Abstract
The synaptonemal complex (SC) is a proteinaceous complex that apparently mediates synapsis between homologous chromosomes during meiotic prophase. In Saccharomyces cerevisiae, the Zip1 protein is the integral component of the SC. In the absence of a DNA double-strand break or the SC initiation protein Zip3, Zip1 proteins aggregate to form a polycomplex (PC). In addition, Zip1 is also responsible for DSB-independent nonhomologous centromere coupling at early meiotic prophase. We report here that Zip3 is a SUMO (small ubiquitin-related modifier) E3 ligase and that Zip1 is a binding protein for SUMO-conjugated products. Our results also suggest that at early meiotic prophase, Zip1 interacts with Zip3-independent Smt3 conjugates (e.g., Top2) to promote nonhomologous centromere coupling. At and after mid-prophase, the Zip1 protein begins to associate with Zip3-dependent Smt3 conjugates (e.g., Red1) along meiotic chromosomes in the wild-type cell to form SCs and with Smt3 polymeric chains in the zip3 mutant to form PCs.
Keywords: Meiosis, synaptomenal complex, Zip1, Zip3, SUMO, Ulp2
Meiosis (or sporulation) in Saccharomyces cerevisiae is a process of cellular differentiation that is initiated in diploid a/a cells in response to nutrient limitation to generate gametes. Following a single round of DNA replication, two successive rounds of chromosomal segregation reduce the diploid chromosome complement by half. Segregation of chromosomes at the reductional division depends on a series of interactions between homologous chromosomes, including pairing, assembly of the synaptonemal complex (SC), genetic recombination, and formation of chiasmata.
SCs consist of paired lateral elements (LEs) perpendicularly connected by transverse filaments (TFs). In addition, a central element (CE) appears as an electron-dense, linear structure running down the center of the SC. In S. cerevisiae, the Zip1 protein is the integral component of the SC and is found throughout the entire length of the SC (Sym et al. 1993). Zip1 is the only known yeast SC component that behaves this way and is believed to be the ortholog of SCP1 in nematodes and mammals (Zickler and Kleckner 1999). Zip1 forms two large, central α-helical coiled-coil domains flanked by N- and C-terminal globular domains (Sym et al. 1993). Zip1 proteins are thought to form a head-to-head dimer or tetramer connected by interaction between the central coiled-coil domains, while the Zip1 C-terminal globular domain connects the TFs to chromosomes or LEs (Tung and Roeder 1998; Dong and Roeder 2000). Several S. cerevisiae proteins on LEs have been implicated in SC assembly, including Red1, Mek1, Hop1, Top2, Pds5, cohesins (e.g., Rec8), and condensins (e.g., Ycs4) (Page and Hawley 2004). The SC-initiating protein Zip3 was presumed to mark crossover-designated recombination sites and recruit Zip2 and Zip1 to form the central region of the SC (Chua and Roeder 1998; Agarwal and Roeder 2000; Borner et al. 2004; Fung et al. 2004; Henderson and Keeney 2004). Two additional proteins, Spo22/Zip4 (Perry et al. 2005) and Spo16 (A. Shinohara, pers. comm.), are also required for SC assembly. Zip1 also mediates centromere coupling between nonhomologous chromosomes during early prophase (Tsubouchi and Roeder 2005); however, whether or not this phenomenon is related to SC assembly remains controversial (Henderson and Keeney 2005).
In many organisms, including S. cerevisiae, aggregates of SC-related material (i.e., polycomplexes, PCs) have been observed either before or during regular SC development. PCs are highly accumulated in yeast mutants where chromosome morphogenesis is aberrant because of a lack of a structural or recombinational component (e.g., spo11, zip2, zip3, red1, hop1) as well as in mutants exhibiting a prophase block (e.g., clb5 clb6). Like the SC, Zip1 is the main structural component of PCs. PCs have morphological characteristics in common with SCs; however, PCs exhibit many different morphologies that indicate that they clearly are not simply stacks of regular SCs discarded from the bivalents (Zickler and Kleckner 1999). The molecular mechanism underlying the formation of SCs and PCs is still unknown. Here we report that SUMO (small ubiquitin-related modifier) modification plays a key role in both processes.
SUMOylation—the covalent linking of SUMO to lysine residues—is a reversible post-translational modification controlled by an enzymatic pathway analogous to the ubiquitin pathway. The addition of SUMO to target proteins has been implicated in cellular functions by a number of mechanisms (Johnson 2004; Potts and Yu 2005; Zhao and Blobel 2005). In S. cerevisiae, a single essential gene, SMT3, encodes the SUMO precursor product. The Smt3 protein is activated by the formation of a thioester bond with a cysteine residue on the E1 activating enzyme (i.e., the Aos1–Uba2 complex). Smt3 is then passed to an E2 conjugation enzyme (Ubc9), with which it also forms a thioester bond with a cysteine residue. Smt3 E3 ligases act as “adapters” to interact with both Ubc9 and substrates and promote transfer of Smt3 from E2 to specific substrates (for reviews, see Gill 2004; Johnson 2004; Muller et al. 2004).
Three Smt3 E3 ligases have been identified in S. cereviseae: Siz1, Siz2/Nfi1, and Mms21/Nse2. Siz1 and Siz2 are not essential proteins, whereas Mms21 is a part of the octameric complex that is essential for vegetative growth and DNA repair (Potts and Yu 2005; Zhao and Blobel 2005). These three E3 enzymes are members of the PIAS (protein inhibitor of activated STAT) family, all of which contain a SP-RING (Siz/PIAS RING) domain. The SP-RING domains interact with Ubc9 and are responsible for their E3 catalytic activities. Structurally, SP-RING domains are similar to the RING domains found in many ubiquitin E3 ligases. Smt3 is often, but not always, attached to most substrates at the lysine in a consensus sequence, ΨKXE, where Ψ is a large hydrophobic amino acid and X can be any amino acid. Removal of Smt3 from substrates is mediated by two yeast Smt3 deconjugating enzymes, Ulp1 and Ulp2, respectively. The S. cerevisiae SMT3, AOS1, UBA2, UBC9, and ULP1 genes are all essential for vegetative growth (for reviews, see Gill 2004; Johnson 2004; Muller et al. 2004). Unlike ULP1, ULP2 is dispensable for vegetative growth. On the other hand, the ulp2-null mutant exhibits a cell cycle arrest phenotype at meiotic prophase (Li and Hochstrasser 2000), indicating that Smt3 modification or removal is likely to have specific meiotic functions.
A special feature of the SUMO system in yeast is that in the absence of E3 ligase, E1 and E2 can catalyze Smt3 polymeric chains or even the Smt3-conjugated E1 and E2 in vitro in an ATP-dependent manner (Bencsath et al. 2002; Takahashi et al. 2003). The reaction rate of Smt3 chain formation is further accelerated upon addition of E3 ligase because Smt3 itself is also a SUMO target and E3 acts an adapter to facilitate transferring additional Smt3 from E2 to the Smt3 chain. Several known Smt3 E3s have been shown to exhibit such activity (Johnson and Gupta 2001; Bencsath et al. 2002; Taylor et al. 2002; Bylebyl et al. 2003; Zhao and Blobel 2005). The Smt3 polymeric chain will not form in a reaction containing a Smt3 mutant lacking either Lys 11, Lys 15, and Lys 19 (i.e., smt3-11, smt3-15, smt3-19R) or all nine lysines (i.e., smt3-allR) (Bylebyl et al. 2003; Takahashi et al. 2003).
Here we used multiple approaches to examine the roles of SUMO modification in yeast meiosis. We show that global Smt3 modifications along meiotic chromosomes are regulated by Zip3 protein at the time SCs form, and Zip1 proteins are colocalized with chromosomal Smt3 conjugates along SCs. Importantly, the zip3 mutant not only exhibits only a few Smt3 modifications along the meiotic chromosome but also accumulates a high level of Smt3 polymeric chains. These Smt3 polymeric chains aggregate with Zip1 proteins at the PC. Subsequent biochemical analysis indicated that Zip3 is an Smt3 E3 ligase and Zip1 is a binding protein for Smt3 conjugated products. These observations have led to a proposed molecular model for the assembly of SCs and PCs.
Results
Zip1 proteins were localized with the immunostaining signals of Smt3 conjugates
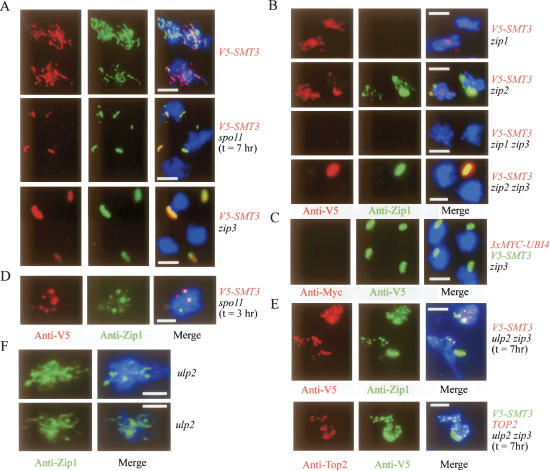
To investigate the roles of Smt3 modifications during meiosis, we constructed SK1 yeast strains that express either the V5-epitope-tagged Smt3 protein (V5-Smt3) and/or the Myc-tagged Ubi4 protein (3Myc-Ubi4). The UBI4 gene encodes a five head-to-tail fusion of ubiquitins and is responsible for polyubiquitination in S. cerevisiae (Finley et al. 1987). The coding sequence of triple Myc tags was added to the 5′ end of the UBI4 gene. Western blotting analysis indicated that both the Myc-tagged Ubi4 monomer and polyubiquitinated products were expressed in mitotic and meiotic yeast cells. Addition of a V5 epitope to Smt3 or triple Myc epitopes to Ubi4 did not result in any apparent defect in vegetative growth, sporulation efficiency, or spore viability (data not shown). Immunostaining of meiotic nuclear spreads revealed that most V5-Smt3 conjugates were colocalized with Zip1 at the pachytene chromosomes (or SCs) of the wild-type cells. On the other hand, in the spo11 or zip3 mutants, the V5-Smt3 conjugates were colocalized with Zip1 at PCs instead of meiotic chromosomes after mid-prophase. Intriguingly, the intensity of the V5-Smt3 signals at PCs was much greater in zip3 than in spo11 (Fig. 1A).
Figure 1.
Colocalization of Smt3 conjugates but not ubiquitin conjugates with Zip1 at SCs and PCs. Representative images of surface nuclei spreads of various sporulating cells stained with DAPI (in blue), anti-Zip1, anti-V5, anti-Myc, or anti-Top2 antibodies, respectively. All white bars represent 5 μm.
Mutant analysis indicated that Zip3 could promote chromosomal Smt3 modifications at and after mid-prophase. In the zip1 single mutants, V5-Smt3 signals appeared as foci during early prophase in a ZIP3-independent manner, and then spread all over chromosomes after mid-prophase in a ZIP3-dependent manner. In the zip2 mutant, both V5-Smt3 and Zip1 signals exhibited patterns similar to those of V5-Smt3 in the zip1 mutant (Fig. 1B). Disappearance of early Smt3 foci and appearance of ZIP3-dependent Smt3 signals along meiotic chromosomes (or SCs) were also observed at mid-prophase of the wild-type cell (Fig. 2; Supplementary Fig. S1).
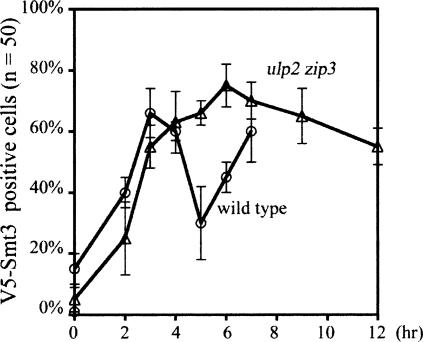
Figure 2.
Time-course analysis of the Smt3 modification along meiotic chromosomes. Surface spreads of the sporulating wild-type and ulp2 zip3 cells were stained with anti-V5 (for V5-Smt3). Percentages of surface spreads showing positive V5-Smt3 signals along meiotic chromosomes were determined. Representative images of the wild-type surface nuclei spreads are provided in Supplementary Figure S1. In the wild-type cell, loss of early Smt3 foci and appearance of extensive chromosomal Smt3 signals occur between 5 and 6 h after the cells were transferred into sporulating medium. The majority of the wild-type cells (>40%) began to exhibit the first nuclear divisions after 8 h of incubation in the sporulating time point. Our results revealed that Smt3 foci gradually accumulated in the ulp2 zip3 mutant at early prophase and were sustained to at least 12 h. No SC-like structure (i.e., extensive line of Smt3 signal) was observed along meiotic chromosomes in the ulp2 zip3 mutant.
Ubiquitin or ubiquitin-conjugated products were not detected at PCs in zip3 either by anti-Myc antibody (for 3Myc-Ubi4) (Fig. 1C) or by two additional polyclonal anti-ubiquitin antibodies (data not shown). The latter were used to detect all four yeast ubiquitins, Ubi1–4 (Finley et al. 1987). We had confirmed by Western blotting and whole-cell immunostaining analysis (Supplementary Fig. S2) that these antibodies can detect ubiquitins or ubiquitinated products. However, these three antibodies exhibited very weak staining signals along meiotic chromosomes of the wild-type cell and at PCs of the zip3 mutant (Fig. 1C). We conclude that Smt3 conjugates, like Zip1, are specific components of SCs and PCs.
Smt3-conjugated products may recruit Zip1 onto meiotic chromosomes
As described above, most Zip1 form foci together with V5-Smt3 conjugates on meiotic chromosomes of wild-type (Fig. 2), spo11 (Fig. 1D), and zip2 and zip3 (data not shown) cells during early meiotic prophase (∼3-h time point). Recently, these early Zip1 foci have been implicated in centromere coupling between nonhomologous chromosomes in a Spo11-independent manner (Tsubouchi and Roeder 2005). At mid-prophase, these early chromosomal Zip1 foci began to disappear in the wild-type cell (Fig. 2) and zip3 mutant (Fig. 1A) but were retained in the ulp2 zip3 mutants (Figs. 1E, 2). Because Ulp2 is an Smt3 deconjugating enzyme, such a ULP2-dependent clearance of Smt3/Zip1 foci from chromosomes suggests that Smt3 conjugates may be involved in recruiting Zip1 to chromosomes (see below). Intriguingly, we also found that the early Smt3 foci were colocalized not only with Zip1 but also with Topoisomerase II (Top2). Top2 staining signals were not uniformly distributed on the chromosomes, and there were clearly several strongly stained Top2 foci or spots (Fig. 1E). This result was consistent with a previous report that Top2 was Smt3 modified and highly accumulated at the centromere (Bachant et al. 2002). Like zip3, the ulp2 zip3 mutant failed to form SCs, indicating that these early Smt3/Zip1 foci were not sufficient for SC assembly. On the other hand, a significant portion of the ulp2 cells (>25%) could form Zip1 lines or SCs along meiotic chromosomes after mid-prophase (Fig. 1F). We conclude that Zip3-dependent SUMO modifications are required for SC formation.
Accumulation of Smt3 polymeric chains in the zip3 mutant after mid-prophase
Western time-course analysis was also carried out to examine the global Smt3 conjugation profiles. We found that the wild-type cell exhibited a dynamic global Smt3 modification profile with several Smt3-conjugated products showing different temporal expression patterns. On the contrary, the majority of Smt3-conjugated products accumulated in the ulp2 mutant (Supplementary Fig. S3). This result is consistent with the role of Ulp2 as an Smt3 deconjugating enzyme in meiosis (Li and Hochstrasser 2000). We next examined the genetic requirements for normal Smt3 profiles, including meiotic cell cycle regulators (i.e., CDC28/CLB5, CLB6), genes involved in DSB formation and repair (i.e., SPO11, DMC1) and genes involved in SC formation (ZIP1–4, SPO16). In all cases, the intensity of V5-Smt3 signals was used as an internal reference for Western analysis. Compared with those in the wild-type cell, the Smt3 conjugation profiles of clb5 clb6, spo11, dmc1 (data not shown), zip1, or zip2 mutants did not display dramatic changes (Supplementary Fig. S4).
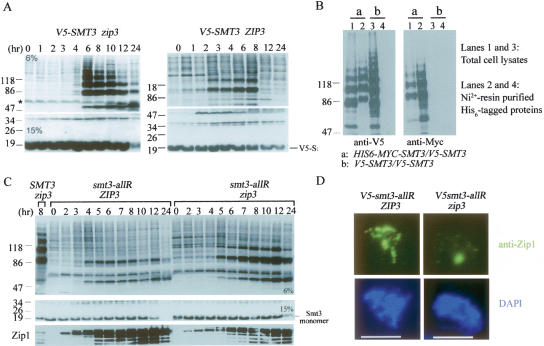
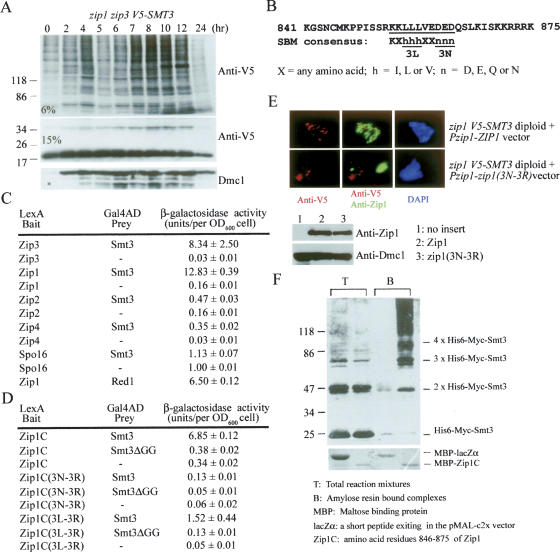
On the other hand, the zip3 mutant exhibited a unique Smt3 modification profile. At 4–6 h (i.e., mid-prophase), it began to accumulate a ladder of V5-Smt3-conjugated products that were not clearly observed either in the wild type or in any other mutants examined here (Fig. 3A; Supplementary Fig. S4). These V5-Smt3-conjugated ladders were not polyubiquitinated products of V5-Smt3, as revealed by the results of immunocytological analysis (Fig. 1C) and Western time-course analysis (Supplementary Fig. S2). Because the differences between their apparent molecular weights are close to the molecular weight of the V5-Smt3 monomer (15,000–20,000), it is likely that these ladders are Smt3 polymeric chains. To examine this hypothesis, we constructed a zip3 strain expressing both V5-Smt3 and His6-Myc-Smt3. Total cellular proteins after mid-prophase were collected by trichloroacetic acid (TCA) precipitation, solubilized in a denaturing buffer containing 8 M urea, and subjected to purification with Ni2+-chelating resins that selectively retained the His6-tagged polypeptides. Upon examination by Western blots using either anti-V5 or anti-Myc antibody, the eluates contained both V5-Smt3 and His6-Myc-Smt3 (Fig. 3B). This result indicated that V5-Smt3 and His6-Myc-Smt3 were covalently linked to one another. A zip3 cell expressing only the V5-Smt3 protein was used here as a negative control for the copurification experiment. In this case, the V5-Smt3-conjugated product was not isolated by Ni2+-chelating resins (Fig. 3B).
Figure 3.
Massive accumulation of Smt3 polymeric chains in zip3 during and after mid-prophase. (A) Western time-course analysis of wild-type and zip3 mutant expressing the V5-tagged Smt3 protein. Total cell lysates were prepared by TCA precipitation, and the proteins were separated either by a 6% or 15% SDS-PAGE as indicated. (B) V5-Smt3 covalently links with His6-Myc-Smt3 in the zip3 mutant. Two different zip3 mutants expressing either both V5-Smt3 and His6-Myc-Smt3 (a) or only the V5-Smt3 protein (b) were constructed. Sporulating cells were harvested at the 8-h time point, TCA-precipitated, and dissolved in a denaturing buffer containing 8 M urea. The His6-tagged polypeptides were purified by the Ni2+ chelating resin. Western analysis of total cell extracts (lanes 1,3) and the purified His6-tagged polypeptides (lanes 2,4) was performed using anti-V5 and anti-Myc antibody as indicated. (C) Western time-course analysis of yeast strains expressing the V5-tagged smt3-allR protein. The smt3-allR mutant protein does not form a polymeric chain. Total cell lysates of zip3 mutant expressing the V5-tagged wild-type Smt3 protein were used here as a positive control for showing the Smt3 polymeric chains (the first lane from the left). Zip1 protein was used to monitor progression of the meiotic cell cycle. (D) Immunostaining of Zip1 in the nuclear surface spreads of yeast strains expressing the V5-tagged smt3-allR protein. All white bars represent 5 μm. The ZIP3 V5-smt3allR cell forms aberrant SCs (i.e., Zip1 foci or short lines), and the zip3 V5-smt3allR cell only forms PCs. These PCs were much smaller in size than those observed in the zip3 V5-smt3allR cells (Fig. 1A).
We also constructed ZIP3 and zip3 strains expressing only the V5-tagged smt3-allR mutant protein. The smt3-allR mutant protein conceivably could not form Smt3 polymeric chains because all nine of its lysine residues were mutated into arginine residues. Because SMT3 is an essential gene and the smt3-allR yeast strain is still viable, the V5-smt3-allR mutant protein is at least partially functional for SUMO modification (Bylebyl et al. 2003). Western time-course analysis revealed that Smt3 ladders or polymeric chains were significantly diminished in the zip3 V5-smt3-allR cell (Fig. 3C); however, a mysterious Smt3 conjugate product with an apparent molecular weight of ∼100,000 also accumulated in this mutant. The apparent molecular weight of this mysterious Smt3 conjugate product indicates that it may be the Smt3-modified product of Uba2, one of the two subunits of the SUMO E1 enzyme. The predicted molecular weight of the Uba2 protein is 71,258. Immunostaining of nuclear surface spreads further revealed that the ZIP3 V5-smt3-allR cell could form Zip1 foci and short lines along meiotic chromosomes but could hardly form normal SCs (Fig. 3D). Moreover, the ZIP3 V5-smt3-allR cells (<5%) hardly form spores. These results suggest that SUMO conjugations with only the Smt3 monomers along meiotic chromosomes are not sufficient for normal SC assembly. Additionally, the zip3 V5-smt3-allR mutant still could form PCs; however, these PCs exhibited much weaker or smaller Zip1 staining signals (Fig. 3D) than those in the zip3 V5-SMT3 mutant (Fig. 1A). Taken together, the results indicate that the zip3 mutant accumulates Smt3 polymeric chains after mid-prophase and that these Smt3 polymeric chains aggregate together with Zip1 proteins to form bigger PCs. Moreover, our data also reveal that SUMO conjugations by Smt3 polymeric chains along meiotic chromosomes may play essential roles in promoting formation of extensive SCs.
Zip3 is an Smt3 E3 ligase
It had been reported previously that in the absence of SUMO E3 ligase, Smt3 together with Aos1–Uba2, Ubc9, and ATP could modify itself into Smt3 polymeric chains (Bencsath et al. 2002; Takahashi et al. 2003). The finding that Smt3 polymeric chains accumulate in the zip3 mutant (Fig. 3) leads to the hypothesis that Zip3 may be an Smt3 E3 ligase. Several different lines of evidence provided here support this view. First of all, our cytological results have indicated that ZIP3-dependent chromosomal Smt3 modifications are required for SC formation (Fig. 1A). Consistent with a previous report that Zip3 acts prior to Zip2 and Zip1 in SC assembly (Agarwal and Roeder 2000), we also showed that Zip3 could promote chromosomal Smt3 modifications in the absence of Zip1 or Zip2 after mid-prophase (Fig. 1B). Second, it was shown that Ubc9 remained covalently attached to Smt3 in the absence of E3 (Bylebyl et al. 2003; Takahashi et al. 2003). We confirmed by Western time-course analysis that the Ubc9-Smt3 conjugates accumulate in the zip3 mutant. In addition, the results of immunocytological experiments revealed that Ubc9 or Ubc9-Smt3 conjugates are located at PCs, but not along meiotic chromosomes in the zip3 mutant (Supplementary Fig. S5). Therefore, Zip3 may have a role in recruiting Ubc9 onto meiotic chromosomes.
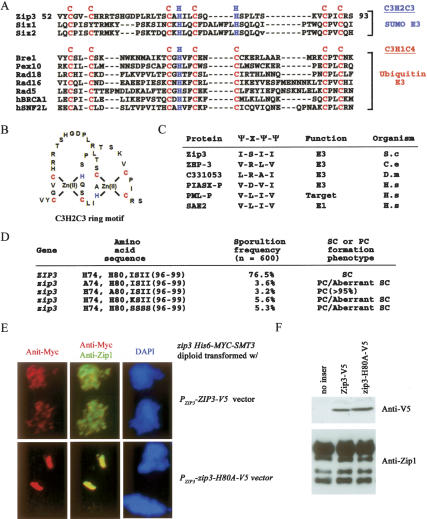
Third, a recent bioinformatics study suggests that Zip3 is a ubiquitin E3 ligase (Perry et al. 2005). This hypothesis was not favored by current findings: Western time-course analysis results in the current study revealed that the global ubiquitin conjugation pattern of the zip3 cell did not significantly differ from that of the wild-type cell at the 4–6-h sporulating time point—in other words, at the time when the Zip3 protein was expected to carry out its meiotic function (Supplementary Fig. S3). Here, we independently found that the Zip3 protein contained a SP-RING domain (residues 52–93) (Johnson and Gupta 2001) and a noncovalent Smt3-binding motif (SBM; residues 96–99, I-S-I-I). The latter was a consensus amino acid sequence (Ψ-X-Ψ-Ψ) identified from several known Smt3-binding proteins or SUMOylating enzymes; Ψ represents I, V, or other large, hydrophobic residues; X could be any amino acid residue (Song et al. 2004). The SP-RING domain contains a conserved C3H2C3 RING-finger motif (RFM), in which C and H represent cysteine and histidine, respectively. C3H2C3 RFMs were also found in Siz1 and Siz2, except that a cysteine residue of Siz1 is replaced by tyrosine. These RFMs are responsible for the E3 catalytic activities of Siz1 and Siz2 (Johnson and Gupta 2001). Similar but slightly different RFMs (i.e., C3HC4) were found in several ubiquitin E3 ligases, including Rad18 and Rad16 (Fig. 4A,B). Intriguingly, both RIM and SBM are evolutionarily conserved among Zip3 homologous proteins found in other organisms, including ZHP-3 of Caenorhabditis elegans and the C331053 protein of Drosophila melanogaster (Fig. 4C; Jantsch et al. 2004). We showed by mutant analysis that both RFM (zip3H74A and zip3H80A) and SBM (zip3I96K and zip3I96S,I99S) were important for Zip3’s function in sporulation efficiency and SC assembly (Fig. 4D–F).
Figure 4.
Zip3 is a putative Smt3 E3 ligase. (A) Amino acid sequence alignments of the RFM in various E3 ligases. The conserved cysteine (C) and histidine (H) residues are indicated. (B) The putative structure of the C3H2C3-RFM in Zip3. (C) Sequence comparison of the SBMs found in yeast Zip3 and other SUMO enzymes, including the Zip3 homologs in C. elegans (ZHP3) and in Drosophila melanogaster (C331053), and human PIAX, SAE2, and PML proteins. (D) The C3H2C3-RFM and SBM are essential for Zip3’s functions. Yeast CEN/ARS6 vectors harboring the wild-type ZIP3 gene or various zip3 mutants were transformed into a zip3 mutant, respectively. The transformants were plated onto the sporulation medium and incubated for 48–72 h at 30°C. Sporulation frequency was determined microscopically. The SCs and PCs were examined by immunostaining the surface nuclei spreads with anti-Zip1 antibody. (E) The zip3 HIS6-MYC-SMT3 diploid cell was transformed with either the V5-tagged wild-type Zip3 or zip3H80A mutant protein. Representative images of surface nuclei spreads of sporulating cells at the 7-h time point stained with DAPI, anti-Zip1 (in green), and anti-Myc (in red), respectively. (F) Western blot analysis revealed that the V5-tagged wild-type Zip3 and zip3H80A mutant proteins were properly expressed in the sporulating cells in E. Zip1 protein was used as a sample loading control.
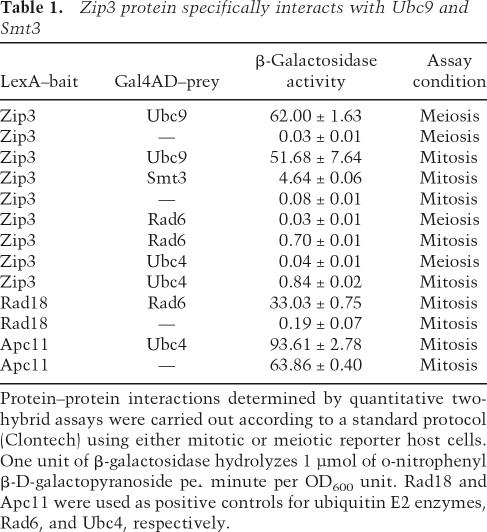
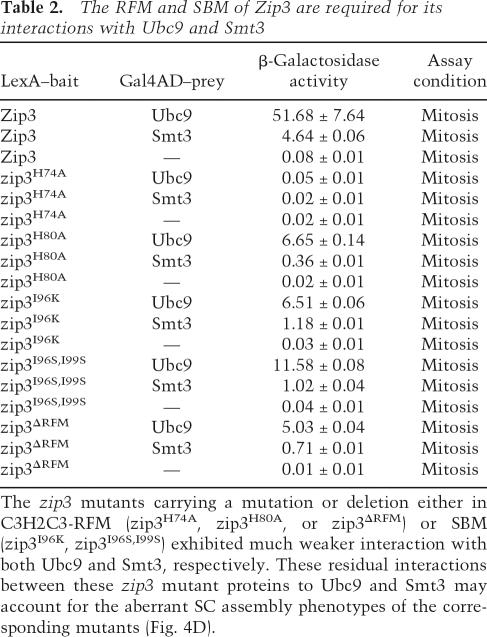
A fourth line of evidence shows that Zip3, as a putative Smt3 E3, exhibited two-hybrid interactions with Smt3 and Ubc9 but not with two ubiquitin E2s—that is, Ubc2/Rad6 and Ubc4 (Table 1). Moreover, mutations or deletions in either SBM (i.e., zip3I96K, zip3I96S,I99S) or RFM (i.e., zip3H74A, zip3H80A, or zip3-ΔRFM) significantly reduced both Zip3–Smt3 and Zip3–Ubc9 interactions (Table 2).
Table 1.
Zip3 protein specifically interacts with Ubc9 and Smt3
Protein–protein interactions determined by quantitative two-hybrid assays were carried out according to a standard protocol (Clontech) using either mitotic or meiotic reporter host cells. One unit of β-galactosidase hydrolyzes 1 μmol of o-nitrophenyl β-D-galactopyranoside per minute per OD600 unit. Rad18 and Apc11 were used as positive controls for ubiquitin E2 enzymes, Rad6, and Ubc4, respectively.
Table 2.
The RFM and SBM of Zip3 are required for its interactions with Ubc9 and Smt3
The zip3 mutants carrying a mutation or deletion either in C3H2C3-RFM (zip3H74A, zip3H80A, or zip3ΔRFM) or SBM (zip3I96K, zip3I96S,I99S) exhibited much weaker interaction with both Ubc9 and Smt3, respectively. These residual interactions between these zip3 mutant proteins to Ubc9 and Smt3 may account for the aberrant SC assembly phenotypes of the corresponding mutants (Fig. 4D).
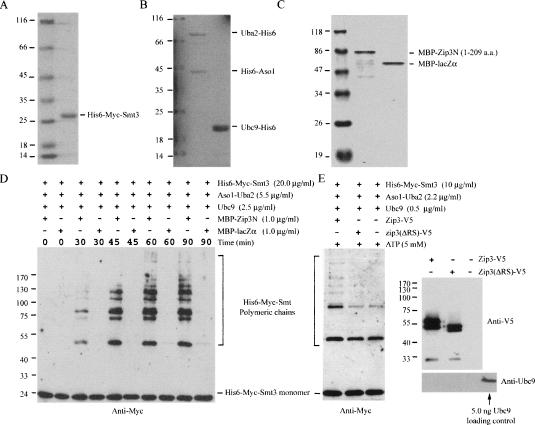
Fifth, Zip3 exhibits Smt3 E3 activity in vitro. We showed here that a purified MBP–Zip3N recombinant protein from Escherichia coli could promote the formation of Smt3 polymeric chains in a condition containing relatively low concentrations of purified Smt3, E1, and E2. MBP–Zip3N is a fusion protein of maltose-binding protein (MBP) and Zip3N. The latter contains the N-terminal portion of the yeast Zip1 protein (amino acid residues 1–209) and includes both RFM and SBM. The MBP–lacZα fusion protein was used here as a negative control for the E3 ligase assay (Fig. 5D). Several other known Smt3 E3s had been shown to exhibit such an activity in promoting formation of Smt3 polymeric chains (Johnson and Gupta 2001; Bencsath et al. 2002; Taylor et al. 2002; Zhao and Blobel 2005). Additionally, we have overexpressed and immunoaffinity-purified the V5-tagged wild-type Zip3 protein (i.e., Zip3-V5) and the zip3-ΔRS-V5 mutant protein from a protease-deficient yeast strain, JEL1 (Worland and Wang 1989). The zip3-ΔRS-V5 is an internal deletion mutant lacking both RFM and SBM (amino acid residues 52–99). We showed that only Zip3-V5 but not zip3-ΔRS-V5 could promote formation of the Smt3 polymeric chain in vitro (Fig. 5E).
Figure 5.
Zip3 exhibits SUMO E3 ligase activity in vitro. Purified SUMOylating His6-Myc-Smt3(A), E1 and E2 (B), and MBP–Zip3N and MBP–lacZα (C) from E. coli were separated by SDS-PAGE and then visualized by Coomassie blue staining. MBP–Zip3N is a fusion protein (molecular weight 73,830) of MBP and the N-terminal portion of yeast Zip3 protein (amino acid residues 1–209). The latter contains both RFM and SBM. (D) Zip3N exhibits SUMO E3 ligase activity in vitro. Purified MBP–Zip3N or MBP–lacZα (1.0 μg/mL) was mixed with SUMOylating His6-Myc-Smt3, E1, and E2 in the presence of ATP (5 mM). The reaction mixtures were taken out at the indicated time points, separated by SDS-PAGE, and visualized by Western blot using anti-Myc antibody. A MBP–lacZα fusion protein (molecular weight 50,840) was used here as a negative control. (E) RFM and SBM are essential for Zip3’s Smt3 E3 ligase activity. The wild-type and mutant Zip3 proteins were separately expressed in a JEL1 yeast strain transformed using either pYC2-PGAL1-Zip3-V5 or pYC2-PGAL1-Zip3-ΔRS-V5 expression vector, respectively. The Zip3-ΔRS-V5, lacking both RFM and SBM (amino acid resides 52–99), is an internal deletion mutant. Both proteins were immunoaffinity-purified using anti-V5-antibody-conjugated agarose (Sigma). A yeast cell harboring only the pYC2 vector was used as a negative control for immunoaffinity purification. The purified V5-tagged proteins were mixed with SUMOylating His6-Myc-Smt3, E1, and E2 (total 10 ng) in the presence of ATP (5 mM). The reaction products were separated by SDS-PAGE and then visualized by Western blot analysis using anti-Myc antibody. The purified V5-tagged proteins were also examined by Western analysis using anti-V5 and anti-Ubc9 antibodies. A portion of Zip3 proteins migrated slower in SDS-PAGE, and these proteins were shown to be phosphorylated by dephosphorylation assay using calf intestinal alkaline phosphatase (data not shown). The wild-type Zip3 protein itself could not carry out autonomous Smt3 modification because no Smt3-conjugated Zip3-V5 product was detected by anti-V5 antibody. Purified E2 or Ubc9 protein (∼5 ng) was loaded in a separate lane (as indicated) to be used as a positive control for anti-Ubc9 Western blot analysis. No Ubc9 protein was detected in all three immunoaffinity-purified products. Because the Smt3 chain formation reactions were carried out in the presence of 5.0 ng of recombinant Ubc9 protein, it is unlikely that the E3 ligase activity of purified Zip3-V5 protein is due to copurification of Ubc9 or other E3 enzymes from host cells.
These results clearly suggest that RFM and SBM are responsible for Zip3’s E3 ligase activity. Taking all of these results together, we conclude that Zip3 is an E3 ligase and that Zip3 mediates Smt3 modifications along meiotic chromosomes at mid-prophase that are required for SC assembly.
Post-translational modifications of Zip3 protein are modulated by Cdc28/Clb5, Clb6 kinase activity, and meiotic DNA recombination
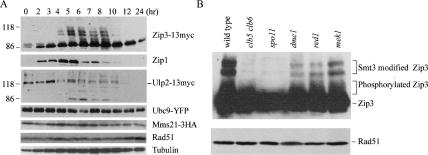
SC assembly depends on the meiotic cell cycle program and DNA double-strand breaks (DSBs) (Agarwal and Roeder 2000; Smith et al. 2001; Henderson and Keeney 2004). Because Zip3 plays an essential role in initiating SC assembly, we examined whether or not Zip3 levels or post-translational modification are modulated by the meiotic cell cycle program and DSB. Western time-course analysis of a wild-type sporulating cell revealed that Zip3-13myc proteins first appeared as a faster migrating form in SDS-PAGE at early prophase (∼2 h). At mid-prophase (∼4 h), a portion of Zip3-13myc proteins began to migrate more slowly and appeared either as two discrete bands or as a smear, respectively (Fig. 6A, top panel). Dephosphorylation analysis using calf intestinal alkinase phosphatase (CIAP) and its inhibitor, β-glycerolphosphate, indicated that the smear represented the phosphorylated Zip3 and that the two discrete bands were likely to be the Zip3-Smt3 conjugates, respectively (Supplementary Fig. S6). Intriguingly, both modifications not only greatly diminished in the clb5 clb6 and spo11 mutants but also were slightly reduced in the dmc1, red1, and mek1 mutants (Fig. 6B).
Figure 6.
Zip3 and Ulp2 are differentially regulated at and after meiotic mid-prophase. (A) Western time-course analysis of Zip3-13myc, Ulp2-13myc, Mms21-3HA, Ubc9-YFP, Rad51, and Zip1 proteins. (B) Post-translational modifications of Zip3 are regulated by Cdc28/Clb5, Clb5 kinase activity, and Spo11 protein (or formation of DSBs in DNA). Western analysis of the total cell lysates from wild-type and various mutants at the 6-h sporulation time point. The unmodified, Smt3-modified, and phosphorylated Zip3 proteins were marked on the right, respectively. Rad51 and tubulin were both used here as protein loading controls.
These results are consistent with the notion that SC assembly mediated by Zip3 depends on initiation of DSBs (Agarwal and Roeder 2000; Henderson and Keeney 2004) and Cdc28 kinase activity (Smith et al. 2001). Two additional lines of evidence further support the notion that Zip3’s functions and/or chromosome localization are controlled by these post-translational modifications: (1) The timing of Zip3 modifications (∼4 h) in the wild-type cell correlates very well with that of Smt3 polymeric chain formation (4–6 h) in the zip3 mutant (Fig. 3A); and (2) Zip3 failed to localize on meiotic chromosomes in the spo11 or clb5 clb6 mutants (data not shown) (Agarwal and Roeder 2000; Henderson and Keeney 2004). Because the purified Zip3 protein could not carry out autonomous Smt3 modification in vitro (Fig. 5E), other E3 ligases may be involved in Smt3 modification of the Zip3 protein in vivo (see Discussion).
Differential regulation of Zip3 and other SUMO enzymes during meiosis
Smt3 polymeric chains massively accumulated only at and after mid-prophase in a zip3 mutant (Fig. 3A). The absence of Smt3 polymeric chains in the wild-type cell or in the zip3 mutant during early prophase is consistent with the following observations: (1) Zip3’s function in SC formation (Fig. 1) and Zip3’s post-translational modifications (Fig. 6A) were only up-regulated at and after mid-prophase. (2) Zip3-independent E3 ligases (i.e., Siz1, Siz2, or Mms21) ought to function at early prophase, and their functions or protein levels will be down-regulated at the time SCs begin to form (mid-prophase). Indeed, we found by Western time-course analysis that the levels of Siz1 and Siz2 proteins began to decrease at mid-prophase (Supplementary Fig. S7). These Zip3-independent E3 activities likely are responsible for the Smt3/Zip1 foci during early prophase for nonhomologous centromere clustering (Tsubouchi and Roeder 2005) as well as for Smt3 modification of the Zip3 protein (see Discussion). (3) The deSUMOylating enzymes, Ulp1 (Supplementary Fig. S7) and Ulp2 (Fig. 6A), both were expressed in premeiotic cells (t = 0 h), sustained throughout early prophase (3 h), and also began to decrease at mid-prophase (t = 4 h). Mutant analysis indicated that Ulp2 degradation was mediated by two ubiquitin E3 ligases—that is, Ubr1 and anaphase-promoting complex (Y.-H. Lo and T.-F. Wang, unpubl.).
In conclusion, opposite regulation of Zip3 and other Smt3 enzymes (i.e., Siz1, Siz2, Ulp1, and Ulp2) at and after mid-prophase in the wild-type cells likely sets up the meiotic chromosomes for global Smt3 modifications and subsequently promotes SC assembly. The same mechanism may also result in a massive accumulation of Smt3 polymeric chains in the zip3 mutant after mid-prophase because Ulp2 is responsible for degradation of Smt3 polymeric chains in vivo (Bylebyl et al. 2003).
Zip1 is a binding protein for Smt3 conjugated products
We next investigated whether or not formation or stability of Smt3 polymeric chains in zip3 was dependent on other SC-initiating proteins (e.g., Zip1, Zip2, Zip4, and Spo16). Smt3 polymeric chains not only formed (Supplementary Fig. S8) but also were colocalized with Zip1 at PCs in zip3 zip2 (Fig. 1B), zip3 zip4, and zip3 spo16 (data not shown), respectively. Therefore, Zip2, Zip4, and Spo16 act after Zip3 in promoting SC formation. On the other hand, few or no Smt3 polymeric chains were detected in the zip3 zip1 mutant either by Western blotting analysis (Fig. 7A) or by immunostaining surface spreads of zip3 zip1 meiotic nuclei (Fig. 1B). One possibility is that Zip1 may directly interact with the Smt3 polymeric chain and subsequently promote the latter’s stability. This hypothesis was supported by the results with the yeast two-hybrid assay. Zip1 exhibited a strong interaction with Smt3. No significant interaction was detected between Smt3 and other SC-initiating proteins (Zip2, Spo16, or Zip4) under identical conditions (Fig. 7C). Zip1 itself is not Smt3 modified during meiosis because the apparent molecular weight (∼100,000) of Zip1 in a sporulating ulp2 cell (Supplementary Fig. S2, right panel) is close to that of the predicted molecular weight (100,034). Zip1–Smt3 association likely is mediated by noncovalent interactions.
Figure 7.
Zip1 is a binding protein for Smt3-conjugated products. (A) Protection of polymeric Smt3 chains by Zip1. Western time-course analysis of the V5-Smt3 conjugates in the zip1 zip3 mutant. The Smt3 monomer (apparent molecular weight ∼20,000) and its conjugated products were detected by anti-V5 antibody. Meiotic cell cycle progression was monitored by induction of the meiosis-specific Dmc1 protein. (B) Zip1 contains an SBM. Sequence comparison of the C-terminal of Zip1 protein with a consensus SBM was recently performed (Hannich et al. 2005). The conserved amino acid residues are indicated. (C) Zip1 interacts with Smt3 and Red1. A quantitative yeast two-hybrid analysis was carried out as described in Table 1. (D) The C-terminal portion of Zip1 preferentially interacts with Smt3-conjugated products but not the Smt3 monomer. Zip1C contains the amino acid residues 846–875 of Zip1. A conjugation-incompetent Smt3 mutant, smt3ΔGG, lacks the terminal pair of glycines of the wild-type Smt3 protein. Zip1C(3L–3R) and Zip1C(3N–3R) are constructed by mutating the 3L or 3N into three arginines. (E) The zip1 V5-SMT3 diploid cell was transformed with either the wild-type ZIP1 or the zip13N–3R mutant gene. Representative images of surface nuclei spreads of sporulating cells at the 7-h time point stained with DAPI, anti-Zip1 (in green), and anti-V5 (in red), respectively. Wild-type Zip1 proteins colocalized with staining signals of V5-Smt3 along meiotic chromosomes. In contrast, the zip13N–3R mutant proteins aggregate together to form PC-like structures. The V5-Smt3 signals were detected along meiotic chromosomes but not with these PC-like structures. (F) In vitro binding analysis of Zip1C with Smt3 polymeric chains. Smt3 polymeric chains were synthesized in vitro using purified His6-Myc-Smt3, E1, and E2 in the presence of ATP. The reaction mixtures were mixed either with a MBP–lacZα or MBP–Zip1C fusion protein in the presence of the amylose resins that specifically recognize MBP. The amylose-resin-bound proteins were separated by SDS-PAGE. MBP–lacZα and MBP–Zip1C were visualized by Coomassie blue staining. Smt3 monomers and polymeric chains were detected by Western blots using the anti-Myc antibody.
It was reported previously that the C-terminal domain of Zip1 is responsible for its localization to meiotic chromosomes or to LEs of SCs (Tung and Roeder 1998; Dong and Roeder 2000). We found that a short stretch of amino acid sequence in Zip1 (residues 853–863; KKLLLVEDEDQ) was closely similar to a new SBM identified by Hannich et al. (2005). This SBM contains a consensus sequence of “K-X1–3-h-h-h-X3-n-n-n”; h represents I, V, and L; n represents D, E, Q, or N; and X can be any amino acid residue (Fig. 7B; Hannich et al. 2005). We then carried out yeast two-hybrid analysis in conjunction with mutant analysis to examine if the C-terminal domain (residues 846–875) of Zip1 (denoted “ZipC”) could associate with Smt3 monomers or Smt3-conjugated products. The results indicate that Zip1C indeed interacts with the wild-type Smt3 protein (Fig. 7C). The Zip1C–Smt3 interaction was greatly diminished if the three conserved “L” or “N” residues (i.e., 3L or 3N) of Zip1C were mutated into three contiguous arginine residues (i.e., 3R). The corresponding Zip1C mutants were referred to here as Zip1C(3L–3R) and Zip1C(3N–3R), respectively. We also carried out yeast mutant analysis to show that the zip13L–3R or zip13N–3R mutant failed to complement the meiotic defects in SC formation of the zip1 mutant (Fig. 7E). These results confirmed that the SBM of the Zip1 protein is indeed crucial for Zip1’s function in meiosis.
To determine if Zip1 recognizes the Smt3 monomer or its conjugated products, we then construct a conjugation-incompetent Smt3 mutant, smt3ΔGG. This smt3ΔGG mutant lacks the terminal pair of glycines of the wild-type Smt3 protein. These two glycines are indispensable for E1-mediated activation of Smt3 from its precursor (for review, see Johnson 2004). The results of two-hybrid analysis here reveal that Zip1C exhibits a much lower binding affinity for smt3ΔGG than for wild-type Smt3 protein (Fig. 7D). Therefore, the Zip1 protein preferentially interacts with the Smt3-conjugated products but not with the Smt3 monomer. This conclusion was further supported here by the results of the in vitro binding assay. MBP–Zip1C (molecular weight 46,656), a fusion protein of MBP and Zip1C, was expressed and purified from E. coli. MBP–lacZα fusion protein (molecular weight 50,840) was used here as a negative control for the binding assay. Smt3 polymeric chains were synthesized in vitro by incubating SUMOylating His6-Myc-Smt3 with E1 and E2 in the presence of ATP and Mg2+. The resulting products, containing both Smt3 monomers and Smt3 polymeric chains, were mixed together with MBP–ZipC or MBP–lacZα in the presence of amylose resins that specifically recognize MBP. The results indicated that MBP–Zip1C exhibits stronger affinity to Smt3 polymeric chains than to Smt3 monomers. This interaction is specific because MBP–lacZα pulled down hardly any Smt3 polymeric chains or Smt3 monomers (Fig. 7F). We have also carried out a binding assay using MBP–Zip1C(3N–3R) and MBP–Zip1C(3L–3R). These results reveal that these two mutant proteins, unlike MBP–Zip1C, exhibit much lower binding affinity to Smt3 polymeric chains (data not shown).
Taken together, we conclude that the C-terminal domain of Zip1 mediates Zip1’s interaction with the Smt3-conjugated products but not Smt3 monomers. This interaction appears to be important for Zip1’s functions in meiosis. Additional two-hybrid analysis revealed that Zip1 can interact with Red1 (Fig. 7C, last row), an essential component of LEs and SCs (Smith and Roeder 1997; Rockmill and Roeder 1998). We also found that Red1 protein is Smt3 modified during meiosis and colocalizes with Smt3 immunostaining signals along meiotic chromosomes at and after mid-prophase (Supplementary Fig. S9). Therefore, SC assembly likely is mediated by protein–protein interaction between Zip1 and Smt3-modified proteins (e.g., Red1) along LE (see below).
Discussion
SUMO modifications and Zip1 functions in meiosis
Zip1, as a head-to-head dimeric or tetrameric protein (Sym et al. 1993) and a binding protein for Smt3 conjugates (this study), has at least three different functions in meiosis. During early prophase, Zip1-dependent centromere coupling between nonhomologous chromosomes likely is positively mediated by Zip3-independent E3 ligases (e.g., Siz1 and Siz2; see below) and negatively regulated by Ulp2 (Fig. 1D). At and after mid-prophase in the wild-type cell, Zip1 associates with Zip3-dependent chromosomal Smt3 conjugates to form SCs between homologous chromosomes. Because Zip1 is unlikely to be the only Smt3-binding protein in vivo, it is reasonable to find that Zip1 staining signals are colocalized with most but not all Smt3 conjugates on meiotic chromosomes of the wild-type cell (Fig. 1A). In the zip3 mutant, Zip1 protein noncovalently cross-links the Smt3 polymeric chains to form larger PCs. The spo11 and clb5 clb6 mutants conceivably produce fewer and shorter Smt3 conjugates (i.e., Smt3 chains, Smt3-E1, or Smt3-E2); these two strains formed only smaller PCs (Fig. 1A).
Several chromosomal proteins have been shown to be Smt3 modified, including Top2 (Bachant et al. 2002), Pds5 (Stead et al. 2003), condensin Ycs4 (Yu and Koshland 2003; D’Amours et al. 2004), and Red1 (Supplementary Fig. S9). These proteins are all located at the axial core of yeast chromosomes (Bachant et al. 2002; Blat et al. 2002; Stead et al. 2003; Yu and Koshland 2003) and have been implicated in SC assembly (Rockmill and Roeder 1988; Klein et al. 1992; van Heemst et al. 2001; Yu and Koshland 2003). Intriguingly, Smt3 modifications of Top2 and Pds5, regulated by Siz1/Siz2 and Ulp2, were important for proper cohesion resolution at centromeric regions (Bachant et al. 2002; Stead et al. 2003; Aguilar et al. 2005). Immunostaining results in the report indicated that most V5-Smt3 signals were colocalized with strong staining spots of Top2 during early meiotic prophase (Fig. 1E). It is of interest to further examine if Smt3-modified Top2 and Pds5 proteins directly associate with Zip1 for centromere coupling at early prophase. Moreover, additional experiments need to be carried out to further confirm whether the Smt3-modified Red1 protein is indeed involved in SC formation at/after mid-prophase.
A general problem in verifying the functions of Smt3-modified proteins is that mutations of the consensus Smt3 target sites (i.e., Ψ-K-X-E/D) on each individual protein often have little or no effect on their biological functions. Therefore, Smt3 target sites might be flexible. For example, we have constructed a yeast strain expressing only a red1K14R,K50R mutant in which the two Ψ-K-X-E/D sites of the wild-type Red1 protein have been mutated into Ψ-R-X-E. This yeast strain exhibits no apparent meiotic defects (data not shown). One possibility is that Smt3 may be transferred to target sites other than Ψ-K-X-E/D sites in the red1K14R,K50R mutant. Therefore, these novel Smt3 target sites need to be explored further. Alternatively, different Smt3-modified proteins may function redundantly in recruiting Zip1 to form the SC, and this activity may be independent of their original function in maintaining chromosome structures (i.e., sister chromatid cohesion and chromosome condensation, etc.). The latter hypothesis may account for the fact that only a small fraction (<5%) of each individual Smt3 target protein (e.g., Red1, Top2, or Pds5) was actually Smt3 modified in vivo.
Possible roles of other SC-initiating proteins
Although the model proposed here could explain the interactions between Zip1 and Smt3-conjugated products, it may be an oversimplification. The model does not account for the perpendicular alignment of Zip1 to AEs. For example, a zip2 mutant shows global Zip1 and Smt3 immunostaining signals along meiotic chromosomes but hardly forms any linear SC structure (Fig. 1B). We suggest that chaperone-like machineries may facilitate perpendicular alignment of Zip1 proteins to form SCs. A recent report indicated that Zip2 and Zip4/Spo22 emerge as a seven-blade WD-40-like repeat protein and a 22-unit TPR repeat protein, respectively (Perry et al. 2005). Both the WD-40 and TPR domains are known to be involved in protein–protein interaction. TPR repeat proteins (e.g., Hip, Hop, and CHIP) were originally identified as the cochaperones of Hsp70 or Hsp90 (D’Andrea and Regan 2003). A proteomic analysis of Saccharomyces cerevisiae, using a tagged ORF to pull down multiple protein complexes, has revealed 16 WD-40 seven-blade proteins (e.g., Cdc20, Cdc55, Cdh1, Rad24, Rad28) that interact with the GroEL-like chaperonin CCT (chaperonin-containing TCP-1; also termed Tric). CCT also has been shown to interact with Hsp70, either directly or through interaction with Hop, the Hsp70/90 organizing protein (Valpuesta et al. 2002). A mouse meiosis-specific Hsp70 protein (i.e., Hsp70-2) has been shown to associate with the SC and plays an essential role in meiosis (Dix et al. 1997; Zhu et al. 1997). Intriguingly, the meiotic phenotypes of zip1–4 mutants became more severe if sporulation were carried out at a higher temperature (Borner et al. 2004; Perry et al. 2005). This is consistent with the fact that many mutants with defective chaperone functions are sensitive to high temperature. Taken all together, we favor the notion that Zip2 and Zip4 likely act as or together with chaperone molecules to promote SC formation.
Epilog
In conclusion, we show that SUMO modifications play important roles in mediating Zip1’s meiotic functions, including centromere clustering at early prophase and SC/PC assembly at/after mid-prophase. The SC was first observed in 1956 by ultrastructural analysis (Fawcett 1956; Moses 1956). For 50 years, many researchers have contributed their efforts to structure and function analysis of the SC. At last, a molecular mechanism underlying SC assembly begins to emerge. While we were preparing this manuscript, we became aware of an independent work on yeast Ubc9 by Gillian Hooker and Shirleen Roeder. These investigators showed, by cytology and genetic analysis, that a ubc9 mutant made aberrant SC and that Zip3 was required to recruit Ubc9 onto meiotic chromosomes (G. Hooker and S. Roeder, pers. comm.).
Zip1 and Zip3, in collaboration with other ZMM proteins (e.g., Zip2, Zip4, Spo16, Msh4, Msh5, and Mer3), were presumed to act at the sites of crossover-designated, axis-associated recombinational interactions to mediate crossover/chiasma formation (Borner et al. 2004; Fung et al. 2004; Perry et al. 2005). Msh4/5 is a meiosis-specific MutS homolog that does not mediate mismatch repair. Mer3 DNA helicase stabilizes the Rad51-mediated nascent joint molecules via 3′-to-5′ DNA heteroduplex extension to permit capture of the second processed end of a double-stranded DNA break, a step that is required for crossover recombinant product formation (Mazina et al. 2004). However, a direct functional link between Zip3-mediated SUMO modifications and crossover DNA recombination during meiosis has not yet been established. Recent data have shown that SUMO modification of PCNA functionally cooperates with Srs2 (Pfander et al. 2005), a DNA helicase that blocks recombinational repair by disrupting Rad51 nucleoprotein filaments (Krejci et al. 2003; Veaute et al. 2003). In this scenario, SUMO-modified PCNA serves to recruit Srs2 as a guarding mechanism to prevent Rad51 filament formation, thereby avoiding unwanted recombination during DNA replication. Genetic analysis also revealed that Srs2 specifically promotes the synthesis-dependent single-strand annealing pathway, subsequently preventing the channeling of recombination substrates into pathways that produce crossovers (Aylon et al. 2003; Ira et al. 2003). Because Zip1 is also a binding protein for Smt3-modified protein, it will be very interesting to determine if Zip1 functions by counteracting Srs2 to promote crossovers during meiosis.
Materials and methods
Yeast strains, meiotic time courses, and spore viability
All strains and yeast vectors used in this study are derivates of SK1 and are available upon request. Several proteins were tagged at the C terminus with different epitopes at the original chromosomal locus by one-step gene targeting (Longtine et al. 1998; Knop et al. 1999). Sporulation frequency and spore viability were comparable to the untagged parental strain. Gene deletion was carried out using PCR-mediated gene replacement (Longtine et al. 1998; Knop et al. 1999). Gene deletion and tagging were confirmed by PCR, Southern blot, and/or Western analysis.
Pregrowth, synchronous meiosis, and DNA isolation were as described previously (Bishop et al. 1992), except that yeast extract/peptone/acetate (YPA) medium used for overnight culture preceding transfer to sporulation medium (SPM) was equilibrated to 30°C prior to inoculation. SPM contained 0.4% potassium acetate, 0.02% raffinose, and antiform 289 (0.01% Sigma). Meiotic divisions were monitored by fluorescence microscopy after staining with 0.2 μg/mL of DAPI. Sporulation frequency in liquid cultures was determined microscopically. Spore viability was determined by dissection of tetrads from SPM plates that had been incubated for 3–4 d at 30°C.
Cytological analysis
Spread nucleoids were prepared and visualized as described (Bishop 1994). Primary antibody staining was carried out using goat anti-Zip1 (Santa Cruz Biotechnology) at 1/250, mouse anti-V5 (Invitrogen) at 1/500, rabbit anti-Myc (Upstate Biotechnology) at 1/250, and anti-ubiquitin and anti-Top2 (Abcam and Santa Cruz Biotechnology) at 1/200, respectively. For secondary antibody staining, fluorochrome-labeled Alexa 488/594 anti-mouse IgG, anti-rabbit IgG, or anti-goat IgG were used at 1/250, respectively (Molecular Probes).
Two-hybrid analysis
The two-hybrid vectors (pBHA and pGADT7) and β-galactosidase plate assay using 5-bromo-4-chloro-3-indolyl-β-galatoside (X-gal) have been described for analysis in a haploid vegetative growing L40 reporter cell (Wang et al. 1999). The diploid ndt80Δ strain used for the meiotic two-hybrid assay was a gift from Scott Keeney (Memorial Sloan-Kettering Cancer Center, New York) (Arora et al. 2004). G1-synchronized ndt80Δ
strain used for the meiotic two-hybrid assay was a gift from Scott Keeney (Memorial Sloan-Kettering Cancer Center, New York) (Arora et al. 2004). G1-synchronized ndt80Δ cells were cultured 6 h in sporulating medium at 30°C with good aeration for the meiotic cell analysis. Quantitative two-hybrid assays were carried out according to a standard protocol (Clontech). One unit of β-galactosidase hydrolyzes
cells were cultured 6 h in sporulating medium at 30°C with good aeration for the meiotic cell analysis. Quantitative two-hybrid assays were carried out according to a standard protocol (Clontech). One unit of β-galactosidase hydrolyzes 
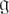 μmol of o-nitrophenyl β-D-galactopyranoside per min per OD600 unit.
μmol of o-nitrophenyl β-D-galactopyranoside per min per OD600 unit.
Preparation of total yeast extracts and Western blotting
Yeast total extracts were prepared according to a trichloroacetic acid (TCA) precipitation method (Knop et al. 1999). Protein separation on SDS-PAGE and Western blotting were performed according to standard procedures. The epitope-tagged proteins were detected by rabbit anti-Myc 9E10 antibody (1/500; Upstate Biotechnology), mouse anti-V5 antibody (1/500; Invitrogen), rabbit anti-GFP antibody (1/500; Invitrogen), goat anti-Zip1 antibody (1/500; Santa Cruz Biotechnology), and rabbit anti-ubiquitin antibodies (1/250; Abcam), respectively. Primary antibodies were detected using the corresponding HRP-conjugated secondary antibodies. Final detection was performed using the ECL plus or ECL advanced Western blotting detection system (Amersham). The emitted chemiluminescent light was recorded by X-ray film.
Dephosphorylation assay
Whole-cell extracts were prepared first by the TCA precipitation method as described above. The protein pellet from 1.5 mL or sporulating cell culture was dissolved in 75 μL of buffer A (50 mM Tris-HCl at pH 7.5, 1 mM MgCl2), and mixed with 4 μL of 2 M Tris-Base to adjust the pH value to 7.5. Dephosphorylationreactions were carried out by mixing 25 μL of whole-cell extractsinto 25 μL of protein phosphatase buffer (50 mM Tris-HCl at pH 7.5, 1 mM MgCl2), 0.1 mM protease inhibitor TPCK, and 50 U of calf intestinal alkaline phosphatase (CIAP; New England Biolabs) at 37°C for 4 h. MG132 was purchased from Calbiochem. In some reactions, phosphatase inhibitor β-glycerolphosphate (20 μM; Sigma) was added to a final concentration of 16 mM. Phosphatase reactions were stopped bymixing an equal volume of 3× Laemmli sample buffer (188 mM Tris-HCl at pH 6.8, 3% SDS, 15% β-mercaptoethanol, 30% glycerol, 0.01% bromophenolblue), and the mixtures were immediately incubated at 95°C for 6 min. SDS-PAGE and Western blotting were carried out as described above.
Purification of Smt3 polymeric chains
To detect Smt3 polymeric chains formed in the sporulating zip3 cells, Smt3 proteins were tagged with V5 or His6-Myc epitope at the original chromosomal locus. Whole-cell extracts were prepared by the TCA precipitation method described above and then dissolved in a denaturing buffer X (8 M urea, 125 mM NaCl, 5 mM imidazole, 0.1% Triton X-100, 50 mM Tris-HCl at pH 8.0). The whole-cell extract of sporulating zip3Δ cells expressing only the V5-Smt3 was used here as a negative control for purification. A tight-fitting Dounce homogenizer (10 strokes) was used to facilitate solubilization. The suspensions were centrifuged at 100,000 × g for 30 min and then subjected to purification on Ni2+-chelating resin (Novagen) that specifically retains the His6-tagged polypeptides. The Ni2+-chelating resins were washed with 80-fold resin volumes of buffer X three times, followed by an additional wash with buffer Y (buffer X with 15 mM imidazole at pH 8.0, added). The His6-tagged polypeptides were then eluted with buffer Z (buffer X with 250 mM imidazole at pH 8.0, added) and subsequently analyzed by Western blotting using either anti-Myc or anti-V5 antibody, as described above.
Purification of recombinant proteins and enzymatic assays
E. coli cells for overexpression of the His6-tagged yeast E1 and E2 were described previously (Zhao and Blobel 2005). E. coli expressing vectors were constructed for expressing the His6-Myc-Smt3 protein and the MBP–Zip3N protein. MBP–Zip3N is a fusion protein of MBP and the N-terminal half of the yeast Zip3 protein (amino acid residues 1–209). Both recombinant proteins were purified either by the Ni2+-chelating resin as described previously (Johnson and Gupta 2001) or by the amylose affinity resin as described by the manufacturer (New England Biolabs). The in vitro sumoylation assay was also performed following the protocol described previously (Johnson and Gupta 2001).
Zip3 protein was expressed in yeast modified from a procedure described previously for yeast topoisomerase II (Worland and Wang 1989). Briefly, low-copy-number yeast pYC2 vectors were used for expressing V5-tagged wild-type Zip3 protein (denoted “Zip3-V5”) or V5-tagged zip3-ΔRS mutant protein (denoted “zip3-ΔRS-V5”) in a protease-deficient haploid host strain JEL1 (Worland and Wang 1989). This vector contained both CEN6/ARSH4 and an inducible promoter of the GAL1 gene. A JEL yeast cell carrying the pYC2 vector was used as a negative control. The cells harboring the pYC2-Zip3-V5, pYC2-ΔRS-V5, or pYC2 vector were grown in medium lacking uracil and supplemented with 2% glucose. At late log phase, the culture was diluted 100-fold into the same medium with 2% raffinose. Growth of cells resumed after an initial lag phase when the culture reached an optical density of 0.8 at OD600nm, measured in a Beckman spectrophotometer; galactose was added to 2% final concentration. Cells were harvested after 4 h, and washed with ice-cold water and then with buffer A plus 25 mM sodium fluoride and 1 mM sodium bissulfite. Buffer A contained 50 mM Tris-HC (pH 7.5), 0.2% (w/v) Triton X-100, 50 mM phosphate buffer (pH 7.5), 2 mM EGTA, 1 mM sodium vanadate, 0.1 mM ZnCl2, 10% glycerol, and a mixture of freshly added protease inhibitors (2 mM TPCK, 2 μg/ml of aprotinin, chymostatin, leupeptin, and pepstatin). The washed cells were resuspended in 1 mL of wash buffer/gram of wet-packed cells, frozen dropwise in liquid nitrogen, and then extracted at 4°C with a glass bead beater with 1 vol glass bead and 1 vol of buffer A. Subsequent steps were all carried out at 4°C. Cell debris were collected at 100,000 × g, and the resulting supernatant was bound in batch for 3 h to anti-V5 agarose resins (Sigma). Anti-V5 agarose resins were then extensively washed in batch with 50 vol of (1) buffer A three times, (2) buffer B (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.1 mM ZnCl2, 2% glycerol, 2 mM TPCK, 2 μg/mL of aprotinin, chymostatin, leupeptin and pepstatin) three times, and (3) buffer C (50 mM Tris-HCl at pH 7.5, 0.1 mM ZnCl2, 5% glycerol, 2 mM TPCK, 2 μg/mL of aprotinin, chymostatin, leupeptin, and pepstatin). The bound V5-tagged protein complexes were resuspended with 2 vol of buffer C and directly used for the in vitro sumoylation assay.
Zip1C-binding assay
Smt3 chains were synthesized by mixing the sumoylating His6-Myc-Smt3 (40 μg/mL), E1 (11 μg/mL), and Ubc9 (20 μg/mL) in the presence of 5 mM ATP and 10 mM Mg2+. For the in vitro binding assay, 40 μg/mL of MBP–lacZα or MBP–Zip1C fusion protein were incubated with a mixture of Smt3 monomers and Smt3 polymeric chains in the presence of amylose resins that specifically recognize MBP. After binding, the amylose resins were extensively washed three times with 20 resin volumes of wash buffer (50 mM Tris-HCl at pH 7.0, 50 mM NaCl, 1 mM EGTA, 10 mM β-mercaptoethanol). The amylose-resin-bound proteins (or protein complexes) were separated by SDS-PAGE. MBP–lacZα and MBP–Zip1C were visualized by Coomassie blue staining. Smt3 monomers and polymeric chains were detected by Western blots using the anti-Myc antibody.
Acknowledgments
We thank Scott Keeney for the yeast two-hybrid reporter strain, Valetine Borner for the zip2 and zip3 stains, James C. Wang for the JEL1 strain, Xiaolan Zhao and Gunter Blobel for the SUMO E1 and E2 expression vectors, Erica Johnson for the smt3-allR mutant, Akira Shinohara for sharing Spo16 results prior to publication, and Jia-Wen Yeh and Jia-Yin Yeh for research assistance. We also thank Chung Wang, Douglas Bishop, Scott Keeney, Thomas Petes, Gillian Hooker, and Shirleen Roeder for their suggestions for this paper. This work was supported by the Academia Sinica (Investigator Award Grant) and the National Science Council (NSC94-2311-B-001-05 and NSC94-2311-B-001-27). We declare no conflict of financial interest.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1430406
References
- Agarwal S., Roeder G.S. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000;102:245–255. doi: 10.1016/s0092-8674(00)00029-5. [DOI] [PubMed] [Google Scholar]
- Aguilar C., Davidson C., Dix M., Stead K., Zheng K., Hartman T., Guacci V. Topoisomerase II suppresses the temperature sensitivity of Saccharomyces cerevisiae pds5 mutants, but not the defect in sister chromatid cohesion. Cell Cycle. 2005;4:1294–1304. doi: 10.4161/cc.4.9.1997. [DOI] [PubMed] [Google Scholar]
- Arora C., Kee K., Maleki S., Keeney S. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol. Cell. 2004;13:549–559. doi: 10.1016/s1097-2765(04)00063-2. [DOI] [PubMed] [Google Scholar]
- Aylon Y., Liefshitz B., Bitan-Banin G., Kupiec M. Molecular dissection of mitotic recombination in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 2003;23:1403–1417. doi: 10.1128/MCB.23.4.1403-1417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachant J., Alcasabas A., Blat Y., Kleckner N., Elledge S.J. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell. 2002;9:1169–1182. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- Bencsath K.P., Podgorski M.S., Pagala V.R., Slaughter C.A., Schulman B.A. Identification of a multifunctional binding site on Ubc9p required for Smt3p conjugation. J. Biol. Chem. 2002;277:47938–47945. doi: 10.1074/jbc.M207442200. [DOI] [PubMed] [Google Scholar]
- Bishop D.K. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Bishop D.K., Park D., Xu L., Kleckner N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Blat Y., Protacio R.U., Hunter N., Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- Borner G.V., Kleckner N., Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- Bylebyl G.R., Belichenko I., Johnson E.S. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J. Biol. Chem. 2003;278:44113–44120. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- Chua P.R., Roeder G.S. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell. 1998;93:349–359. doi: 10.1016/s0092-8674(00)81164-2. [DOI] [PubMed] [Google Scholar]
- D’Amours D., Stegmeier F., Amon A. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell. 2004;117:455–469. doi: 10.1016/s0092-8674(04)00413-1. [DOI] [PubMed] [Google Scholar]
- D’Andrea L.D., Regan L. TPR proteins: The versatile helix. Trends Biochem. Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Dix D.J., Allen J.W., Collins B.W., Poorman-Allen P., Mori C., Blizard D.R., Brown P.R., Goulding E.H., Strong B.D., Eddy E.M. HSP70-2 is required for desynapsis of synaptonemal complexes during meiotic prophase in juvenile and adult mouse spermatocytes. Development. 1997;124:4595–4603. doi: 10.1242/dev.124.22.4595. [DOI] [PubMed] [Google Scholar]
- Dong H., Roeder G.S. Organization of the yeast Zip1 protein within the central region of the synaptonemal complex. J. Cell Biol. 2000;148:417–426. doi: 10.1083/jcb.148.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett D.W. The fine structure of chromosomes in the meiotic prophase of vertebrate spermatocytes. J. Biophys. Biochem. Cytol. 1956;2:403–406. doi: 10.1083/jcb.2.4.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D., Ozkaynak E., Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- Fung J.C., Rockmill B., Odell M., Roeder G.S. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell. 2004;116:795–802. doi: 10.1016/s0092-8674(04)00249-1. [DOI] [PubMed] [Google Scholar]
- Gill G. SUMO and ubiquitin in the nucleus: Different functions, similar mechanisms? Genes & Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- Hannich J.T., Lewis A., Kroetz M.B., Li S.J., Heide H., Emili A., Hochstrasser M. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- Henderson K.A., Keeney S. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc. Natl. Acad. Sci. 2004;101:4519–4524. doi: 10.1073/pnas.0400843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson K.A., Keeney S. Synaptonemal complex formation: Where does it start? Bioessays. 2005;27:995–998. doi: 10.1002/bies.20310. [DOI] [PubMed] [Google Scholar]
- Ira G., Malkova A., Liberi G., Foiani M., Haber J.E. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch V., Pasierbek P., Mueller M.M., Schweizer D., Jantsch M., Loidl J. Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Mol. Cell. Biol. 2004;24:7998–8006. doi: 10.1128/MCB.24.18.7998-8006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.S. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Johnson E.S., Gupta A.A. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- Klein F., Laroche T., Cardenas M.E., Hofmann J.F., Schweizer D., Gasser S.M. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J. Cell Biol. 1992;117:935–948. doi: 10.1083/jcb.117.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: More tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Krejci L., Van Komen S., Li Y., Villemain J., Reddy M.S., Klein H., Ellenberger T., Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- Li S.J., Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 2000;20:2367–2377. doi: 10.1128/mcb.20.7.2367-2377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mazina O.M., Mazin A.V., Nakagawa T., Kolodner R.D., Kowalczykowski S.C. Saccharomyces cerevisiae Mer3 helicase stimulates 3′–5′ heteroduplex extension by Rad51; implications for crossover control in meiotic recombination. Cell. 2004;117:47–56. doi: 10.1016/s0092-8674(04)00294-6. [DOI] [PubMed] [Google Scholar]
- Moses M.J. Chromosomal structures in crayfish spermatocytes. J. Biophys. Biochem. Cytol. 1956;2:215–218. doi: 10.1083/jcb.2.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S., Ledl A., Schmidt D. SUMO: A regulator of gene expression and genome integrity. Oncogene. 2004;23:1998–2008. doi: 10.1038/sj.onc.1207415. [DOI] [PubMed] [Google Scholar]
- Page S.L., Hawley R.S. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 2004;20:525–558. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- Perry J., Kleckner N., Borner G.V. Bioinformatic analyses implicate the collaborating meiotic crossover/chiasma proteins Zip2, Zip3, and Spo22/Zip4 in ubiquitin labeling. Proc. Natl. Acad. Sci. 2005;102:17594–17599. doi: 10.1073/pnas.0508581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B., Moldovan G.L., Sacher M., Hoege C., Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- Potts P.R., Yu H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell. Biol. 2005;25:7021–7032. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B., Roeder G.S. RED1: A yeast gene required for the segregation of chromosomes during the reductional division of meiosis. Proc. Natl. Acad. Sci. 1988;85:6057–6061. doi: 10.1073/pnas.85.16.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B., Roeder G.S. Telomere-mediated chromosome pairing during meiosis in budding yeast. Genes & Dev. 1998;12:2574–2586. doi: 10.1101/gad.12.16.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.V., Roeder G.S. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell Biol. 1997;136:957–967. doi: 10.1083/jcb.136.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.N., Penkner A., Ohta K., Klein F., Nicolas A. B-type cyclins CLB5 and CLB6 control the initiation of recombination and synaptonemal complex formation in yeast meiosis. Curr. Biol. 2001;11:88–97. doi: 10.1016/s0960-9822(01)00026-4. [DOI] [PubMed] [Google Scholar]
- Song J., Durrin L.K., Wilkinson T.A., Krontiris T.G., Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead K., Aguilar C., Hartman T., Drexel M., Meluh P., Guacci V. Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J. Cell Biol. 2003;163:729–741. doi: 10.1083/jcb.200305080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M., Engebrecht J.A., Roeder G.S. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Toh E.A., Kikuchi Y. Comparative analysis of yeast PIAS-type SUMO ligases in vivo and in vitro. J. Biochem. 2003;133:415–422. doi: 10.1093/jb/mvg054. [DOI] [PubMed] [Google Scholar]
- Taylor D.L., Ho J.C., Oliver A., Watts F.Z. Cell-cycle-dependent localisation of Ulp1, a Schizosaccharomyces pombe Pmt3 (SUMO)-specific protease. J. Cell Sci. 2002;115:1113–1122. doi: 10.1242/jcs.115.6.1113. [DOI] [PubMed] [Google Scholar]
- Tsubouchi T., Roeder G.S. A synaptonemal complex protein promotes homology-independent centromere coupling. Science. 2005;308:870–873. doi: 10.1126/science.1108283. [DOI] [PubMed] [Google Scholar]
- Tung K.S., Roeder G.S. Meiotic chromosome morphology and behavior in zip1 mutants of Saccharomyces cerevisiae. Genetics. 1998;149:817–832. doi: 10.1093/genetics/149.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valpuesta J.M., Martin-Benito J., Gomez-Puertas P., Carrascosa J.L., Willison K.R. Structure and function of a protein folding machine: The eukaryotic cytosolic chaperonin CCT. FEBS Lett. 2002;529:11–16. doi: 10.1016/s0014-5793(02)03180-0. [DOI] [PubMed] [Google Scholar]
- van Heemst D., Kafer E., John T., Heyting C., van Aalderen M., Zickler D. BimD/SPO76 is at the interface of cell cycle progression, chromosome morphogenesis, and recombination. Proc. Natl. Acad. Sci. 2001;98:6267–6272. doi: 10.1073/pnas.081088498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X., Jeusset J., Soustelle C., Kowalczykowski S.C., Le Cam E., Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- Wang T.F., Kleckner N., Hunter N. Functional specificity of MutL homologs in yeast: Evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc. Natl. Acad. Sci. 1999;96:13914–13919. doi: 10.1073/pnas.96.24.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worland S.T., Wang J.C. Inducible overexpression, purification, and active site mapping of DNA topoisomerase II from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1989;264:4412–4416. [PubMed] [Google Scholar]
- Yu H.G., Koshland D.E. Meiotic condensin is required for proper chromosome compaction, SC assembly, and resolution of recombination-dependent chromosome linkages. J. Cell Biol. 2003;163:937–947. doi: 10.1083/jcb.200308027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Dix D.J., Eddy E.M. HSP70-2 is required for CDC2 kinase activity in meiosis I of mouse spermatocytes. Development. 1997;124:3007–3014. doi: 10.1242/dev.124.15.3007. [DOI] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]