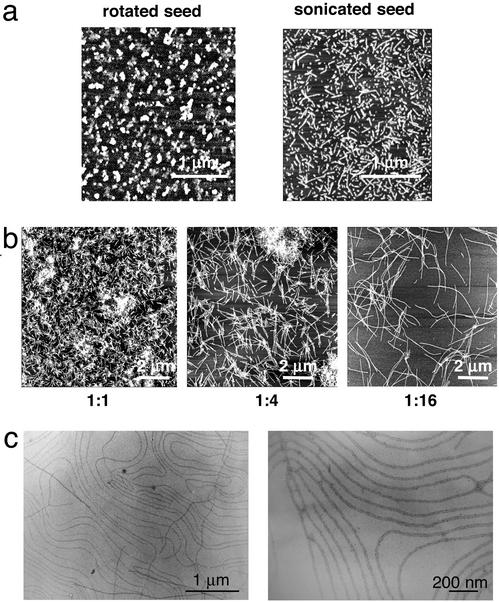
Our studies of the NM amyloid fibers have provided insights into how fibers assemble and how assembly can be controlled (28–30). The rate of fiber formation by purified soluble NM is dramatically increased by the addition of preformed NM fibers, which seed assembly from their ends (31, 32). We were able to generate pools of fibers with different average lengths by simple manipulation of the assembly conditions. First we produced extremely short fibers (60–200 nm, Fig. 1a Left) by rotating solutions with high NM protein concentrations (>25 μM) at high speeds to increase turbulence and surface area. These conditions produced short fibers by greatly increasing the efficiency of seeding (such that it dominates over assembly), rather than by simply shearing fibers after they had assembled. Indeed, when preformed fibers were sheared by the much more physically disruptive force of sonication, the resulting fibers had longer average lengths and a much more heterogeneous distribution (Fig. 1a Right). The resulting fibers showed lengths varying from 100 to 500 nm (32). The short fibers produced by vigorous rotation of high concentrations of NM were used to seed further soluble NM. By simply changing the ratios of seed to soluble NM and by controlling the assembly temperatures we were able to generate fibers of different average length (Fig. 1b). At seed to soluble NM ratios of 1:1 (wt/wt), fibers showed an average length of 500 ± 100 nm (Fig. 1b). Increasing the soluble NM concentration increased fiber lengths. At ratios of 1:16 of seed to soluble NM, fibers were ≈5 ± 1 μm long (Fig. 1b). Ratios of 1:64 led to even longer fibers but these had more variable lengths (10 μm up to several hundred micrometers; data not shown). A remarkable phenomenon that was sometimes observed when long fibers were prepared for microscopy was their alignment next to each other without any external manipulation (Fig. 1c). This alignment varied with the buffers in which fibers were suspended and the manner in which the surfaces were prepared in a fashion that has not been completely deciphered. Once the relevant parameters have been better defined, this unusual characteristic of the amyloid fibers will undoubtedly prove valuable in manufacturing processes, where the intrinsic tendency of more typical amyloids to form intractable clumps presents a significant barrier.
Figure 1.
NM fiber formation. (a Left) NM fibers assembled at a protein concentration of >25 μM with fast rotation, which results in many short fibers (average fiber length from 60 to 200 nm). (Right) NM fibers assembled at a protein concentration of 5 μm with fast rotation, which results in long fibers. Fibers were sonciated as described (32) and both were imaged on mica by AFM. (b) The conditions of a Left led to controlled, reproducible formation of amyloid fibers with defined fiber length as visualized on mica by AFM. At seed:soluble NM ratios of 1:1 (wt/wt), resulting fibers showed an average length of 500 ± 100 nm. Increasing the soluble NM concentration led to increased fiber lengths. At ratios of 1:16 of seed:soluble NM the resulting fibers were ≈5 ± 1 μm long. (c) Natural alignment of NM fibers. (Left) Lower magnification. (Right) Higher magnification (imaged by TEM).
NM Fibers Are Insulators.
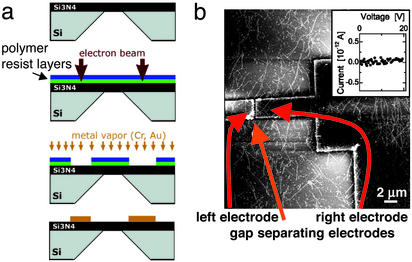
To examine the electrical behavior of the protein fibers, Si3N4 membrane substrates were prepared, which allowed for in-plane electrode fabrication, low-temperature transport measurements, and direct visualization by TEM (33). The electrodes were constructed by spinning polymer resist layers onto Si3N4 substrates and exposing them to a scanned electron beam. The electron beam demarcated the electrode sites (Fig. 2a). The exposed polymer was etched away, and gold vapor was applied to fill the resulting gaps. Finally, the remaining polymer was dissolved away, leaving the gold in the pattern inscribed by the electron beam. Typically, gaps between electrodes were 2–10 μm. NM fibers with polydispersed lengths (>2 μm) were randomly deposited on the electrodes. Binding of the protein fibers to the electrodes and bridging of the gap between the electrodes were confirmed by AFM (Fig. 2b). Current (I) and voltage (V) readings were taken as electricity was applied to the electrodes and the I–V curve for bare fibers showed a very high resistance (R > 1014Ω), with no measurable conductivity (Fig. 2b Inset). Thus, NM amyloid fibers are by themselves good insulators.
Figure 2.
NM fiber electrical properties. (a) Electrodes were prepared by spinning polymer resist layers onto a silicon nitride substrate. Theses layers were exposed to a scanned electron beam to pattern the sites of the electrodes. The resulting exposed sites were filled by applying gold vapor. Finally, the remaining polymer was dissolved away, leaving the gold electrodes. (b) In-plane electrodes were prepared on Si3N4 membrane substrates (ref. 33 and a). Nonlabeled fibers with lengths >2 μm were randomly deposited on substrates with lithographically patterned electrodes imaged by AFM. With these conditions, the I–V curve shows no measurable conductivity with a high resistance of R > 1014 Ω (Inset), indicating that NM amyloid fibers are insulators.
NM Fibers Can Be Converted into Conducting Nanowires with Low Ohmic Resistance.
NM fibers were converted to conducting nanowires by a multistep process. We used a derivative of NM that was genetically engineered to contain a cysteine residue that remained accessible after fiber formation (32). This derivative, NMK184C, assembled in vitro with kinetics that were indistinguishable from those of the wild-type protein and led to fibers with the same physical properties (32). Monomaleimido Nanogold (Nanoprobes), which has the chemical specificity to form covalent links with the sulfhydrl groups of cysteine residues, was covalently cross-linked to NMK184C fibers. The gold particles had a diameter of 1.4 nm and their distribution along the surface of the NMK184C fibers was confirmed by TEM (32). Importantly, linking Nanogold covalently to NM fibers affected neither fiber stability nor fiber morphology (data not shown).
As the distance between the cysteine residues is ≈3–5 nm and the Nanogold particles have a diameter of only 1.4 nm, it was necessary to bridge the particles with metal to gain conductivity. First, we used GoldEnhance LM (Nanoprobes), by which gold ions are deposited from the solution onto the preexisting particles of Nanogold, followed by chemical reduction of the gold ions to form metallic gold. This process itself was inefficient in gaining conductivity, because binding and reducing the soluble gold ions did not fill all of the gaps between the covalently linked Nanogold particles as determined by TEM and AFM (data not shown).
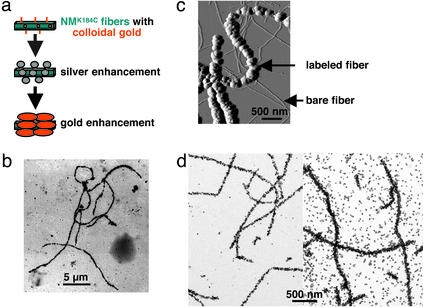
A different enhancement protocol (gold toning, Fig. 3a) proved much more efficient. There, the Nanogold particles on the labeled fibers acted as promoters for reducing silver ions (LI Silver, Nanoprobes) from a solution. The resulting silver-coated fiber-bound Nanogold particles were then gold-enhanced with GoldEnhance LM. This gold-toning technique led to fibers with densely packed gold particles (Fig. 3b). The gold-toned fibers showed a significant increase in diameter/height, from 9–11 nm (bare fibers) to 80–200 nm (labeled fibers) (Fig. 3c), with the diameter/height of the resulting fiber strictly depending on the length of exposure time of both the silver and the gold enhancement solution (Fig. 3d and data not shown). The diameters of the metal wires varied somewhat with different batches of fibers and gold- and silver-toning solutions but were extremely consistent within reactions, i.e., all were within a 10% range (Fig. 3d). Gold toning was remarkably specific for fibers that had been covalently labeled with Nanogold particles. When NMK184C fibers that were linked to Nanogold were incubated together with a large excess of unlabeled NMK184C fibers, the toning process was restricted to labeled fibers (Fig. 3c). Furthermore, the diameters of the wires were consistent within single experiments with fixed exposure times (Fig. 3d and data not shown). Therefore, controlling the enhancement exposure time controlled the thickness for the resulting gold wires.
Figure 3.
Gold and silver enhancement of NM fibers. (a) Long NMK184C fibers were assembled by seeding soluble NMK184C with short NMK184C fibers. Monomaleimido Nanogold was covalently cross-linked and the 1.4-nm Nanogold particles were subjected to gold toning. (b) Fibers were gold-toned on Si3N4 membrane substrates (33) by enhancing with LI Silver and GoldEnhance LM each for 5 min and imaged by TEM. (c) Gold toning is specific to labeled fibers. The resulting gold-toned fibers show a significant increase in height from 9–11 nm (bare fibers) to 80–200 nm (labeled fibers), imaged by AFM. (d) The height of the resulting fiber strictly depends on the length of exposure time of either the silver and/or the gold enhancement solution, imaged by AFM. (Left) Gold-labeled fibers on Si3N4 subjected to 3 min of silver enhancement and 5 min of gold enhancement, leading to a fiber diameter of 50 ± 5 nm. (Right) Fibers after 5 min of silver enhancement and 5 min of gold enhancement, leading to a fiber diameter of 100 ± 7 nm.
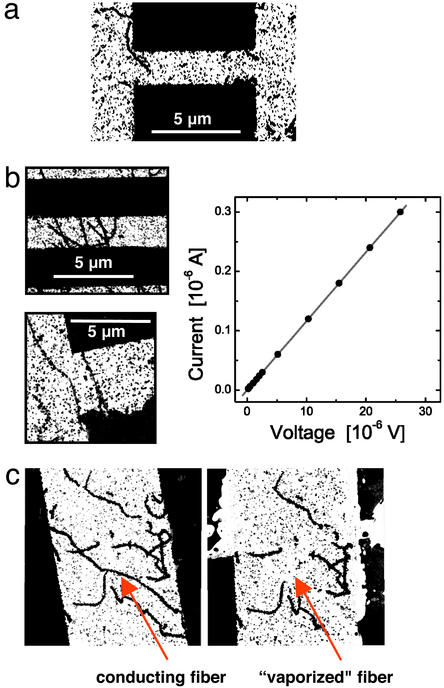
The electrical behavior of NM-templated metallic fibers was assessed by randomly depositing fibers with a length >2 μm and covalently attached Nanogold particles on patterned electrodes, followed by gold toning to form metallically continuous gold nanowires (Fig. 4). Although no background deposition of gold had been detected on unlabeled NM fibers deposited on mica, some gold deposition did occur when enhancement was performed on the Si3N4 electrodes. No conductivity was detected in cases where the gold nanowires did not bridge the electrode gap (Fig. 4a). In contrast, conductivity was readily detected when single or multiple gold-toned nanowires crossed the gap. I–V curves were linear, exhibiting ohmic conductivity with low resistance (R = 86 Ω for fibers with diameters of ≈100 nm; this resistance was exhibited in each of six repeated measurements with <1 Ω variation, and with one to four bridging nanowires). The resistance measurements were stable within tenths of ohms within any given fiber (Fig. 4b). Such an ohmic response indicates continuous, metallic connections across the sample. The low resistance is that expected for grain-boundary-dominated transport in a polycrystalline metal. In most cases the current was independent of the voltage scan direction and experiments could be repeated several times with the same pair of electrodes and the same nanowire (data not shown). Notably, in some instances fibers were vaporized from the electrodes when the voltage was increased after the initial conductivity measurements were finished (Fig. 4c). Bridging fibers were vaporized and did not reassemble but nonbridging fibers remained. In such cases conductivity was lost on remeasurement. This loss of conductivity confirmed that the bridging fibers were the active nanowires and demonstrated that they can act as fuses at higher voltages and currents.
Figure 4.
Electrical behavior of NM-templated metallic fibers. Gold- and gold-toned fibers were randomly deposited on patterned electrodes and imaged by TEM. (a) Gold nanowires that did not bridge the gap between two electrodes did not conduct. (b) Gold nanowires bridging the gap between two electrodes (Left) exhibit linear I–V curves (Right), demonstrating ohmic conductivity with low resistance of R = 86 Ω (the same for each). Such ohmic response is indicative of continuous, metallic connections across the sample. (c) In some experiments, after ramping up the voltage, reversing the scan direction no longer showed conductivity. Imaging revealed that the conducting nanowires were vaporized. Before (Left) and after (Right) conductivity measurement.
Discussion
A major obstacle in the manufacture of nanoscale electronics is identifying materials that provide properties amenable to industrial production. Biomaterials are at the forefront of this search; however, few have the diversity of positive characteristics required. In this study, we demonstrated that NM protein fibers are excellent candidates for nanocircuit construction. They are exceedingly good insulators without metal coating (R > 1014 Ω) and have very good electrical conductivity with gold and silver coating (R = 86 Ω) and linear I–V curves. Previously the least resistance achieved with metallized proteinaceous material was of the order of 200 kΩ, >1,000 times greater than the resistance for metallized NM fibers (3).
The width of the wires we produced was 80–200 nm, well below the dimensions accessible by standard electronic manufacturing methods. Having achieved the construction of wires with these dimensions, it is easy to envision methods to produce even thinner ones. The thickness of our wires was dictated by the relatively large amounts of silver and gold enhancement that were required to fill the gaps between the Nanogold particles attached to cysteine residues (Fig. 3 a and c). The sizes of these gaps could be reduced by introducing additional cysteines into NM (or using other residues), thus providing more frequent binding sites for the gold particles. As the gaps between gold particles become smaller, less enhancement should be necessary to make contacts continuous, and the resulting wire would be thinner. This smaller diameter would allow the manufacture of more intricate circuits and could potentially provide a new model system for quantum confinement and single-electron charging effects when electrons tunnel through restricted pathways (34–37).
While this article was in preparation, nanowires were produced by electroless gold plating of DNA and reported to have conductive properties similar to those of metallized NM fibers (23). Just as plastic films and glass may share an important physical property (transparency) but differ in other ways that make them suitable for different purposes, wire templates formed by DNA and amyloid protein fibers have similar conductive properties but are so fundamentally different in other ways that they will be suited to different purposes. DNA provides an extraordinary degree of programmable assembly specificity, all within the common framework of hydrogen-bonding between nucleotide pairs. Proteins have many inherent specificities to drive assembly but are less programmable. They are, however, based on a much broader platform of chemical diversity. Moreover, among the various DNA and protein fibers that have been described, NM fibers are unusual in that they are highly resistant to extended periods at high temperatures, exposure to high and low salt, strong denaturants, strong alkalis and acids, and 100% ethanol. These properties will allow them to withstand the harsh conditions in industrial processes. Depending on the conditions, NM fibers can nucleate spontaneously or self-assemble from preformed nuclei (30), which may be a crucial property for the practical assembly of circuits on a large scale. Our studies of NM fiber formation (28–30) enabled us to develop protocols for the rapid production of fibers with roughly predetermined lengths ranging from 500 nm up to several hundred micrometers. This ability to manipulate the fiber length will give great flexibility in designing nanostructures.
As described (32), bidirectional growth from NM seeded fibers can be used to incorporate NM derivatives with different modifications, interspacing them along individual fibers, e.g., with and without exposed cysteines. As different substrates can be prepared to bind to cysteine and to native lysine residues (lysine residue data not shown), these alternative binding sites provide flexibility and diversity in the patterning and mixing of substrates covalently bound to the fiber. More remarkably genetic engineering can be used to fuse a wide array of protein domains to the C terminus of NM during its initial in vivo synthesis in such a way that the domains are tethered laterally, external to the surface of assembled fibers. Thus they remain functional even when NM is in its fibrous form. This functionality is demonstrated by the ability of GFP to fluoresce green when fused to NM in its prion form (38) and by the ability of luciferase to function when attached to the C terminus of fibrous NM (M. Patino, J. Moslehi, and S.L.L., unpublished observation). Because many enzymes can function when attached to protein fibers (39), it should be possible to incorporate more complex reaction centers into NM nanocircuity, thereby creating electronic circuits that can take advantage of biological capacities. Indeed it can be imagined that mechanisms such as the vaporization of NM fibers with high voltages that we observed could act as a fuse or a switch to permanently activate or inactivate specific reaction centers within the circuitry.
There is a great opportunity to expand further the potential interconnections in these circuits by exploiting the natural diversity and strength of protein–protein interactions (25–27). Protein–protein interactions can be extremely specific and strong, as can the interactions of protein–ligand–protein. Protein properties can be used as a mechanism to bring premetallized wires into juxtaposition in response to changes in physical conditions, the presence of ligands, and the appearance of partner proteins, etc. These connections are readily reversible (e.g., refs. 40 and 41).
Complex circuit schematics could be generated with NM fibers, initiated by patterned surface modifications (independently or in combination) such as lithography, growth in flows or magnetic field gradients, alignment by electrical fields, active patterning with optical tweezers, dielectrophoresis and 3D patterning using hydrogels or microfluidic channels (refs. 42–47 and D. G. Grier, personal communication). The feasibility of such maneuvers is demonstrated by the natural tendency of NM fibers to align with each other rather than to form dense intractable clumps characteristic of other protein amyloids (Fig. 1c). Determining the exact conditions that produce such alignments should prove extremely useful. Attachment of NM to patterned surfaces can be mediated via covalent bonds to native lysine residues, genetically engineered cysteine residues, or other novel residues or modifications.
Our work provides a mechanism for generating robust nanowires that meet the needs of industrial processes with the potential to couple powerful combinations of biological processes and functionalities with electronic circuitry.
Acknowledgments
We thank A. Kowal for EM. This research was supported by the National Institutes of Health, the W. M. Keck Foundation, the University of Chicago Materials Research Science and Engineering Center (Materials Research Science and Engineering Centers Program of the National Science Foundation), the Howard Hughes Medical Institute, and a postdoctoral fellowship of the Deutsche Forschungsgemeinschaft (to T.S.).
Abbreviations
- NM
N-terminal and middle region
- CR
Congo red
- AFM
atomic force microscopy
- TEM
transmission electron microscopy
References
- 1.Whitesides G M, Mathias J P, Seto C T. Science. 1991;254:1312–1319. doi: 10.1126/science.1962191. [DOI] [PubMed] [Google Scholar]
- 2.Braun E, Eichen Y, Sivan U, Ben-Yoseph G. Nature. 1998;391:775–778. doi: 10.1038/35826. [DOI] [PubMed] [Google Scholar]
- 3.Fritzsche W, Böhm K, Unger E, Köhler J M. Appl Phys Lett. 1999;75:2854–2856. [Google Scholar]
- 4.Boal A K, Ilhan F, DeRouchey J E, Thurn-Albrecht T, Russell T P, Rotello V M. Nature. 2000;404:746–748. doi: 10.1038/35008037. [DOI] [PubMed] [Google Scholar]
- 5.Niemeyer C M. Curr Opin Chem Biol. 2000;4:609–618. doi: 10.1016/s1367-5931(00)00140-x. [DOI] [PubMed] [Google Scholar]
- 6.Quake S R, Scherer A. Science. 2000;290:1536–1540. doi: 10.1126/science.290.5496.1536. [DOI] [PubMed] [Google Scholar]
- 7.Richter J, Seidel R, Kirsch R, Mertig M, Pompe W, Plaschke J, Schackert H K. Adv Mater. 2000;12:507–510. [Google Scholar]
- 8.Thurn-Albrecht T, Schotter J, Kastle G A, Emley N, Shibauchi T, Krusin-Elbaum L, Guarini K, Black C T, Tuominen M T, Russell T P. Science. 2000;290:2126–2129. doi: 10.1126/science.290.5499.2126. [DOI] [PubMed] [Google Scholar]
- 9.Whaley S R, English D S, Hu E L, Barbara P F, Belcher A M. Nature. 2000;405:665–668. doi: 10.1038/35015043. [DOI] [PubMed] [Google Scholar]
- 10.Yu J-S, Kim J Y, Lee S, Mbindyo J K N, Martin B R, Mallouk T E. Chem Commun. 2000;24:2445–2446. [Google Scholar]
- 11.Huang Y, Duan X, Cui Y, Lauhon L J, Kim K-H, Lieber C M. Science. 2001;294:1313–1317. doi: 10.1126/science.1066192. [DOI] [PubMed] [Google Scholar]
- 12.Lopes W A, Jaeger H M. Nature. 2001;414:735–738. doi: 10.1038/414735a. [DOI] [PubMed] [Google Scholar]
- 13.Lee S W, Mao C, Flynn C E, Belcher A M. Science. 2002;296:892–895. doi: 10.1126/science.1068054. [DOI] [PubMed] [Google Scholar]
- 14.Kirsch R, Mertig M, Pompe W, Wahl R, Sadowski G, Böhm K J, Unger E. Thin Solid Films. 1997;305:248–253. [Google Scholar]
- 15.Kim M L, Wong K W, Mann S. Chem Mater. 1999;11:23–26. [Google Scholar]
- 16.Mirkin C A, Taton T A. Nature. 2000;405:626–627. doi: 10.1038/35015190. [DOI] [PubMed] [Google Scholar]
- 17.Mbindyo J K, Reiss B D, Martin B R, Keating C D, Natan M J, Mallouk T E. Adv Mater. 2001;13:249–254. [Google Scholar]
- 18.Schnur J M, Proce R, Schoen P, Yager P, Clavert J M, Georger J, Singh A. Thin Solid Films. 1987;152:181–206. [Google Scholar]
- 19.Mathias J P, Simanek E E, Whitesides G M. J Am Chem Soc. 1994;116:4326–4340. [Google Scholar]
- 20.Fowler C E, Shenton W, Stubbs G, Mann S. Adv Mater. 2001;13:1266–1269. [Google Scholar]
- 21.Rudolph A S, Calvert J M, Schoen P E, Schnur J M. Adv Exp Med Biol. 1988;238:305–320. doi: 10.1007/978-1-4684-7908-9_24. [DOI] [PubMed] [Google Scholar]
- 22.Pazirandeh M, Campbell J R. J Gen Microbiol. 1993;139:859–864. doi: 10.1099/00221287-139-4-859. [DOI] [PubMed] [Google Scholar]
- 23.Harnack O, Word W E, Yasuda A, Wessels J M. Nanosci Lett. 2002;2:919–923. [Google Scholar]
- 24.Patolsky F, Weizmann Y, Lioubashevski O, Willner I. Angew Chem Int Ed Engl. 2002;41:2323–2327. doi: 10.1002/1521-3773(20020703)41:13<2323::AID-ANIE2323>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 25.Begley T J, Rosenbach A S, Ideker T, Samson L D. Mol Cancer Res. 2002;1:103–112. [PubMed] [Google Scholar]
- 26.Uetz P, Giot G, Cagney G, Mansfield T A, Judson R S, Knight J R, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 27.Marcotte E, Pellegrini M, Thompson M J, Yeates T O, Eisenberg D. Nature. 1999;402:83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- 28.Glover J R, Kowal A S, Schirmer E C, Patino M M, Liu J J, Lindquist S L. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 29.Serio T R, Cashikar A G, Kowal A S, Sawicki G J, Moslehi J J, Serpell L, Arnsdorf M F, Lindquist S L. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 30.Scheibel T, Lindquist S L. Nat Struct Biol. 2001;8:958–962. doi: 10.1038/nsb1101-958. [DOI] [PubMed] [Google Scholar]
- 31.DePace A H, Weissman J S. Nat Struct Biol. 2002;9:389–396. doi: 10.1038/nsb786. [DOI] [PubMed] [Google Scholar]
- 32.Scheibel T, Kowal A, Bloom J, Lindquist S L. Curr Biol. 2001;11:366–369. doi: 10.1016/s0960-9822(01)00099-9. [DOI] [PubMed] [Google Scholar]
- 33.Morkved T L, Lopes W A, Hahm J, Sibener S J, Jaeger H M. Polymer. 1998;39:3871–3875. [Google Scholar]
- 34.Halperin W P. Rev Mod Phys. 1986;58:533–606. [Google Scholar]
- 35.Kastner M A. Rev Mod Phys. 1992;64:849–858. [Google Scholar]
- 36.Grabert H, Devoret M H, editors. Single Charge Tunneling. New York: Plenum; 1992. [Google Scholar]
- 37.Timp G L, editor. Nanotechnology. New York: Springer; 1999. [Google Scholar]
- 38.Patino M M, Liu J J, Glover J R, Lindquist S. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 39.Baxa U, Speransky V, Steven A C, Wickner R B. Proc Natl Acad Sci USA. 2002;99:5253–5260. doi: 10.1073/pnas.082097899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schreiber S L, Crabtree G R. Harvey Lect. 1995–1996;91:99–114. [PubMed] [Google Scholar]
- 41.Spencer D M, Wandless T J, Schreiber S L, Crabtree G R. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 42.Korda P, Spalding G C, Dufresne E R, Grier D G. Rev Sci Instrum. 2002;73:1956–1957. [Google Scholar]
- 43.Kane R S, Takayama S, Ostuni E, Ingber D E, Whitesides G M. Biomaterials. 1999;20:2363–2376. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 44.Inouye H, Fraser P E, Kirschner D A. Biophys J. 1993;64:502–519. doi: 10.1016/S0006-3495(93)81393-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luther P W, Peng H B, Lin J J. Nature. 1983;303:61–64. doi: 10.1038/303061a0. [DOI] [PubMed] [Google Scholar]
- 46.Kubista M, Hagmar P, Nielsen P E, Norden B. J Biomol Struct Dyn. 1990;8:37–54. doi: 10.1080/07391102.1990.10507788. [DOI] [PubMed] [Google Scholar]
- 47.Hermanson K D, Lumsdon S O, Williams J P, Kaler E W, Velev O D. Science. 2001;294:1082–1086. doi: 10.1126/science.1063821. [DOI] [PubMed] [Google Scholar]