Abstract
Yeast endoplasmic reticulum (ER) vesicle protein Erv46p is a novel membrane protein involved in transport through the early secretory pathway. Investigation of mammalian Erv46 (mErv46) reveals that it is broadly expressed in tissues and protein-secreting cells. By immunofluorescence microscopy, mErv46 displays a crescent-shaped perinuclear staining pattern that is characteristic of the Golgi complex. Quantitative immunoelectron microscopy indicates that mErv46 is restricted to the cis face of the Golgi apparatus and to vesicular tubular structures between the transitional ER and cis-Golgi. Minor amounts of mErv46 reside in ER membranes and later Golgi cisternae. On Brefeldin A treatment, mErv46 redistributes to punctate structures that costain for ERGIC53. Depletion of mErv46 protein by RNA interference caused no apparent structural changes in the intermediate compartment or Golgi complex. These findings place mErv46 in a group of itinerant proteins that cycle between the ER and Golgi compartments such as ERGIC53 and the p24 proteins.
In mammalian cells, secretory protein transport from the endoplasmic reticulum (ER) to the Golgi complex proceeds through the morphologically distinct membrane structures of the transitional ER to vesicular tubular elements that then appear to migrate toward and merge with the cis face of the Golgi (1, 2). Transport through these early compartments of the secretory pathway relies on a series of protein sorting and membrane fusion events that are mediated by coat protein complexes and fusion catalysts (3, 4). Coat protein complex II (COPII) generates anterograde vesicles at transitional ER sites that then fuse with or form vesicular tubular structures adjacent to ER exit sites. In a counter membrane flow, coat protein complex I (COPI) mediates retrograde transport from the Golgi and from vesicular tubular elements back to the ER. These vesicular tubular elements, also known as the ERGIC (ER–Golgi intermediate compartment; ref. 5), appear to be formed by fusion of ER-derived vesicles with one another or with existing vesicular tubular structures. Peripheral segments of the ERGIC then appear to mature into new cis-Golgi cisternae as COPI recycles vesicle proteins back to the ER (6). Although morphologically described, many questions remain concerning the molecular mechanisms that govern overall organization of the early secretory pathway.
Genetic and biochemical studies in yeast have contributed significantly to our understanding of the components and mechanisms underlying transport between the ER and Golgi compartments (7). Indeed, COPII was first identified and characterized in yeast (3). Subsequent isolation of in vitro-generated COPII vesicles from yeast ER has allowed for the identification of abundant ER vesicle (Erv) proteins (8). These proteins include members of the p24 protein family, such as Emp24p (9) and Erv25p (10), as well as SNARE proteins, the sorting factors Erv14p and Erv29p, and the Erv41p–Erv46p complex (8, 11, 12). Erv proteins are integral membrane species that cycle between the ER and Golgi compartments and are conserved among eukaryotes. Many of the ERV genes are dispensable for yeast growth but, when mutated, influence protein sorting and/or transport in the early secretory pathway.
To date, the ultrastructural localization of only a few proteins that cycle between the mammalian ER and Golgi has been examined in detail. Specifically, the subcellular distribution patterns for the KDEL receptor, ERGIC53, the p24 complex, and some of the ER–Golgi SNARE proteins have been documented (13–16). In contrast, none of the mammalian homologs of the newly described yeast Erv proteins has been characterized. Given that Erv proteins are widespread in nature and influence ER–Golgi transport in yeast, we hypothesize that they are important components of the early secretory pathway in mammalian cells. Here we investigate a representative mammalian Erv protein, mouse Erv46 (mErv46), to determine its tissue distribution, subcellular localization, and function. We find that mErv46 is not required for the overall structural integrity of the early secretory pathway and speculate that mErv46 influences movement of cargo through this pathway.
Materials and Methods
Antibodies.
The mERV46 cDNA in the pT7T3D-Pac vector was obtained from Research Genetics (Huntsville, AL), I.M.A.G.E. Consortium (LLNL) clone ID 480257 (17). A fragment encoding the luminal domain of mErv46 (amino acid residues 98–272) was amplified from this plasmid by using primers mERV46-pQE-F2 (5′-CGCGGATCCCTGGATGTGGAACACAACCTGTTC-3′) and mERV46-pQE-R (5′-CCCAAGCTTGGCTTGGGGTGCAGTCACGTTGG-3′). The product was inserted into the BamHI and HindIII restriction sites of vector pQE-30 (Qiagen). After induction by isopropyl β-d-thiogalactoside, the 6× histidine-tagged amino-terminal fusion protein was purified on a Ni-nitrilotriacetic acid agarose column (Qiagen) as recommended by the manufacturer. This recombinant protein was used to immunize rabbits according to standard procedures (18). For affinity purification, mErv46 fusion protein was coupled to an Affigel-15 matrix (Bio-Rad), and mErv46-specific antibodies were purified as described (18).
The Sec23p antibody has been described (19). Mouse monoclonal antibodies were purchased from Sigma (tubulin and actin), BD Transduction Laboratories (GM130), and StressGen Biotechnologies (Victoria, BC, Canada; Ig heavy chain binding protein, BiP). Mouse monoclonal ERGIC53 antibody (no. G1/93) was a gift from H. P. Hauri (Biozentrum, Basel), and mouse monoclonal trans-Golgi network (TGN)38 antibody (no. 2F7.1) was a gift from G. Banting (University of Bristol, Bristol, U.K.). Secondary antibodies for immunocytochemistry were from Biosys (Compiegne, France), Abcam (Cambridge, U.K.), Molecular Probes, and Vector Laboratories. Protein A-gold conjugates were prepared in our laboratory.
RNA Interference Experiments.
The mErv46 mRNA was targeted as described (20) with the sequence 5′-AAGGUGGCCGGAAACUUCCACdTdT-3′. Purified and annealed double-stranded RNA oligonucleotides were obtained from Dharmacon Research (Lafayette, CO). HeLa cells were maintained in DMEM containing 10% FCS, 50 units/ml penicillin, and 50 μg/ml streptomycin and seeded onto glass coverslips or dishes and grown to 60–80% confluence. For each transfection, 4 μl of oligofectamine reagent (Invitrogen) was mixed with 15 μl of Optimem (Invitrogen). In addition, 10 μl of a 20-μM double-stranded RNA oligonucleotide solution or Optimem was mixed with 175 μl of Optimem. These solutions were incubated for 10 min at room temperature, and 19 μl of the diluted oligofectamine solution was mixed with 185 μl of diluted RNA oligonucleotide and incubated for 20 min. This transfection solution was then added dropwise to the culture dish containing 800 μl of antibiotic-free DMEM with 10% FCS. Cells were incubated for 3 h, and an additional 500 μl of antibiotic-free media was added to each dish, followed by 48-h incubation. Coverslips were then processed for immunofluorescence or the cells scraped into SDS sample buffer for Western blotting.
Western Blotting.
Various mouse tissue lysates were obtained from Santa Cruz Biotechnology. In addition, mouse livers were homogenized in 40 mM Hepes, pH 7.5/150 mM NaCl/1 mM PMSF/1 μg/ml leupeptin/5 μg/ml pepstatin/1 mM EDTA in a glass/Teflon homogenizer. The homogenate was centrifuged for 5 min at 1,000 × g, and the supernatant was centrifuged for 1 h at 100,000 × g to separate the soluble fraction (S1) from insoluble material. A portion of the pellet fraction (P1) was solubilized with 2% Triton X-100 and centrifuged again for 1 h at 100,000 × g to obtain a detergent extract (S2) and detergent-insoluble pellet (P2). Confluent cultured cells were lysed in 50 mM Tris/HCl (pH 7.4)/150 mM NaCl/1% Triton X-100/5 mM EDTA/1 mM PMSF/200 kallikrein units aprotinin/1 μg/ml pepstatin A. Samples were resolved by standard PAGE and transferred to poly(vinylidene difluoride) or nitrocellulose membranes. Blots were developed by using the enhanced chemiluminescence method (ECL, Amersham Pharmacia).
Light Microscope Immunofluorescence.
Cell lines were grown on glass coverslips and incubated at 37°C. In brefeldin A (BFA) experiments, the cells were incubated with 6 μg/ml BFA for 1 h. Control and BFA-treated cells were fixed with 4% paraformaldehyde and permeabilized by dehydration with ethanol. Single labeling was carried out with mErv46 antibodies followed by FITC-conjugated goat anti-rabbit IgG. Affinity-purified (final dilution 1/20) or immune serum (dilution 1/100) were both used with the same results. For double labeling, cells were first incubated with the mErv46 antibody mixed with either ERGIC53 (1/500), TGN38 (1/10), or BiP (1/50) antibodies. The labeling was visualized with a mixture of anti-rabbit and anti-mouse antibodies conjugated to FITC (green) and tetramethylrhodamine B isothiocyanate (red), respectively. Cells were observed with a laser scanning confocal microscope (LSM 510, Zeiss).
For RNA interference experiments, cells were fixed in 100% methanol at −20°C for 10 min and then blocked in PBS with 1% BSA for 10 min. Cells were then incubated for 30 min at room temperature with the mErv46 antibody (1/1,000) mixed with either GM130 (1/1,000) or ERGIC53 (1/1,000). Cells were then washed for 5 min in PBS and incubated with Alexa594-conjugated goat anti-mouse and FITC-conjugated goat anti-rabbit antibodies for 30 min. Cells were washed in PBS for 30 min, DNA-stained with diamidophenylindole, and mounted with VECTASHIELD (Vector Laboratories). Immunofluorescent images were collected with a Hamamatsu (Bridgewater, NJ) Orca II cooled charge-coupled device camera mounted on a Zeiss Axioplan 2 microscope, using openlab software (Improvision, Lexington, MA).
Electron Microscope Immunogold Labeling.
Cell lines cultured in plastic dishes were fixed with 1% paraformaldehyde and 0.5% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4. Fixed cells were washed with PBS, gently scraped from the dish, and pelleted in 12% gelatin. Gelatin-embedded pellets were cooled on ice, cut on small blocks, infused with 2.3 M sucrose, and processed for ultrathin cryosectioning and immunolabeling according to the protein A-gold method (21). The sections were observed with a 410 LS electron microscope (Philips, Eindhoven, The Netherlands).
Quantitative Immunoelectron Microscopy.
The distribution of mErv46 was quantified on ultrathin cryosections of NRK and INS-1 cells. The following cellular compartments were evaluated: (i) The ERGIC, defined as the tubular vesicular membrane profiles located in the area delimited by transitional elements of the rough ER and the cis-most Golgi cisterna; (ii) the Golgi stack, defined as the area containing stacked ribosome-free cisternae. The lateral rims of the cisternae and the buds were included in the Golgi stack compartment. The cis-to-trans distribution of the labeling was evaluated on cross-sectioned Golgi stacks showing a well defined cis–trans polarity. The cisternae were numbered from cis (C1) to trans (C5). The number of gold particles present on each cisterna was expressed as the percentage of the total number of gold on the stack; (iii) lateral Golgi vesicles, defined as 50- to 70-nm circular membrane profiles not connected to cisternae on the section, and located within 200 nm of the lateral rim of Golgi cisternae; and (iv) the TGN, defined as the tubulo-vesicular membrane profiles at the trans-most side of the Golgi stack. The surface density of labeling was quantified as described (20). All evaluations were performed on micrographs of Golgi areas enlarged 77,000 times. Data are means ± SEM, obtained on 30 different Golgi areas.
Results
Database searches with the yeast Erv46p sequence revealed one Erv46p homolog each in Caenorhabditis elegans, Drosophila melanogaster, and mammals (8). These proteins share a conserved membrane topology, a dilysine COPI binding motif (22, 23), and eight completely invariant cysteine residues. Most of Erv46p is luminally oriented with two membrane spanning segments and short N and C termini that are cytoplasmically exposed and are required for COPII binding and sorting into COPII vesicles (24). Overall, mErv46 and yeast Erv46p sequences consist of 37% identical and 52% similar amino acid residues. From the characterization of yeast Erv46p, we hypothesized that mErv46 is located in the early secretory pathway and involved in secretory processes in mammalian cells (8).
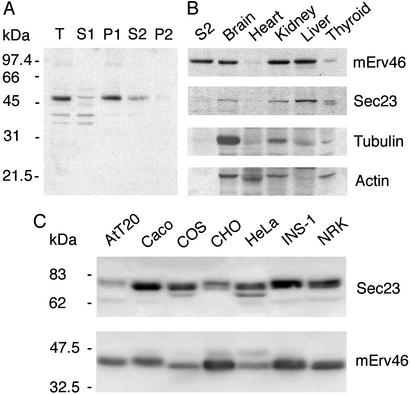
Preliminary studies with antibodies directed against yeast Erv41p and Erv46p showed a weak perinuclear staining pattern in CHO and NRK cells. The number of reactive cells was very small and not sufficient for quantitative experiments. Therefore, high affinity antibodies were raised against the luminal domain of mErv46. The antiserum was tested for specificity on Western blots of mouse liver fractions (Fig. 1A) and purified by affinity chromatography. A strong band of the anticipated electrophoretic mobility was detected in total liver homogenates. This immunoreactive species was highly enriched in the membrane pellet fraction. Moreover, a detergent extract generated from the membrane pellet fraction contained the immunoreactive species as anticipated for an integral membrane protein. Based on these results and depletion experiments (Figs. 4 and 5), we conclude that this antiserum can be used for specific detection of the mErv46 protein.
Figure 1.
(A) mErv46 fractionates as a detergent-soluble membrane protein. Total liver homogenate (T), soluble extract (S1), total membranes (P1), a 2% Triton X-100 extract (S2), and Triton X-100-insoluble material (P2) were resolved on a 12.5% polyacrylamide gel and detected with the polyclonal antibody against mErv46. (B) Tissue distribution of mErv46. Tissue homogenates (16 μg per lane) were resolved on a 12.5% polyacrylamide gel and detected with antibodies against mErv46 (affinity purified), Sec23, tubulin, and actin. The detergent liver extract (S2) from A is included as a positive control. (C) mErv46 expression in cultured cell lines. Total cell extracts (50 μg per lane) were resolved on a 12% polyacrylamide gel and probed with antisera against Sec23 and mErv46.
Figure 4.
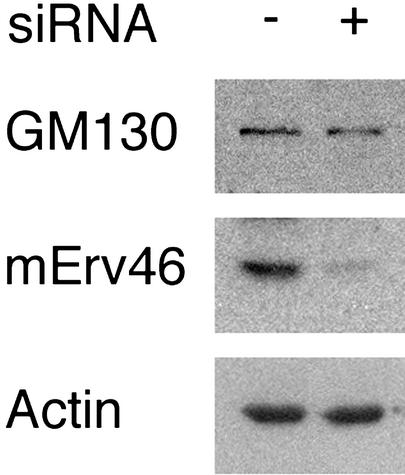
Depletion of mErv46 protein by RNA interference. HeLa cells were lysed 48 h posttransfection with a double-stranded RNA oligonucleotide, and total cell lysates were analyzed by Western blot with the mErv46 antiserum. GM130 and actin are shown as loading controls.
Figure 5.
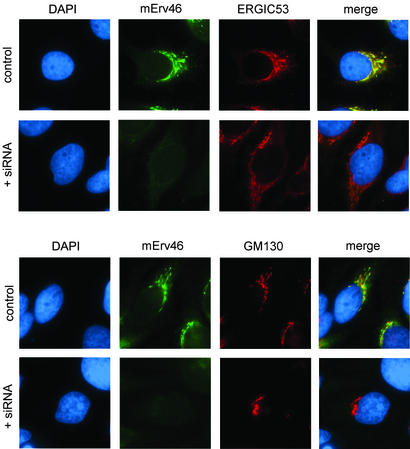
Depletion of mErv46 by RNA interference does not affect the expression and subcellular localization of ERGIC53 and GM130 in HeLa cells. Transfected and control cells were fixed 48 h posttransfection and stained for DNA (diamidophenylindole, DAPI), mErv46, and ERGIC53 (Upper) or for DNA (DAPI), mErv46, and GM130 (Lower).
We hypothesized that, as a component of the secretory pathway, mErv46 would be differentially expressed in various tissues and cultured cell lines depending on their level of secretory activity. Western blots of tissue homogenates (Fig. 1B) and cultured cells (Fig. 1C) showed that mErv46 expression level was indeed tissue specific and, in a majority of cases, similar to the expression of the COPII subunit Sec23. mErv46 expression was particularly strong in liver, kidney, and brain but almost undetectable in heart tissue. Expression was also differential in various commonly used cell lines, in accordance with mErv46 representation in cDNA libraries used for serial analysis of gene expression (25). Importantly, mErv46 was readily detected in INS-1, NRK, COS, CHO, and HeLa cells, enabling us to use these cell lines for further immunocytochemical studies.
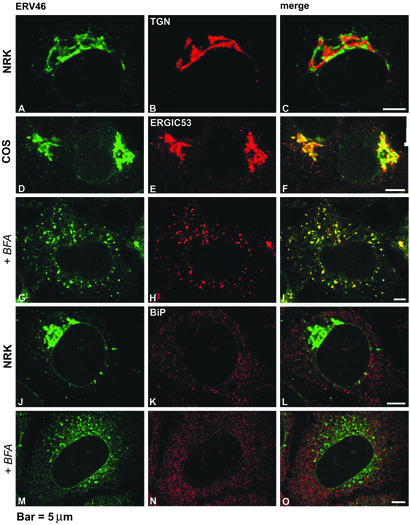
To investigate the subcellular location of mErv46, we first performed double-labeling immunofluorescence experiments with the mErv46 antibody on NRK and COS cells (Fig. 2 A–F). Antibodies against TGN38 and ERGIC53 were used as markers for the TGN and ERGIC, respectively. The mErv46 antibody specifically labeled perinuclear structures in NRK cells (Fig. 2 A and D). mErv46 and TGN38 labeling did not coincide, but staining was observed in adjacent perinuclear compartments of NRK cells (Fig. 2 A–C). In contrast, the mErv46 and ERGIC53 staining patterns showed an extensive but incomplete overlap (Figs. 2 D–F and 5 Upper). Moreover, in double-labeling experiments with the Golgi matrix marker protein GM130, significant but not total overlap was observed (Fig. 5 Lower). These observations suggest that mErv46 localizes to the ERGIC and to the cis pole of the Golgi stack.
Figure 2.
mErv46 is localized to the cis side of the Golgi complex. NRK and COS cell lines, cultured on glass coverslips, were incubated at 37°C or treated with BFA (+BFA) at 6 μg/ml for 1 h. Cells were fixed, permeabilized, and processed for double-labeling immunofluorescence. mErv46 was stained in combination with well characterized markers of the cis and trans sides of the Golgi complex, ERGIC53 and TGN38, respectively, and with the ER marker BiP. Rabbit anti-mErv46 antibody was tagged with FITC-conjugated anti-rabbit IgG (A, D, G, J, and M). Mouse monoclonal antibodies against TGN38 (B), ERGIC53 (E and H), or BiP (K and N) were probed with tetramethylrhodamine B isothiocyanate (TRITC)-conjugated anti-mouse IgG. (Left and Middle) Single-slice confocal images. (Right) Superimposed red and green labeling. mErv46 and TGN38 signals, shown in A–C, are distinctly separated from each other. In contrast, mErv46 (D) and the cis-side Golgi marker ERGIC53 (E) extensively overlap, as shown in the merge image (F). In BFA-treated cells, mErv46 redistributes into spot-like structures, which still colocalize with ERGIC53 (G–I) but do not overlap with the ER marker BiP (M–O). (Bar, 5 μm.)
Because the yeast Erv41p–Erv46p complex is dynamically distributed between ER and Golgi membranes, we tested the effects of COPI inhibition by BFA on the distribution of mErv46 in mammalian cells. BFA treatment causes Golgi resident enzymes to redistribute to the ER (reviewed in ref. 26), whereas cycling proteins such as the p24 proteins (27), KDEL receptor (28), and ERGIC53 (29) are observed in dispersed structures after BFA treatment. BFA treatment caused a striking disruption of the Golgi complex, and both mErv46 and ERGIC53 redistributed to scattered punctate structures that showed extensive overlap (Fig. 2 G–I). Notably, the ER marker BiP and mErv46 did not colocalize before or after BFA treatment (Fig. 2 J–O). These results demonstrate that the cycling behavior of mErv46 is comparable to other ER/Golgi itinerant proteins such as ERGIC53 and the p24 proteins.
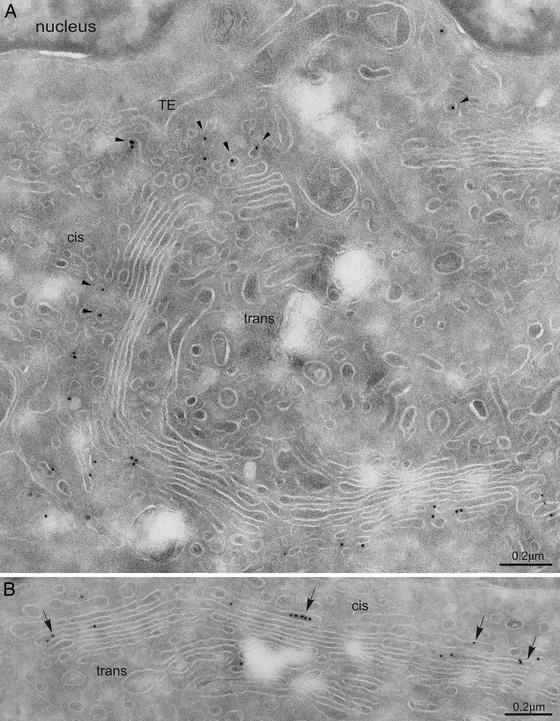
To more precisely quantify the distribution of mErv46 between membrane compartments of the early secretory pathway, immunogold electron microscopy studies were performed (Fig. 3). Gold labeling was frequently seen on vesicular tubular structures near the cis face of the Golgi complex and on the cis-most cisternae of the Golgi. Labeling was also observed in transport vesicles budding from the ER. These observations were corroborated by quantitative immunoelectron microscopy on NRK and INS-1 cells. Comparing the distribution over the ERGIC, the Golgi stack, vesicles lateral to the Golgi stack, and the TGN, ≈70% of labeling was observed within the early Golgi stack. In addition, a significant percentage (≈20%) localized to the ERGIC (Table 1). Regarding distribution over the five Golgi cisternae, the vast majority of mErv46 labeling was observed in the cis-most cisternae of both cell lines. In contrast, cisternae 3–5 contained very minor amounts (Table 2). Taken together, these data demonstrate that the bulk of mErv46 localizes to the ERGIC and to cis-Golgi compartments.
Figure 3.
Immunogold localization of mErv46 on ultrathin cryosections of NRK cells. Shown are Golgi areas in a cis–trans orientation. The region between the transitional elements (TE) of the ER and the cis side of the Golgi contains numerous vesicular tubular profiles. Gold immunolabeling is observed on vesicular tubular elements (arrowheads in A) and on the first two cisternae at the cis-Golgi side (arrows in B). The quantitative evaluation of the labeling for NRK and insulin cells is shown in Tables 1 and 2.
Table 1.
Relative distribution of mErv46 immunogold
| Surface density, gold/μm2
|
% distribution of total gold
|
|||
|---|---|---|---|---|
| NRK | INS-1 | NRK | INS-1 | |
| ERGIC | 30 ± 5 | 51 ± 7 | 20 ± 4 | 22 ± 3 |
| Golgi stack (cisterna 1–5 plus buds) | 36 ± 3 | 27 ± 3 | 71 ± 4 | 71 ± 3 |
| Vesicles lateral to cisternae | 9 ± 3 | 7 ± 3 | 3 ± 1 | 1 ± 1 |
| TGN | 5 ± 1 | 4 ± 1 | 5 ± 2 | 5 ± 1 |
See Quantitative Immunoelectron Microscopy for the detailed methodological steps. Data are means ± SEM.
Table 2.
Percent distribution of immunogold particles on Golgi cisternae C1 (cis-most) to C5 (trans-most)
| Cells | C1 | C2 | C3 | C4 | C5 |
|---|---|---|---|---|---|
| NRK | 72 ± 4 | 21 ± 3 | 4 ± 1 | 1 ± 1 | 1 ± 1 |
| INS-1 | 65 ± 4 | 20 ± 3 | 6 ± 1 | 8 ± 2 | 1 ± 1 |
See Quantitative Immunoelectron Microscopy for the detailed methodological steps. Data are means ± SEM.
To investigate the influence of mErv46 depletion on ERGIC and Golgi structure, mErv46 mRNA was targeted for degradation by RNA interference (20). Typically, transfection efficiencies of 90% were observed in HeLa cells 48 h post transfection with the double-stranded RNA oligonucleotide. Untransfected and transfected cells were homogenized and analyzed for mErv46 protein expression by Western blotting using actin as a loading control (Fig. 4). mErv46 protein was reduced ≈10-fold in the transfected cells, whereas the expression level of the Golgi matrix protein GM130 was unchanged. In immunofluorescence experiments, these cells did not exhibit any detectable staining with the mErv46 antibody (Fig. 5), confirming that mErv46 protein expression was efficiently suppressed. Moreover, this result demonstrated the high specificity of the mErv46 antibody. Double labeling with the ERGIC53 antibody did not show any significant difference in expression and subcellular localization of this protein in the transfected cells. Likewise, the localization pattern of the Golgi matrix protein GM130 was unchanged. Taken together, these results demonstrate that mErv46 expression can be efficiently inhibited by RNA interference. However, loss of mErv46 function does not appear to have an obvious effect on the maintenance of ERGIC or Golgi matrix structure.
Discussion
Our localization studies support the hypothesis that mErv46 operates in the early secretory pathway of mammalian cells. Compared with other integral membrane proteins that act in these early transport steps, mErv46 displays a similar distribution pattern as reported for ERGIC53, the KDEL receptor, and p24 proteins (14, 15, 21). Furthermore, BFA causes a similar relocalization of this set of cycling proteins to distinct punctate structures. Costaining experiments revealed a striking separation of mErv46 from the trans-Golgi-localized protein, TGN38. Quantitative immunoelectron microscopy further documents this separation because the concentration of mErv46 falls sharply from cis- to trans-Golgi cisternae. This boundary, also delineated by ERGIC53 and the KDEL receptor, likely represents a functional division of the ERGIC and cis-Golgi from later Golgi compartments (6).
The subcellular distribution of ERGIC53 and the p24 proteins are controlled by cytoplasmically exposed C-terminal amino acid residues (30, 31). In yeast cells, Erv46p is in tight association with a related protein, termed Erv41p, and this complex cycles between the ER and Golgi complex. Erv41–Erv46 localization depends on the C-terminal tails of both subunits, which are recognized by the COPI and COPII coats. Erv46p contains a canonical COPI binding motif for retrograde transport, a feature that is conserved among all of its homologs in other species. Anterograde transport by COPII requires a hydrophobic COPII binding motif on Erv41p in concert with a diaromatic motif on Erv46p (24). Therefore, we propose that the C-terminal tail sequences of a mammalian mErv41–mErv46 complex are responsible for its observed localization through dynamic associations with COPI and COPII. Notably, many of the localization properties of the mErv41–mErv46 complex are reminiscent of ERGIC53 and the p24 complex, which have been characterized in both yeast and mammalian cells (14, 27, 31–33).
The subcellular localization of mErv46 reported in this study points to a conserved function in yeast and mammalian cells. Although the precise function of Erv46p in yeast is unclear, biochemical and genetic experiments suggest a role for this protein at the Golgi complex (8). Our RNA interference data indicate that although mErv46 localizes to the ERGIC and cis-Golgi compartments, loss of mErv46 protein does not alter the expression levels or localization of marker proteins for these compartments. Therefore, mErv46 is apparently not required for the maintenance of the ERGIC compartment or the Golgi matrix, at least under the time frame we examined. Similarly, Erv46p is not essential for secretory pathway function in yeast (8).
The parallel properties of mErv46 with ERGIC53 and the p24 proteins suggest related functions. Studies on ERGIC53 indicate it functions as a transport receptor for soluble secreted glycoproteins (33, 34), whereas the p24 proteins act in transport of specific glycosylphosphatidylinositol-anchored proteins (35). These activities are apparently not essential for operation of the secretory pathway but rather influence transport of specific secretory cargo through this pathway (33, 36). Similarly, our RNA interference data indicate that mErv46 is not required for the maintenance of the ERGIC and Golgi compartments in animal cells. Based on these observations, we propose that mErv46 acts on protein or lipid species at the ER–Golgi interface, perhaps in their transport or retrieval within the early secretory pathway. Further studies in yeast will provide tests of this proposal. Regardless, the demonstration that mErv46 expression is widespread and dynamic in animal cells suggests that it and other Erv proteins could emerge as important components of the eukaryotic secretory pathway.
Acknowledgments
This work was supported by grants from the Swiss National Science Foundation (to L.O.), the National Institutes of Health (to C.B.), and the Hitchcock Foundation (to C.B.). S.O. was supported by a fellowship from Deutsche Forschungsgemeinschaft.
Abbreviations
- ER
endoplasmic reticulum
- COPI
coat protein complex I
- COPII
coat protein complex II
- ERGIC
ER–Golgi intermediate compartment
- Erv
ER vesicle
- mErv46
mouse Erv protein 46
- BFA
brefeldin A
- TGN
trans-Golgi network
- BiP
Ig heavy chain binding protein
References
- 1.Palade G. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- 2.Presley J F, Cole N B, Schroer T A, Hirschberg K, Zaal K J, Lippincott-Schwartz J. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- 3.Schekman R, Orci L. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- 4.Rothman J E, Wieland F T. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 5.Hauri H-P, Schweizer A. Curr Opin Cell Biol. 1992;4:600–608. doi: 10.1016/0955-0674(92)90078-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klumperman J. Curr Opin Cell Biol. 2000;12:445–449. doi: 10.1016/s0955-0674(00)00115-0. [DOI] [PubMed] [Google Scholar]
- 7.Kaiser C A, Gimeno R E, Shaywitz D A. In: Yeast III. Pringle J R, Broach J R, Jones E W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 91–227. [Google Scholar]
- 8.Otte S, Belden W J, Heidtman M H, Liu J, Jensen O N, Barlowe C. J Cell Biol. 2001;152:503–517. doi: 10.1083/jcb.152.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schimmöller F, Singer-Krüger B, Schröder S, Krüger U, Barlowe C, Riezman H. EMBO J. 1995;14:1329–1339. doi: 10.1002/j.1460-2075.1995.tb07119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belden W J, Barlowe C. J Biol Chem. 1996;271:26939–26946. doi: 10.1074/jbc.271.43.26939. [DOI] [PubMed] [Google Scholar]
- 11.Powers J, Barlowe C. Mol Biol Cell. 2001;13:880–891. doi: 10.1091/mbc.01-10-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belden W J, Barlowe C. Science. 2001;294:1528–1531. doi: 10.1126/science.1065224. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths G, Ericsson M, Krijnse-Locker J, Nilsson T, Goud B, Söling H-D, Tang B L, Wong S H, Hong W. J Cell Biol. 1994;127:1557–1574. doi: 10.1083/jcb.127.6.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rojo M, Pepperkok R, Emery G, Kellner R, Strang E, Parton R G, Gruenberg J. J Cell Biol. 1997;139:1119–1135. doi: 10.1083/jcb.139.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klumperman J, Schweizer A, Clausen H, Tang B L, Hong W, Oorschot V, Hauri H-P. J Cell Sci. 1998;111:3411–3425. doi: 10.1242/jcs.111.22.3411. [DOI] [PubMed] [Google Scholar]
- 16.Hay J C, Klumperman J, Oorschot V, Steegmaier M, Kuo C S, Scheller R H. J Cell Biol. 1998;141:1489–1502. doi: 10.1083/jcb.141.7.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lennon G G, Auffray C, Polymeropoulos M, Soares M B. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 18.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 312–319. [Google Scholar]
- 19.Hicke L, Schekman R. EMBO J. 1989;8:1677–1684. doi: 10.1002/j.1460-2075.1989.tb03559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elbashir S M, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 21.Orci L, Stamnes M, Ravazzola M, Amherdt M, Perrelet A, Söllner T H, Rothman J E. Cell. 1997;90:335–349. doi: 10.1016/s0092-8674(00)80341-4. [DOI] [PubMed] [Google Scholar]
- 22.Jackson M R, Nilsson T, Peterson P A. EMBO J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cosson P, Letourneur F. Science. 1994;263:1629–1631. doi: 10.1126/science.8128252. [DOI] [PubMed] [Google Scholar]
- 24.Otte S, Barlowe C. EMBO J. 2002;21:6095–6104. doi: 10.1093/emboj/cdf598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Velculescu V E, Zhang L, Vogelstein B, Kinzler K W. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 26.Klausner R, Donaldson J G, Lippincott-Schwartz J. J Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Füllekrug J, Suganuma T, Tang B L, Hong W, Storrie B, Nilsson T. Mol Biol Cell. 1999;10:1939–1955. doi: 10.1091/mbc.10.6.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Füllekrug J, Sönnichsen B, Schäfer U, Nguyen Van P, Söling H D, Mieskes G. FEBS Lett. 1997;404:75–81. doi: 10.1016/s0014-5793(97)00097-5. [DOI] [PubMed] [Google Scholar]
- 29.De Lemos-Chiarandini C, Ivessa N E, Black V H, Tsao Y S, Gumper I, Kreibich G. Eur J Cell Biol. 1992;58:187–201. [PubMed] [Google Scholar]
- 30.Kappeler F, Klopfenstein D R, Foguet M, Paccaud J-P, Hauri H-P. J Biol Chem. 1997;272:31801–31808. doi: 10.1074/jbc.272.50.31801. [DOI] [PubMed] [Google Scholar]
- 31.Dominguez M, Dejgaard K, Füllekrug J, Dahan S, Fazel A, Paccaud J-P, Thomas D Y, Bergeron J J M, Nilsson T. J Cell Biol. 1998;140:751–765. doi: 10.1083/jcb.140.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schröder S, Schimmöller F, Singer-Krüger B, Riezman H. J Cell Biol. 1995;131:895–912. doi: 10.1083/jcb.131.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato K, Nakano A. Mol Biol Cell. 2002;13:2518–2532. doi: 10.1091/mbc.E02-01-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Appenzeller C, Andersson H, Kappeler R, Hauri H-P. Nat Cell Biol. 1999;1:330–334. doi: 10.1038/14020. [DOI] [PubMed] [Google Scholar]
- 35.Muniz M, Nuoffer C, Hauri H P, Riezman H. J Cell Biol. 2000;148:925–930. doi: 10.1083/jcb.148.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Springer S, Chen E, Duden R, Marzioch M, Rowley A, Hamamoto S, Merchant S, Schekman R. Proc Natl Acad Sci USA. 2000;97:4034–4039. doi: 10.1073/pnas.070044097. [DOI] [PMC free article] [PubMed] [Google Scholar]