Abstract
The androgen receptor (AR), a nuclear transcription factor, mediates male sexual differentiation, and its excessive action is associated with prostate cancer. We have characterized a negative regulatory domain in the AR hinge region, which interacted with filamin A (FLNa), an actin-binding cytoskeletal protein. FLNa interfered with AR interdomain interactions and competed with the coactivator transcriptional intermediary factor 2 to specifically down-regulate AR function. Although full-length FLNa was predominantly cytoplasmic, a C-terminal 100-kDa fragment of FLNa colocalized with AR to the nucleus. This naturally occurring FLNa fragment repressed AR transactivation and disrupted AR interdomain interactions and transcriptional intermediary factor 2-activated AR function in a manner reminiscent of full-length FLNa, raising the possibility that the inhibitory effects of cytoplasmic FLNa may be transduced through this fragment, which can localize to the nucleus and form part of the pre-initiation complex. This unanticipated role of FLNa adds to the growing evidence for the involvement of cytoskeletal proteins in transcription regulation.
The androgen receptor (AR), a member of the steroid/nuclear receptor superfamily, mediates male morphogenesis in utero, gametogenesis and prostate growth at puberty, and the development of prostatic cancer in older men. The AR has four principal domains: a large N-terminal transactivation domain (ARTAD), a DNA-binding domain (ARDBD), and a hinge domain, followed by the C-terminal ligand-binding domain (ARLBD). In the absence of androgen, it is generally accepted that AR is cytoplasmic. Androgens bind specifically to a ligand-binding pocket in the lower half of the LBD, causing a conformation change (1), the release of heat-shock proteins, and translocation of the ligand–AR complex to the nucleus, where the DBD interacts with specific response elements on the promoters of target genes. Transactivation functions reside mainly in the ARTAD, and very minimal activity can be demonstrated in the ARLBD (2, 3). Unlike other steroid receptors, transactivity depends on ligand-induced interactions between the ARTAD and ARLBD, and mutations that reduce TAD–LBD interactions affect AR activity, causing androgen insensitivity syndromes (4–6). Like other transcription regulators, the promoter-bound AR serves as a nidus to recruit cofactors that up-regulate (coactivators) or down-regulate (corepressors) AR activity. The most clearly defined class of coactivators is the p160/steroid receptor coactivator (SRC) family. They include SRC1, transcriptional intermediary factor 2 (SRC2/TIF2), and SRC3/TRAM1/AIB1/pCIP/ACTR/RAC3, who bind hydrophobic grooves in the LBDs of steroid receptors via LXXLL motifs in their nuclear-receptor-interacting boxes, draw in cAMP-response element-binding protein (CBP/p300), pCAF, and other cofactors to modulate chromatin and initiate transcription by RNA polymerase II (7). Of the p160/SRC proteins, TIF2 interacts most strongly with the AR (8) and, in concert with CBP/p300, brings enhancer and promoter elements together to increase AR transactivation function (9).
On the other hand, interaction with corepressors like NcoR/SMRT (10); cell cycle control protein D-type cyclins (11); HBO1, a protein that interacts with the human origin recognition complex (12); PAK6, a novel p21-activated kinase (13); Smad3, a member of the transforming growth factor β intracellular signal transducers (14); and the tumor suppressors TSG101 (15) and p53 (16), are thought to inhibit AR function. The manner in which repressors interact with coactivators to regulate AR function is not clear. Although recruitment of coactivators to the ARLBD is a ligand-induced process, the existence of the independent activation function of ARLBD has been difficult to prove, unlike for the estrogen, progesterone, and glucocorticoid receptors, where strong LBD activation functions can be demonstrated. Because an AR hinge deletion mutant has been reported to have increased LBD transactivity (17), we reasoned that accessory factors that interact with the AR hinge may inhibit receptor function. Here we describe the identification of filamin A (FLNa), a 280-kDa actin-binding protein, as an AR-hinge-interacting protein that acts as transcriptional inhibitor of the AR. Deletion studies were performed to understand the mechanisms whereby FLNa, originally identified as a cytoplasmic protein that participates in orthogonal actin formation, can mediate transcription repression by a nuclear hormone receptor. Our data support a transcriptional model in which a fragment of FLNa translocates to the nucleus and competes with the coactivator TIF2 to modulate AR activity.
Materials and Methods
Yeast Two-Hybrid System.
The yeast two-hybrid system was used to identify clones that interact with the human AR hinge region. The bait, pGBKT7-AR(aa622–670) generated by inserting a PCR fragment encoding residues 622–670 of the AR in-frame into the EcoRI/BamHI site of pGBKT7, was used to screen an adult human prostate cDNA library (CLONTECH). Positive clones were selected by growth on a synthetic dropout minimal medium lacking tryptophan, leucine, histidine, and adenine, and by activation of α-galactosidase function in the presence of X-α-Gal chromogenic substrate. Curing of the bait plasmid and testing against empty pGBKT7 vector and negative control pGBKT7 containing lamin C by a mating strategy verified all potential clones interacting with the AR hinge. Positive cDNA clones were recovered from the host cells and identified by DNA sequencing.
Construction of Plasmids.
The AR hinge deletion mutant AR(Δ628–646) was constructed as described (17). The primer extension method of Higuchi et al. (18) was used to create AR hinge mutants AR(Δ622–670) and AR(Δ647–670). pCMV-FLNa was created by first transferring a fragment containing amino acid residues 21–2647 from the full-length FLNa plasmid, kindly provided by T. P. Stossel (Harvard Medical School, Boston) into the pFlag-CMV expression vector. The first 20-aa residues of FLNa were then generated by PCR and cloned in-frame into pCMV-FLNa(aa 21–2647) to create the full-length pCMV-FLNa expression plasmid. FLNa repeats 16–24 [pCMV-FLNa(16–24), aa1788–2647] was generated by PCR and inserted into pFlag-CMV. pSG5-TIF2, encoding full length TIF2, was provided by P. Chambon (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Strasbourg, France). ARE2-Luc, containing two tandem copies of the androgen response element (ARE) from the aminotransferase gene driving the luciferase reporter gene, was obtained from G. Jenster (Erasmus University, Rotterdam, The Netherlands). Mouse mammary tumor virus–luciferase (MMTV-Luc) and the 1.6-kb prostate-specific antigen–luciferase (PSA-LUC) were described previously (5). Construction of GAL-ARLBD(628–919), GAL-ARLBD(647–919), VP16-ARTAD(1–565), and pG5Luc has been described (17). For confocal experiments, amino acid residues 21–2647 of FLNa were inserted into the pEGFPc1 vector (CLONTECH) to create pEGFP-FLNa. To create pEGFP-FLNa(16–24), amino acid residues 1788–2647 were excised from pCMV-FLNa(16–24) and inserted into the pEGFPc1 vector. All constructs were sequenced to confirm the fidelity of the enzymatic manipulations.
Cell Culture and Transfection Assays.
HeLa cells were maintained in RPMI supplemented with 7% FBS (GIBCO/BRL) at 37°C under 5% CO2. Cells were cultured at 4 × 104 cells per well in 24-well plates and grown for 24 h before transfection. DNA transfection was carried out by using Lipofectamine (GIBCO/BRL) following the manufacturer's protocol. For normalization purposes, the empty vectors of the respective clones were used. The luciferase reporter gene assays were measured as relative light units (RLU) 48 h posttransfection, and each data point represented the mean ± SE of at least three replicates (17).
Western Blot Analysis.
Cells were treated according to experimental designs, and immunoblotting of total cell lysates was performed according to standard protocol by using anti-AR 441 antibody (Santa Cruz Biotechnology), anti-FLNa antibody (Chemicon MAB1680), and anti-paxillin antibody (Chemicon MAB3060). AR-deficient prostatic carcinoma cell lines PC3 and DU145 were obtained from American Type Culture Collection. Genital skin fibroblasts containing intact AR (LDC) and nonfunctional truncated AR (TBL) were cultured from genital skin biopsies (19). Filamin-deficient M2 and filamin-repleted A7 cells were gifts of T. P. Stossel (Harvard Medical School).
Confocal Microscopy.
HeLa cells were plated on glass coverslips in 24-well plates at 2.5 × 105 cells per coverslip for 24 h before transfection. One hundred nanograms of pSV-AR was cotransfected with 80 ng of pEGFP-FLNa or pEGFP-FLNa(16–24) into the cells by using Lipofectamine (GIBCO/BRL). Five hours after transfection, the cells were grown for 24 h in charcoal-treated RPMI media containing 1 nM 5α-dihydrotestosterone (DHT). Indirect immunofluorescence was performed according to standard protocol.
Results
Unexpected Repressor Function of AR Hinge Subdomain.
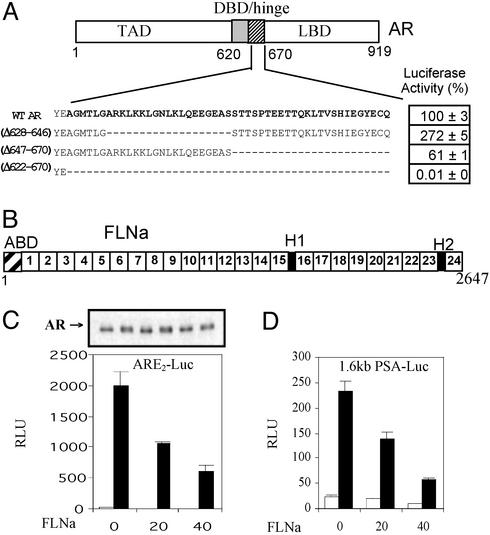
To explore the functions of the AR hinge, we deleted portions of this domain and tested the transactivation activity of the resultant AR mutants (Fig. 1A) in androgen-driven reporter gene assays. Deletion of the entire hinge region (residues 622–670) resulted in total abrogation of AR activity (Fig. 1A). In contrast, deletion of the C-terminal half of the hinge (residues 647–670) resulted in a mutant AR that was partially active. Most strikingly and consistent with our previous findings (17), deletion of the N-terminal half resulted in a fragment that was transcriptionally twice as active as the WT AR, suggesting the presence of an inhibitory function in the subdomain defined by residues 628–646. This, together with transcription inhibition assays by using chimeric AR LBD fragments (Fig. 6 and Supporting Materials and Methods, which are published as supporting information on the PNAS web site, www.pnas.org), suggests an unanticipated repressive function residing in residues 628–646, which directly or indirectly exerts an inhibitory effect on the activation function of the ARLBD.
Figure 1.
Transactivity of AR hinge deletion mutants and effect of FLNa on AR function. (A) The AR consists of a TAD and a DBD, linked to the LBD by a hinge region. The AR hinge (amino acids 622–670) used as bait in the yeast two-hybrid screen is in bold. WT AR or the deletion mutants [(Δ628–646), (Δ647–670), and (Δ622–670)] were tested for their ability to transactivate the ARE2-Luc reporter in the presence of 1 nM DHT (WT AR set at 100% ± SEM). (B) Structure of monomeric FLNa. Indicated in this schematic representation are the actin-binding domain (ABD), hinge-1 and -2 (H1, H2), and the 24 Ig-like repeats. (C and D) WT AR was coexpressed with increasing amounts of FLNa, and androgenic activity with (filled bars) or without (open bars) 1 nM DHT was measured with ARE2-Luc (C) or prostate-specific antigen–luciferase (D). The arrow (C Upper) indicates immunoblot analyses of AR protein from representative cell lysates.
The Actin-Binding Protein FLNa Interacts with the AR Hinge Domain.
To understand the mechanism(s) by which the AR hinge exerts its negative effect on receptor function, a yeast two-hybrid screening strategy was used to identify proteins that interact specifically with the AR hinge. A total of 8 × 105 individual colonies from a human prostate library were screened with a chimeric bait containing the GAL4 DNA-binding domain and AR amino acid residues 622–670 encoding the entire AR hinge region. Seven of the positive cDNA clones encoded polypeptides that corresponded specifically to the C-terminal domain (spanning amino acids 2363–2647) of the actin-binding protein FLNa, but not the other members of the filamin protein family (20). FLNa (280 kDa, ABP280), the principal human nonmuscle isoform, contains an N-terminal actin-binding domain followed by 24 Ig-like repeats, the last of which represents the self-dimerization domain of the protein (Fig. 1B). Previously, FLNa (amino acids 1788–2121) was identified as an AR-interacting protein by using the entire AR ligand-binding domain and AR constructs containing the hinge region were able to interact with FLNa in immunoprecipitation studies (21). Our results suggest that the AR hinge region is a bona fide interactor for FLNa.
FLNa Specifically Represses AR Transactivation Activity.
To test whether FLNa has any influence on AR transcriptional activity, full-length FLNa and AR were coexpressed in HeLa cells and their transactivation function tested with synthetic and natural androgen-responsive promoters. The synthetic ARE2-Luc reporter was strongly induced by the AR in the presence of DHT, and this activation was dose-dependently repressed by cotransfection of increasing amounts of FLNa (Fig. 1C). This repression by FLNa was not associated with a change in the steady-state levels of the AR protein, as demonstrated by Western blotting of the cell lysates with an anti-AR antibody (Fig. 1C Upper). To understand the role of FLNa in a physiologically relevant context, transactivation studies were repeated by using the naturally occurring MMTV-long terminal repeat (LTR) and human PSA androgen-responsive promoters driving the luciferase gene. The MMTV-LTR promoter contains several copies of the consensus ARE, and human PSA gene 1.6-kb promoter (with two putative AREs) controls expression of PSA, an androgen-regulated protein that is widely used as a marker for prostate cancer progression (5). A dose-dependent inhibitory effect of FLNa was observed on AR-mediated transcription from both the MMTV-LTR (data not shown) and PSA (Fig. 1D) reporter constructs, demonstrating that FLNa can repress androgen signaling in naturally occurring androgen-responsive promoter elements. In addition, this inhibitory effect was specific for the AR because FLNa did not have any inhibitory effects on the other nuclear receptors such as the estrogen, progesterone, and glucocorticoid receptors (Fig. 7 and Supporting Materials and Methods, which are published as supporting information on the PNAS web site). Furthermore, FLNa had no inhibitory effect on the constitutive thymidine kinase promoter, which is unrelated to the nuclear hormone receptors (results not shown). Taken together, our results indicate that the inhibitory effect of FLNa on nuclear receptor-mediated transcriptional activation is specific to the AR.
FLNa Disrupts Interactions Between N- and C-Terminal Domains of the AR, and Its Repressive Action Can Be Reversed by the Coactivator TIF2.
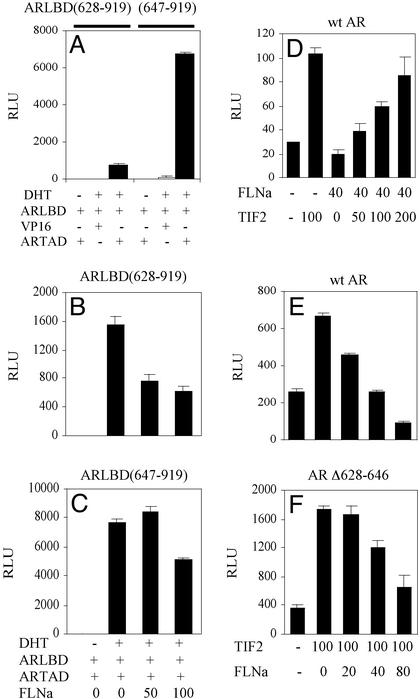
Interdomain protein–protein interactions between N- and C-terminal domains are essential for normal transactivation function of the AR. To test the effects of FLNa on AR N–C interactions, we used a modified mammalian two-hybrid strategy. The AR N-terminal TAD linked to a VP-16 activation domain (VP16-ARTAD) was coexpressed with GAL4DBD-ARLBD constructs (with or without amino acids 628–646) and the GAL4-Luc reporter gene. GAL4DBD alone did not interact with VP16-ARTAD, and no reporter gene activity was observed (data not shown). Weak interactions were detected between the ARTAD and the fragment consisting of the hinge and ARLBD (residues 628–919) (Fig. 2A). Strikingly, deletion of hinge residues 628–646 from the ARLBD increased N–C interactions to an order of magnitude higher than that observed for the intact LBD fragment, consistent with the repressive function of this AR subdomain. The presence of FLNa reduced interactions between ARTAD and the hinge-LBD (628–919) fragment, suggesting that one mechanism that contributes to the inhibitory function of the FLNa may involve the disruption of AR N–C interactions (Fig. 2B). Interestingly, this repressor action of FLNa was less obvious when hinge residues 628–646 were deleted (Fig. 2C), supporting the hypothesis that this hinge subdomain is the region through which FLNa acts, at least partially, to inhibit AR activity.
Figure 2.
Effect of FLNa on AR N–C interaction, and TIF2-mediated coactivation. (A) The two-hybrid system was used to measure the interaction between AR N and C termini. The ARTAD (amino acids 1–565) fused in-frame to the activation domain of VP16 was coexpressed with GAL-ARLBD (628–919) or GAL-ARLBD (647–919), and N–C interactions were measured by using the GAL4-Luc reporter gene with and without 10 nM DHT. (B and C) ARTAD and ARLBD (628–919) (B) or ARLBD (647–919) (C) chimeric proteins were coexpressed with indicated doses (nanograms) of FLNa and 10 nM DHT. N–C interactions were measured with the GAL4-Luc reporter gene. (D–F) Effect of TIF2 on FLNa repression. Increasing amounts of TIF2 were coexpressed with WT AR and FLNa (D). Increasing amounts of FLNa were coexpressed with TIF2 and WT AR (E) or AR(Δ628–646) (F).
To investigate the possibility that FLNa-mediated repression of AR may also interfere with binding of the p160 coactivator TIF2, we tested whether overexpression of TIF2 can relieve the inhibitory effects of FLNa on AR activity. Consistent with previous reports (17), TIF2 alone augmented WT AR activity by 100% (Fig. 2D). Although the presence of FLNa alone inhibited AR-dependent transactivation function, overexpression of TIF2 was able to dose-dependently reverse the FLNa-mediated down-regulation of AR activity. Conversely, the presence of FLNa dose-dependently repressed AR-coactivator activity (Fig. 2E). Consistent with our hypothesis that AR residues 628–646 harbor the inhibitor domain, AR lacking these residues are inhibited to a lesser degree by FLNa (Fig. 2F). Collectively, these experiments suggest that FLNa may act by constraining full activation of the AR by destabilizing receptor–coactivator and interdomain interactions of the AR.
Endogenous Expression of FLNa Fragments and Localization of the C-Terminal Fragment to the Nucleus.
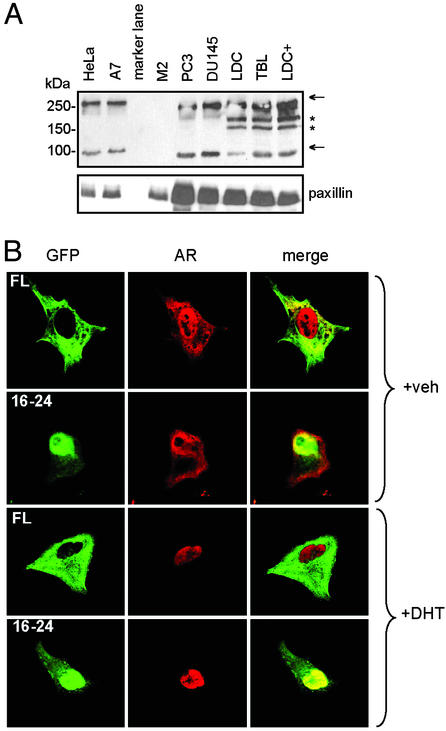
To date, it is thought that FLNa resides and executes most of its functions in the cytoplasm (20). However, recent data (21, 22) and our current findings suggest that FLNa also participates in nuclear-related functions. How then does a largely cytoplasmic protein mediate nuclear events? Interestingly, members of the filamin protein family are highly susceptible to proteolysis. There are two calpain-cleavage sites, designated as H1 and H2, in FLNa (Fig. 1B). Cleavage at the H1 site leads to the formation of 190- and 100-kDa fragments, corresponding to the N terminus of FLNa terminating at repeat 15 and C-terminus-encompassing repeats 16–24 [FLNa(16–24)], respectively (23). The precise role of FLNa cleavage remains unknown. To study the natural expression of filamin fragments in untransfected cells, we probed a variety of transformed and primary cell cultures with an antibody for FLNa(16–24). Besides full length FLNa, a 100-kDa fragment corresponding to FLNa(16–24) was consistently observed in all but one of the seven cell lines tested, the exception being filamin-deficient M2 cells (Fig. 3A). Expression of FLNa(16–24) fragment did not appear to depend on androgen action, because it was expressed in both AR-intact (LDC) and AR-deficient (PC3, DU145, and TBL) cells.
Figure 3.
Expression of endogenous FLNa fragments and localization of C-terminal fragment of FLNa. (A) Immunoblot of FLNa fragments from HeLa, FLNa-repleted A7, FLNa-deficient M2, prostate carcinoma PC3, and DU145, primary genital skin fibroblasts containing normal AR [LDC, cultured with (+) or without DHT] or nonfunctional mutant AR (TBL) cell cultures. Cell extracts were probed with an antibody that recognizes FLNa repeats 16–20. Arrows indicate full-length FLNa (280 kDa) and a fragment (100 kDa) corresponding to FLNa(16–24). Immunoreactive bands (≈185 and 160 kDa, marked by *) were observed only in extracts from genital fibroblasts. The cytoskeletal paxillin was used to indicate the presence of proteins in each lane. (B) Subcellular localization of GFP-FLNa constructs. Chimeric full-length filamin (FL) or FLNa(16–24, aa1788–2647) linked to GFP was coexpressed with WT AR in the absence (+vehicle) or presence of DHT. Representative green fluorescence images for GFP-FLNa constructs (GFP), red immunofluorescence images for AR (AR), and merged images after superimposition of GFP and AR (merge) are shown. Note that yellow in the merged images indicate colocalization of AR and FLNa(16–24) to the nucleus.
To ascertain the biological significance of the 100-kDa FLNa fragment, we made GFP chimeras of full length FLNa and FLNa(16–24), which harbors the AR interaction domains, and observed their cellular localization. GFP-FLNa chimeras were transfected into HeLa cells, and their cellular localization were observed by using confocal microscopy (Fig. 3B). Consistent with previous reports, the majority of full-length GFP-FLNa was located in the cytoplasm. Unexpectedly and intriguingly, GFP-FLNa(16–24) was observed mainly in the nucleus, both in the presence and absence of androgen. The nuclear localization of GFP-FLNa(16–24) was consistently demonstrated in over five independent experiments. Immunostaining with an AR-specific antibody indicates that ligand-activated AR and GFP-FLNa(16–24) were observed simultaneously in the nucleus. This previously undescribed finding that FLNa(16–24) colocalizes with liganded AR to the nucleus suggests a direct nuclear regulatory function for what was generally assumed to be a cytoplasmic protein. In addition, both GFP-FLNa and GFP-FLNa(16–24) retained the capacity to inhibit AR transcriptional activity when cotransfected into HeLa cells, with the latter exhibiting a somewhat more potent inhibition of AR activity (data not shown). This may reflect increased availability of the FLNa(16–24) fragment, because only a proportion of full-length FLNa is expected to be processed by proteolysis.
FLNa(16–24) Is Sufficient to Mediate FLNa Effects on AR Function.
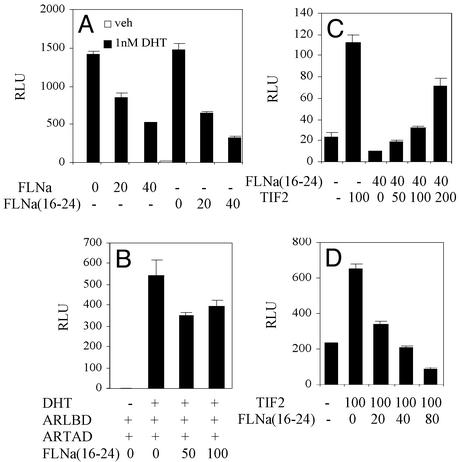
Our results that GFP-FLNa(16–24) not only localizes to the nucleus but also inhibits AR activity suggest that this FLNa fragment is sufficient to modulate the AR transcription complex. To test this, an expression vector containing FLNa(16–24) was coexpressed with the AR in HeLa cells, and ARE-driven luciferase activity was measured. FLNa(16–24) exhibited an inhibitory effect on ligand-induced AR transcriptional activity that was similar to that exerted by the full-length FLNa (Fig. 4A). In addition, the C-terminal fragment was able to interfere with AR interdomain N–C interaction (Fig. 4B). Furthermore, TIF2 dose-dependently relieved AR repression by FLNa(16–24), and vice versa, in a manner reminiscent of that of full-length FLNa (comparing Figs. 2 D and E and 4 C and D). Thus, our results suggest that the nuclear repressor effects of FLNa may be exerted through its calpain-cleavage fragment, FLNa(16–24), because this naturally present FLNa fragment is predominantly nuclear unlike full-length FLNa, which is mainly cytoplasmic.
Figure 4.
Effect of FLNa(16–24) on AR N–C interaction and TIF2-mediated coactivation. (A) Indicated doses of full-length FLNa or FLNa(16–24) were coexpressed with WT AR in the presence or absence of DHT and androgenic activity measured with ARE2-Luc. (B) Effect of FLNa(16–24) on AR N–C interaction. Increasing amounts of FLNa(16–24) were coexpressed with chimeric ARTAD and ARLBD and protein–protein interactions measured with GAL4-Luc as in Fig. 2C. (C and D) Effect of TIF2 on FLNa(16–24) repression. Increasing amounts of TIF2 were coexpressed with WT AR and FLNa(16–24) (C). Increasing amounts of FLNa(16–24) were coexpressed with TIF2 and WT AR (D). Transactivation activity was measured with ARE2-Luc as in Fig. 2 D and E.
Discussion
Our data suggest the concept that the actin-crosslinking protein FLNa, generally regarded as a cytoplasmic architectural molecule, may act as a nuclear transcription modulator to directly and specifically repress AR function. FLNa could exert this inhibitory effect through an endogenous 100-kDa proteolytic fragment, FLNa(16–24), which localizes to the nucleus and interferes with interactions between the N- and C-terminal domains, and coactivator functions of the AR. This role of FLNa as an AR repressor is supported by converging strands of evidence, namely the characterization of a negative regulatory function in the N-terminal subdomain of the AR hinge, the identification of FLNa as an AR hinge-interacting protein that can selectively repress AR transactivation activity, the consistent presence of an endogenous 100-kDa FLNa fragment in all transformed and primary cell lines tested, the discovery that this fragment resides mainly in the nucleus and can exert a repressor function on interdomain and coactivator functions of the AR.
Our discovery of FLNa as a nuclear regulator of AR was entirely unexpected, because FLNa is known as a cytoplasmic scaffolding protein that dimerizes and orthogonally crosslinks actin through its N-terminal actin-binding domains (20). The filamentous nature of FLNa is due to the presence of 24 repeats consisting of pleated β-sheets made up of ≈96 amino acids each, interrupted by two short hinge segments that are susceptible to calpain cleavage (Fig. 1B). The C-terminal end of FLNa also binds a variety of proteins on the cell surface like the transmembrane β-integrins (24) and the Rho family of small GTPases and their guanine nucleotide exchange factor, Trio (25). The resultant interwebbing of actin-filamin scaffolds with cell surface receptors provides mechanical stability, maintains cell–cell and cell–matrix connections, and transduces stress signals to the actin skeleton, shaping the cytoskeleton into configurations useful to the cell (20). Our data indicate another role for FLNa, that of a nuclear coregulator for transcription. FLNa had profound and specific dose-dependent inhibitory actions on AR transactivation function, in the context of both artificial and natural androgen-driven promoters.
Known repressors of the AR such as calreticulin (26), cyclins (11), HBO1 (12), Smad3 (14), TSG101 (15), and p53 (16) are all nuclear proteins. Ozanne et al. (21) had earlier proposed that FLNa may serve to regulate the tethering of AR to the cytoskeleton via the hsp90, a chaperone protein that plays a key role in the conformational change and transcriptional activity of the AR (27) and that has been reported to interact with the motor protein dynein (28). Although this scheme is reminiscent of the role of cytoskeletal components in the hsp90-mediated cellular trafficking of the glucocorticoid receptor into the nucleus (29), it does not explain how cytoplasmic FLNa can directly mediate AR function in the nucleus. Our finding that a naturally generated filamin fragment residing in the nucleus provided a tantalizing clue. A plausible model envisages full-length FLNa being bound to the actin cytoskeleton on the cell surface and perinuclear areas of the cell through its N-terminal actin-binding domain. AR in the absence of ligand is predominantly cytoplasmic and is tethered to the C-terminal end of FLNa through its hinge domain and the LBD (Fig. 5). Full-length FLNa, when cleaved at the protease-cleavage site between repeats 15 and 16, releases the FLNa(16–24). FLNa(16–24) colocates with liganded AR to the nucleus. It is interesting to note that a putative nuclear localizing signal (RRRR) is present in FLNa repeat 20 (residues 2146–2149). This model is also consistent with the observation that nuclear localization of the AR does not occur in FLNa-deficient cells (21). In the nucleus, FLNa(16–24) disrupts interactions between the N and C termini of the AR and interferes with the binding of the coactivator, TIF2. There is evidence that interaction between the FXXLF motif of the TAD and the LBD reduces coactivator recruitment and the binding of LXXLL motifs of TIF2 (3). Because FLNa(16–24) also interacts with the LBD, it is plausible that the TAD, FLNa(16–24), and TIF2 are in dynamic competition. Deletion of the AR hinge region loosens FLNa(16–24) binding to the LBD, allowing a more stable coactivator–AR configuration and consequently up-regulation of AR activity (Fig. 5). Final AR transactivation activity would depend on the relative quantities of coactivator or repressor that are recruited to the promoter. Our data that the inhibitory effects of FLNa(16–24) can be reversed by TIF2 and vice versa support this model. It is interesting to note that substitutions within the AR hinge region are associated with the androgen-driven tumor prostate cancer (AR mutations database: www.mcgill.ca/androgendb). Indeed, mutation of hinge residues bordering the LBD result in a 2- to 4-fold increase in transactivation activity in transgenic adenocarcinoma of the mouse prostate model (30). Alternatively, FLNa(16–24) could also directly recruit transcriptional repressors onto the target promoter or possess intrinsic histone deacetylase activity to inhibit transcription initiation. However, the latter is less likely, because FLNa or various fragments of FLNa fused to the heterologous GAL4-DBD did not display any notable intrinsic transcription repression from a constitutively active promoter containing GAL4-binding sites (data not shown).
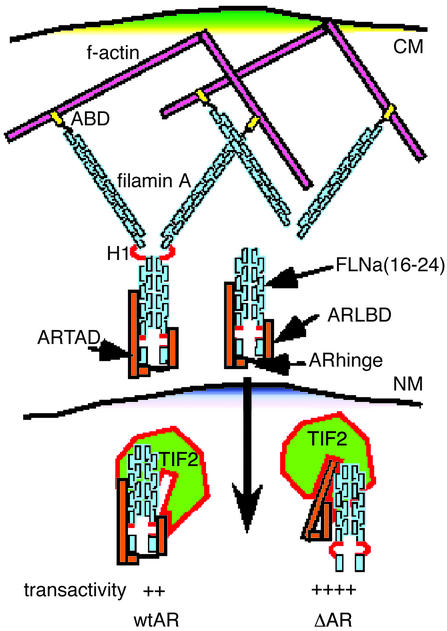
Figure 5.
Model of FLNa/TIF2-mediated AR transcription. Dimeric FLNa (blue), in the cytoplasm, binds the f-actin cytoskeleton (pink) orthogonally through its N-terminal actin-binding domain (ABD, yellow). AR (orange bars) interacts with the C-terminal end of FLNa through the AR hinge (black loop) and the ARLBD. Cleavage of FLNa at H1 (red loop) releases FLNa(16–24), which colocalizes with the AR into the nucleus. Here, FLNa(16–24) and TIF2 (green) compete for binding to the ARLBD and AR hinge. When the N-terminal subdomain of the AR hinge region is deleted (ΔAR), one of the binding sites for FLNa(16–24) is lost, leading to enhanced TIF2 binding and improved TAD–LBD interactions and consequently increased AR transactivity. CM, NM, cytoplasmic and nuclear membranes.
The generation of C-terminal FLNa(16–24) may be achieved through cleavage at the H1 site by calpain or via an alternative proteolytic mechanism. Interestingly, the C-terminal portion of FLNa acts as a GTPase docking site by binding several RhoGTPases including RalA, Cdc42, Rac1, and RhoA (31). The recent report of the Rho-regulated PAK6 as an AR hinge-interacting kinase (13) suggests that the FLNa(16–24)–AR hinge complex may serve as an integrator for the many cytoskeletal signaling cascades converging on the AR. Our findings provide general insights on the role of cytoskeleton-associated proteins in transcription regulation and support the growing appreciation of the essential role of cytoskeletal proteins in gene transcription regulation in general, and nuclear receptor translocation in particular (29, 32–34). Furthermore, the role of FLNa is reminiscent of β-catenin, which has dual functions as a structural protein and a transcription regulator (35, 36). Thus β-catenin links the actin cytoskeleton to adherens junctions on the cell surface formed by E-cadherin and α-catenin. In addition to this role as a skeletal protein, free cytoplasmic β-catenin is part of the Wingless/Wnt signaling pathway that interacts with TCF transcription factors and that can enhance AR transactivation. These two functions of β-catenin are mediated through separate domains, and its signaling function depends on the levels of available cytoplasmic β-catenin, akin to our model whereby the function of FLNa(16–24) on AR depends on its relative concentration vis-à-vis the coactivator TIF2.
Although the physiological importance of filamin interactions to AR function in vivo remains to be determined, our findings may help conceptualize the nature of FLNa interactions with other nuclear proteins such as the tumor suppressor protein, BRCA2 (22). The control of AR activity is likely to involve multiple regulatory processes whose signals impinge on receptor function (7), and this unanticipated role for FLNa contributes to the possible mechanisms whereby mutations of the AR hinge region result in prostate cancer.
Supplementary Material
Acknowledgments
This work was supported by the Singapore National Medical Research Council Grant NMRC/0361/1999.
Abbreviations
- AR
androgen receptor
- FLNa
filamin A
- DHT
5α-dihydrotestosterone
- TAD
transactivation domain
- LBD
ligand-binding domain
- TIF2
transcriptional intermediary factor 2
- MMTV-Luc
mouse mammary tumor virus–luciferase
- DBD
DNA-binding domain
- SRC
steroid receptor coactivator
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Wurtz J M, Bourguet W, Renaud J P, Vivat V, Chambon P, Moras D, Gronemeyer H. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 2.Berrevoets C A, Doesburg P, Steketee K, Trapman J, Brinkmann A O. Mol Endocrinol. 1998;12:1172–1183. doi: 10.1210/mend.12.8.0153. [DOI] [PubMed] [Google Scholar]
- 3.He B, Kemppainen J A, Wilson E M. J Biol Chem. 2000;275:22986–22994. doi: 10.1074/jbc.M002807200. [DOI] [PubMed] [Google Scholar]
- 4.Ghadessy F J, Lim J, Abdullah A A, Panet-Raymond V, Choo C K, Lumbroso R, Tut T G, Gottlieb B, Pinsky L, Trifiro M A, et al. J Clin Invest. 1999;103:1517–1525. doi: 10.1172/JCI4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim J, Ghadessy F J, Abdullah A A, Pinsky L, Trifiro M, Yong E L. Mol Endocrinol. 2000;14:1187–1197. doi: 10.1210/mend.14.8.0499. [DOI] [PubMed] [Google Scholar]
- 6.Thompson J, Saatcioglu F, Janne O A, Palvimo J J. Mol Endocrinol. 2001;15:923–935. doi: 10.1210/mend.15.6.0647. [DOI] [PubMed] [Google Scholar]
- 7.Heinlein C A, Chang C. Endocr Rev. 2002;23:175–200. doi: 10.1210/edrv.23.2.0460. [DOI] [PubMed] [Google Scholar]
- 8.Ding X F, Anderson C M, Ma H, Hong H, Uht R M, Kushner P J, Stallcup M R. Mol Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 9.Shang Y, Myers M, Brown M. Mol Cell. 2002;9:601–610. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 10.Dotzlaw H, Moehren U, Mink S, Cato A C, Iniguez Lluhi J A, Baniahmad A. Mol Endocrinol. 2002;16:661–673. doi: 10.1210/mend.16.4.0798. [DOI] [PubMed] [Google Scholar]
- 11.Knudsen K E, Cavenee W K, Arden K C. Cancer Res. 1999;59:2297–2301. [PubMed] [Google Scholar]
- 12.Sharma M, Zarnegar M, Li X, Lim B, Sun Z. J Biol Chem. 2000;275:35200–35208. doi: 10.1074/jbc.M004838200. [DOI] [PubMed] [Google Scholar]
- 13.Lee S R, Ramos S M, Ko A, Masiello D, Swanson K D, Lu M L, Balk S P. Mol Endocrinol. 2002;16:85–99. doi: 10.1210/mend.16.1.0753. [DOI] [PubMed] [Google Scholar]
- 14.Hayes S A, Zarnegar M, Sharma M, Yang F, Peehl D M, ten Dijke P, Sun Z. Cancer Res. 2001;61:2112–2118. [PubMed] [Google Scholar]
- 15.Sun Z, Pan J, Hope W X, Cohen S N, Balk S P. Cancer. 1999;86:689–696. doi: 10.1002/(sici)1097-0142(19990815)86:4<689::aid-cncr19>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 16.Shenk J L, Fisher C J, Chen S Y, Zhou X F, Tillman K, Shemshedini L. J Biol Chem. 2001;276:38472–38479. doi: 10.1074/jbc.M103652200. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Lu J, Yong E L. J Biol Chem. 2001;276:7493–7499. doi: 10.1074/jbc.M009916200. [DOI] [PubMed] [Google Scholar]
- 18.Higuchi R, Krummel B, Saiki R K. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim J, Ghadessy F J, Yong E L. Mol Cell Endocrinol. 1997;131:205–210. doi: 10.1016/s0303-7207(97)00109-3. [DOI] [PubMed] [Google Scholar]
- 20.Stossel T P, Condeelis J, Cooley L, Hartwig J H, Noegel A, Schleicher M, Shapiro S S. Nat Rev Mol Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 21.Ozanne D M, Brady M E, Cook S, Gaughan L, Neal D E, Robson C N. Mol Endocrinol. 2000;14:1618–1626. doi: 10.1210/mend.14.10.0541. [DOI] [PubMed] [Google Scholar]
- 22.Yuan Y, Shen Z. J Biol Chem. 2001;276:48318–48324. doi: 10.1074/jbc.M102557200. [DOI] [PubMed] [Google Scholar]
- 23.Gorlin J B, Yamin R, Egan S, Stewart M, Stossel T P, Kwiatkowski D J, Hartwig J H. J Cell Biol. 1990;111:1089–1105. doi: 10.1083/jcb.111.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calderwood D A, Shattil S J, Ginsberg M H. J Biol Chem. 2000;275:22607–22610. doi: 10.1074/jbc.R900037199. [DOI] [PubMed] [Google Scholar]
- 25.Bellanger J M, Astier C, Sardet C, Ohta Y, Stossel T P, Debant A. Nat Cell Biol. 2000;2:888–892. doi: 10.1038/35046533. [DOI] [PubMed] [Google Scholar]
- 26.Dedhar S, Rennie P S, Shago M, Hagesteijn C Y, Yang H, Filmus J, Hawley R G, Bruchovsky N, Cheng H, Matusik R J, et al. Nature. 1994;367:480–483. doi: 10.1038/367480a0. [DOI] [PubMed] [Google Scholar]
- 27.Georget V, Terouanne B, Nicolas J C, Sultan C. Biochemistry. 2002;41:11824–11831. doi: 10.1021/bi0259150. [DOI] [PubMed] [Google Scholar]
- 28.Galigniana M D, Harrell J M, Murphy P J, Chinkers M, Radanyi C, Renoir J M, Zhang M, Pratt W B. Biochemistry. 2002;41:13602–13610. doi: 10.1021/bi020399z. [DOI] [PubMed] [Google Scholar]
- 29.Galigniana M D, Housley P R, DeFranco D B, Pratt W B. J Biol Chem. 1999;274:16222–16227. doi: 10.1074/jbc.274.23.16222. [DOI] [PubMed] [Google Scholar]
- 30.Buchanan G, Yang M, Harris J M, Nahm H S, Han G, Moore N, Bentel J M, Matusik R J, Horsfall D J, Marshall V R, et al. Mol Endocrinol. 2001;15:46–56. doi: 10.1210/mend.15.1.0581. [DOI] [PubMed] [Google Scholar]
- 31.Ohta Y, Suzuki N, Nakamura S, Hartwig J H, Stossel T P. Proc Natl Acad Sci USA. 1999;96:2122–2128. doi: 10.1073/pnas.96.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michigami T, Suga A, Yamazaki M, Shimizu C, Cai G, Okada S, Ozono K. J Biol Chem. 1999;274:33531–33538. doi: 10.1074/jbc.274.47.33531. [DOI] [PubMed] [Google Scholar]
- 33.Coppolino M G, Woodside M J, Demaurex N, Grinstein S, St-Arnaud R, Dedhar S. Nature. 1997;386:843–847. doi: 10.1038/386843a0. [DOI] [PubMed] [Google Scholar]
- 34.Ting H J, Yeh S, Nishimura K, Chang C. Proc Natl Acad Sci USA. 2002;99:661–666. doi: 10.1073/pnas.022469899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Truica C I, Byers S, Gelmann E P. Cancer Res. 2000;60:4709–4713. [PubMed] [Google Scholar]
- 36.Mulholland D J, Cheng H, Reid K, Rennie P S, Nelson C C. J Biol Chem. 2002;277:17933–17943. doi: 10.1074/jbc.M200135200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.