Abstract
The environment provided by conspecifics is often the most important component of the environment experienced by individuals, frequently having profound effects on fitness and trait expression. Although these social effects on fitness and trait expression may appear to be purely environmental, they differ from other sorts of environmental influences, because they can have a genetic basis and thus can contribute to evolution. Theory has shown that these effects modify the definition of genetic architecture by making the phenotype the property of the genotypes of multiple individuals and alter evolutionary dynamics by introducing additional heritable components contributing to trait evolution. These effects suggest that genetic and evolutionary analyses of traits influenced by social environments must incorporate the genetic components of variation contributed by these environments. However, empirical studies incorporating these effects are generally lacking. In this paper, I quantify the contribution of genetically based environmental effects arising from social interactions during group rearing to the quantitative genetics of body size in Drosophila melanogaster. The results demonstrate that the genetic architecture of body size contains an important component of variation contributed by the social environment, which is hidden to ordinary genetic analyses and opposes the direct effects of genes on body-size development within a population. Using a model of trait evolution, I show that these effects significantly alter evolutionary predictions by providing hidden constraints on phenotypic evolution. The importance of relatedness of interactants and the potential impact of kin selection on the evolution of body size are also examined.
Since its origin, one of the major goals of genetics has been to understand the relative contribution of heritable and environmental factors to trait variation. Quantitative genetic (QG) methods have been developed as the primary means to achieve this goal, generally with the ultimate goal of understanding the evolutionary potential of traits (1). QG analyses use statistical approaches that rely on hypothetical constructs, devised to reflect causative influences producing variation in traits (e.g., ref. 2), to partition phenotypic variation into heritable components that contribute to trait evolution and nonheritable components that do not. The success of the QG approach for this purpose depends critically on the validity of the underlying model (3). Because of the primary interest in trait evolution, most analyses focus on the genetic components, casting environmental influences aside as sources of nonheritable random variation. However, in the case of the social environment (i.e., the environment provided by conspecifics), there can be a genetic component to the environment, because it is created by traits expressed by individuals. This genetic component of the environment blurs the distinction between genetic and environmental effects and thereby complicates the definition of genetic architecture. Because the environment itself can have a genetic basis, it can evolve. As a result, it must be included in genetic analysis if one wishes to gain a thorough understanding of trait evolution (4–8).
A modeling scheme that incorporates indirect genetic effects (IGEs) [also known as associate effects (9)] has been developed to investigate the genetic and evolutionary consequences of social influences on trait expression (8). IGEs are best understood by contrasting them with direct genetic effects (DGEs). DGEs occur when genes possessed by an individual directly influence that individual's phenotype. In contrast, IGEs occur when genes expressed in one individual have phenotypic effects in another (6, 8). IGE models solve the duality of these effects, as both environmental and genetic, by explicitly including the genetic basis of the environment in the definition of the individual phenotype (6, 9, 10). By using this approach, these models have demonstrated that IGEs can alter our view of genetic architecture, because the phenotype becomes the property of the genotypes of multiple individuals (6). The mapping of the individual phenotype to multiple genotypes can make the genetics of these traits particularly complex, because genetic analysis requires some understanding of interactions between individuals (9). The altered genetic architecture can also lead to very different evolutionary dynamics, such as accelerated or retarded responses to selection (6, 9, 10). Although IGEs can result from any sort of social interaction, with few exceptions (6, 9), IGE models have focused on the particular influence that maternal genotypes have on the expression of traits in their offspring (so-called maternal genetic effects; see ref. 10).
IGE models are analogous to, but more general than, models derived to examine kin selection (11–15). These models focus on the effect that genes in one individual have on the fitness of related individuals, and thus they are implicitly IGE models (e.g., equation 1 in ref. 11). These models have primarily been developed to gain an understanding of the conditions under which altruism can evolve. Cheverud (12) made the relationship between kin selection and IGE models explicit by using a QG model of maternal effects on a fitness-related trait of progeny. The QG model of Cheverud, and other more general related models (13, 14, 16), can be used to model evolution by kin selection when IGEs directly affect either fitness or fitness-related traits. Like kin-selection models, IGE models predict that the evolutionary dynamics of traits can be strongly influenced by the degree of relatedness of interactants.
Despite the fact that theoretical models have demonstrated that evolutionary dynamics can be quite different when IGEs are present, there is an absence of empirical studies that have quantified the occurrence of IGEs outside of the parent–offspring interaction. There have been, however, a number of experiments that have implied the presence of IGEs (17–20), although none of these studies explicitly quantified their importance. To investigate the importance of IGEs arising from other types of social interactions, this study focuses on the development of body size in Drosophila melanogaster. Many aspects of developmental and quantitative genetics are well understood in D. melanogaster. This species has been one of the main model organisms for studies of developmental genetics (21) and has been extensively studied from a QG perspective (1). Despite this enormous body of work, no previous investigation has directly examined the contribution of IGEs to genetic architecture, although previous work suggests that they should play an important role in trait expression (22–25). Because flies develop under high densities in an environment largely created by conspecifics [through the excretion of biotic residues (24), egestion of digestive enzymes (26), mechanical processing of medium, and direct competition for nutrients], there is considerable opportunity for IGEs to affect development. To examine the importance of IGEs in this system, I begin by presenting a simple model of trait development and evolution that incorporates IGEs (6). This model also forms the foundation for QG analysis by providing the expected components of variation used in partition phenotypic variation into IGEs, DGEs, and their covariance.
A Model of Trait Expression and Evolution
Influences on the development of body size can be modeled as a linear function of the direct effects of an individual's genes and the effects of the environment provided by conspecifics. Other functions are possible, but the simple additive model appears adequate based on the analysis of Drosophila pupa size (i.e., epistatic, dominance, and maternal effects are nonsignificant).
With individuals interacting in pairs (i.e., each individual provides an environment for its partner), the phenotypic value of an individual (zf) can be expressed as (after ref. 6):
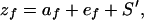 |
1 |
where zf is the phenotypic value of trait f in the focal individual, af is the additive DGE, ef is a random environmental effect (the subscript f indicates that this is the focal trait), and S′ is the effect of the environment provided by an individual's social partner, which can be negative (the prime indicates that this component is a characteristic of a different individual). The social effect of the individual's social partner (S′) is, likewise, modeled as a linear equation:
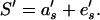 |
2 |
a′s is the additive genetic effect on the quality of the social environment provided by an interacting partner, and e′s is the random environmental effect on the social environment provided. The trait S is analogous to “maternal performance” (the effect of the mother on the phenotype of her progeny) considered in classic maternal-effect models (see ref. 27) and will be referred to as “social performance.” Under this model of inheritance, the response to selection (cross-generation change in the mean phenotype) (Δz̄f) is predicted by the equation (see ref. 6 for details on the derivation):
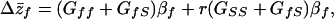 |
3 |
where Gff is the direct additive genetic variance of the trait, GSS is the additive indirect genetic variance, βf is the selection gradient acting on the phenotype (28), GfS is the genetic covariance between direct and indirect genetic effects, and r is the coefficient of relatedness of individuals. The genetic covariance, GfS, differs from the usually considered genetic covariance, because it measures the degree to which genes simultaneously affect the phenotypes of individuals and the phenotypes of their social partners. A positive value of GfS indicates that alleles that make an individual larger also make their partner larger. A negative value indicates that alleles that make individuals larger make their partners smaller. In the absence of IGEs, Eq. 3 simplifies to a version of the classic “breeder's equation,” Δz̄f = Gffβf (29).
The first composite term on the right side of Eq. 3 [(Gff + GfS)βf] gives the response to selection when social partners are unrelated. This term illustrates that the evolution of a character described by Eq. 1 is determined by both changes in average contribution of DGEs (Gffβf) and correlated changes in the average contribution of IGEs via the social environment (GfSβf). The term also demonstrates that, when interacting individuals are unrelated, selection cannot act directly on the IGE component, because IGEs do not directly map to the individual phenotype (i.e., GSS does not contribute to Δz̄f). However, the IGE component can evolve as a result of a correlated response to selection when there is a genetic covariance between DGEs and IGEs. The change in the IGE component can be seen as an evolutionary change in the mean social environment (ΔS̄), which contributes to the cross-generational change in the mean phenotype. Traditional QG models do not include this last term and predict response to selection solely on the basis of strength of selection and direct additive genetic variance. Thus, the response to selection can be greater or less than expected from the traditional QG model. When GfS is greater than Gff, the response to selection can even be in the direction opposite that predicted by the model that lacks IGEs.
The second composite term in Eq. 3 [r(GSS + GfS)βf] points out how relatedness and kin effects modify the evolutionary dynamics of traits affected by of IGEs. Interactions among kin modify the response to selection, because they alter the genotype-phenotype relationship (10) by introducing a sort of “genotype-environment” covariance (with “environment” referring to the social environment). The degree of this covariance is determined by the coefficient of relatedness of interactants (14, 30). When interacting with kin, individuals experience predictable social environments, because they are related to the individuals providing those environments. This means that there is a correlation between individual DGE values (af) and the IGE values that they experience (a′S), leading to a covariance between individual phenotypic (zf) and IGE values (aS). As a result, selection on a trait influenced by IGEs can act on both DGEs and IGEs simultaneously [because cov(zf, aS) = GfS + rGSS]. Depending on the relative magnitude of the IGE variance (GSS) and the sign and magnitude of the IGE–DGE covariance (GfS), the net response to selection can be increased or decreased when interactants are related (27).
The evolutionary dynamics of social performance (S) can be analyzed by using a social selection (16, 31–33) or kin-selection framework (12, 13, 15). Although most kin selection models focus on the evolution of altruism, where a trait decreases the individual's fitness but increases the fitness of the individual's relatives, they can be applied to any type of trait (e.g., competition) (11, 12, 27, 30, 34–36). Because fitness was not directly measured in this experiment, to model kin selection, it is necessary to assume that body size is a fitness correlate (i.e., βf ≠ 0) (12). Because the individual phenotype explains all variance in fitness, this is not true social or kin selection (16). However, because the characteristics of one individual influence the fitness of another (with the individual phenotype mediating the mapping from the traits of one individual to the fitness of another), the evolutionary consequences of these social effects can still be interpreted in the framework of social or kin selection (12, 16).
To model the evolution of IGEs by kin selection, Cheverud (12) considered a case where mothers affect the phenotypes of their offspring. He focused on conditions under which maternal performance is expected to evolve in an altruistic direction. Using that same model, Cheverud also presented a more general equation that can be applied to other types of kin effects (see equation 16 in ref. 12). To relate these models, it is necessary to first introduce an equation predicting response of the social-performance trait to selection on body size:
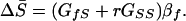 |
4 |
Note that the terms in Eq. 4 are all present in Eq. 3, because the evolution of mean social performance directly contributes to the evolution of mean body size. As pointed out above, when r = 0, selection cannot act directly on IGEs, and social performance evolves only as a correlated response to selection on body size. However, when individuals are related, selection can act directly on genetic variation affecting social performance.
Eq. 4 can be related to the kin selection models by rearranging it to examine the conditions under which social performance shows an evolutionary increase (i.e., ΔS̄ > 0) due to selection on body size (i.e., evolves in an altruistic direction):
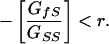 |
5 |
This equation is equivalent to the Cheverud model (equation 16 of ref. 12) under the assumptions used here. From Eq. 5, it is clear that, when the covariance is positive, these conditions are always met. The conditions become more interesting when the covariance is negative (i.e., DGEs and IGEs are antagonistic), because the term on the left is positive. The conditions are met when the ratio of genetic covariance to indirect genetic variance is less than the coefficient of relatedness. The ratio in Eq. 5 is analogous to the cost–benefit ratio (k) in “Hamilton's rule” (11), or what he called the “ratio of diminution” when referring to competition. The negative covariance (GfS) can be viewed as the selective component due to negative impacts on fitness of kin and genetic variance (GSS) as the selective component associated with the positive effects on fitness (12).
Eq. 3 can also be rearranged to examine conditions for positive or negative responses to selection under the various combinations of positive or negative selection and positive or negative genetic covariances (as in section 5 of ref. 11). The evolutionary dynamics in the presence of nonkin social selection can be examined by setting r equal to zero in the above equations (e.g., equation 14 in ref. 16).
Methods
Study Population.
The focal population (LH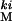 ) is a wild-type highly variable laboratory adapted population segregating for a codominant marker (kinked [ki] on the third chromosome). This population is adapted to a 2-week generation cycle, with larvae developing under controlled intermediate densities on standardized cornmeal molasses medium seeded with yeast in 10-dram vials. The LH
) is a wild-type highly variable laboratory adapted population segregating for a codominant marker (kinked [ki] on the third chromosome). This population is adapted to a 2-week generation cycle, with larvae developing under controlled intermediate densities on standardized cornmeal molasses medium seeded with yeast in 10-dram vials. The LH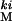 population was created by introgressing ki from a marker stock (Bloomington Stock Center no. 2975, Bloomington, IN) into 120 lineages derived from the LHM population by 18 generations of selective backcrossing to LHM (see ref. 37 for details on the LHM population). The retention of the 2975 genome in each of these lineages was estimated by testcrosses back to 2975 and scoring of the nine linked recessive third-chromosome markers that flank the ki mutation in 2975. Lines retaining a large portion of the 2975 genome were discarded, leaving 101 lines that were combined to found the LH
population was created by introgressing ki from a marker stock (Bloomington Stock Center no. 2975, Bloomington, IN) into 120 lineages derived from the LHM population by 18 generations of selective backcrossing to LHM (see ref. 37 for details on the LHM population). The retention of the 2975 genome in each of these lineages was estimated by testcrosses back to 2975 and scoring of the nine linked recessive third-chromosome markers that flank the ki mutation in 2975. Lines retaining a large portion of the 2975 genome were discarded, leaving 101 lines that were combined to found the LH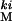 population. The resulting population contains between 97.5% and 99% of its genome from the laboratory population with the remaining 1–2.5% coming from the marker stock.
population. The resulting population contains between 97.5% and 99% of its genome from the laboratory population with the remaining 1–2.5% coming from the marker stock.
Experimental Crosses.
Virgin females were collected during the first 4 days of adult emergence and were mixed together to create an all-female population with a representation of genotypes from the entire period of normal adult emergence. Females that were either homozygous wild-type at the ki locus (w/w) or homozygous kinked (ki/ki) were chosen at random from this mixed population of virgins. w/w males (sires) were chosen at random from the LH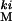 population and were placed in a vial with one w/w and one ki/ki female (dams) and were left to mate for 3 days.
population and were placed in a vial with one w/w and one ki/ki female (dams) and were left to mate for 3 days.
Experimental Rearing Scheme.
Eggs were collected from individual females every 2 h for 48 h. Eggs from the first 2-h interval were discarded to avoid eggs that may have been held by females. Five pairs of dams mated to five sires were randomly assigned to an experimental block. Each replicate of the experiment contained five blocks, and the entire experiment was replicated three times over a 4-mo period. All replicates together included a total of 150 dams and 75 sires. A total of 2,930 progeny from these families were used in the analysis.
To partition IGEs from DGEs, this study uses a simple social structure that corresponds to the structure assumed in the above model. Same-age progeny from each family were reared from egg to pupation with, and thus experience an environment provided by, either their full siblings (sibs), half sibs, or individuals from an unrelated family. Eggs from each wi/wi dam (which have the genotype w/w) were reared with eggs from every ki/ki dam (which have the genotype w/ki) in the same block, and each dam combination was replicated six times. These combinations allowed parentage to be assigned based on the ki phenotype. Eggs derived from a single female were also reared together. A single egg from a dam was placed in a rearing tube (details below) along with an egg, collected within the same time period, from one other dam from her block. Pairs were reared under competitive conditions in 0.4-ml polypropylene microcentrifuge tubes containing 50 μl of cornmeal molasses medium (the same recipe as in the population tubes) and 5 μl of a yeast solution (1 g of yeast per 10 ml of distilled water). Tubes were placed in random locations in humidified boxes inside an environmentally controlled chamber at 25°C on a 12:12-h light/dark cycle and were randomly repositioned every day to diminish the influence of environmental differences among locations. At pupation, pupae were placed individually in 0.5-ml microcentrifuge tubes and held in the environmental control chamber. Images of each pupa were taken under a stereomicroscope by using nih image software. Length was measured to the nearest 5 μm (yielding values with four significant digits). Pupae were held until eclosion, at which time parentage was determined based on the ki genotype. Dry body weights, including the pupal case, were measured on a small subsample of 175 male and 175 female flies to establish the relationship between pupa size and dry weight. These individuals were dried for 2 h at 60°C and then kept in a dessication chamber until being weighed. Dry weights were taken to the nearest 1.0 μg by using a microbalance (yielding values with three significant digits).
Genetic Analysis.
QG analysis of pupa length was performed by using restricted maximum likelihood to fit a mixed-model ANOVA (38, 39). Separate analyses were done for the three social structures: (i) pairs of unrelated individuals reared in combination (analysis U; n = 724), where each paternal half-sib family is combined with just one other family; (ii) individuals reared with half sibs (analysis H; n = 708), and (iii) full sibs reared together (analysis F; n = 1498). All three analyses included two random effects: sire and dam(sire), where the parentheses indicate a nested effect. The sire component estimates the covariance of half-sibs (40), whereas dam(sire) estimates the covariance of full-sibs minus the covariance of half-sibs. These two terms correspond to the ordinary sire and dam(sire) terms from a paternal half-sib breeding design (40). All models also included the fixed effect of sex. Other fixed effects were tested (e.g., block and effects of the ki) but were not significant and were not included in the final models.
Expected covariances for each of the analyses were derived by using the model for trait expression presented above (Eqs. 1 and 2) and are given in Table 1. The expected covariances of full and half-sibs (i.e., the sire and dam[sire] terms) under the IGE model differ for each of the three social structures. The expected covariances were calculated by using a model that matches this particular social system (pairwise social interactions) and thus would differ for other possible social structures. This further illustrates the potential problems that arise when considering the role of IGEs in genetic analyses and trait evolution; for any single experiment, it may be necessary to derive expected covariances by using a hypothetical construct that matches the expected pattern of phenotypic effects and social structure being used.
Table 1.
Expected covariances for individuals reared with individuals that are their full sibs or half sibs, or are unrelated
| Relationship of social partners | ANOVA term | Expected covariance |
|---|---|---|
| Full sibs | Sire | σ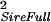 = ¼ Gff + ½ GfS + ¼ GSS = ¼ Gff + ½ GfS + ¼ GSS
|
| Dam(Sire) | σ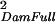 = ¼ Gff + ½ GfS + ¼ GSS = ¼ Gff + ½ GfS + ¼ GSS
|
|
| Half sibs | Sire | σ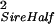 = ¼ Gff + ½ GfS + ½ GSS = ¼ Gff + ½ GfS + ½ GSS
|
| Dam(Sire) | σ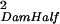 = ¼ Gff = ¼ Gff
|
|
| Unrelated | Sire | σ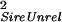 = ¼ Gff + ¼ GSS = ¼ Gff + ¼ GSS
|
| Dam(Sire) | σ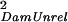 = ¼ Gff + ¼ GSS = ¼ Gff + ¼ GSS
|
For the case where individuals are reared with individuals they are not related to, each half-sib family is reared with individuals from one other unrelated half-sib family. Covariances were derived by using Eqs. 1 and 2. Gff is the additive genetic variance of the trait measured in the focal individual; GSS is the additive genetic variance of the indirect genetic effect (i.e., genetic variance of the environment provided by another individual); and GfS is the genetic covariance of direct and indirect effects. Dam(Sire) indicates that dams are nested within sires in the half-sib design.
Because the expected covariances for the three social structures have different contributions from DGEs (Gff), IGEs (GSS), and their covariance (GfS), they can be used to solve for each of these components (after ref. 41). There are multiple ways to solve for each component, but only results from a single approach are presented here (all approaches yield similar results). The three sire terms were used, because they are more likely to conform to the model (i.e., they contain no potential contribution of dominance or maternal effects). Covariance terms were solved for as follows:
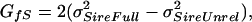 |
6a |
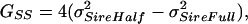 |
6b |
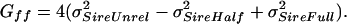 |
6c |
Separate models were fitted for each block of the experiment, providing 15 independent estimates of each covariance parameter. Means and confidence intervals for all covariances were estimated from these replicated estimates.
Results
The partial correlation between pupa length and dry weight, corrected for sex differences in these two characters, is 0.71, P < 0.0001, indicating that length is a very good substitute for total body size.
The parameter estimates and confidence intervals of the direct additive genetic variance (Gff), the additive indirect genetic variance (GSS), and the additive direct–indirect genetic covariance (GfS) are given in Table 2. The largest component of variance, direct additive genetic variance, accounts for 34.4% of phenotypic variation. This value corresponds to the narrow sense heritability, h2. The component attributed to additive IGEs accounts for 17.9% of phenotypic variation. Note that DGEs account for just less than twice as much phenotypic variation as IGEs. The genetic covariance between direct and indirect effects is large and negative, corresponding to a genetic correlation (RfS) of −0.85, with an approximate 95% confidence interval of ±0.20 (see ref. 1). This indicates a large degree of nonindependence of these effects (due to either pleiotropy or linkage disequilibrium).
Table 2.
Estimated quantitative genetic variance components
| Parameter | Estimate | 95% C.I. |
|---|---|---|
| Direct additive genetic variance (GFF) | 11,290 | ±1,497 |
| Indirect additive genetic variance (GSS) | 5,865 | ±1,520 |
| Direct–indirect genetic covariance (GFS) | −6,955 | ±1,488 |
| Total phenotypic variance | 32,815 | ±1,199 |
Values are the mean of 15 individual estimates (in μm2) taken from the 15 independent replicates of the experimental design with 95% confidence intervals (C.I.).
The magnitude of the narrow-sense heritability is in accord with the general findings of other studies that have estimated the heritability of body size in Drosophila. Most heritability estimates for body size (weight or thorax length) in other populations fall within the range of ≈0.2–0.4 (reviewed in ref. 42). The heritability estimated here is also very close to the mean heritability (which equals 0.32) estimated from a large suite of morphological traits in Drosophila (43).
Discussion
The results of this experiment demonstrate that the view of genetic architecture, derived from an analysis that includes both DGEs and IGEs, is very different from what we would have found had we taken the traditional approach and ignored IGEs. Although the direct genetic variation would be apparent in a traditional QG analysis, the contribution of IGEs would be missed or confounded with DGEs, depending on the social structure used (Table 1). The presence of a significant IGE variance (GSS) indicates that a considerable portion of the genetic variation for body size results from interactions among individuals. In these interactions, individuals are in part determining each other's size. Because these effects have a genetic basis, they result in IGEs. This component of genetic variation is hidden to ordinary genetic analyses that are focused exclusively on direct effects. This is significant, because the IGE component represents real heritable variation in a population that can contribute to evolutionary changes (6, 9, 44). The presence of IGEs means that the genetic architecture of any phenotype influenced by social environments may be even more complex than previously thought (45), because genetic influences can arise from the genes of the individual as well as from genes in other individuals. As a result, a thorough understanding of genetic architecture requires an understanding of the various pathways, including indirect ones, through which genetic variation leads to phenotypic variation in populations.
Perhaps the most significant finding in this study is the very large negative genetic correlation between direct and indirect genetic effects (RfS = −0.85), indicating that the direct effects that genes have on body size are negatively associated with indirect effects on body size. This antagonistic relationship is likely due to pleiotropy, where genes that make individuals larger create an environment that makes other individuals smaller due to social competition for limited resources. In this scenario, genetic variation for competitive ability appears as genetic variation for body size, because differential competitive success leads to different resource accumulation, which is reflected in body size. As a result, loci affecting competitiveness should appear as part of the genetic architecture of body size. However, the impact of this sort of genetic variation on evolution is very different from other types of variation; the influence on the phenotype depends on the context provided by social interactions. Because these loci influence body size through their effect on competitiveness, they can also affect the size of individuals' social partners that are competing for those same limited resources. Thus, social competition necessarily makes the direct and indirect effects of genotypes negatively correlated. Because competition is widespread and likely to influence trait expression, it seems probable that this sort of antagonistic relationship between DGEs and IGEs is common.
The genetic covariance parameters from the QG analysis (Table 2) can be combined with the evolutionary model presented above to examine how IGEs modify the predicted evolutionary dynamics of body size. The genetic parameters can be substituted into Eq. 3 to predict response to selection as a function of strength of selection. The resulting equation tells us how the genetic system translates selection into evolutionary change. Starting with the assumption that interactions are among unrelated individuals, we would predict response of body size (pupa length in micrometers) to selection by using the equation:
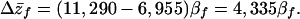 |
7 |
In contrast, if we ignore the contribution of IGEs and use the standard QG model (Δz̄f = Gff βf), we predict:
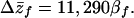 |
8 |
Thus, under the IGE model, the predicted response to selection is less than half that expected under a traditional model.
When individuals that develop together are related, the expected response to selection is further diminished due to kin effects. Adding relatedness to Eq. 8 yields
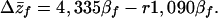 |
9 |
Eq. 9 demonstrates that the more closely individuals are related to their competitors, the lower the expected response to selection on body size. For example, if individuals develop with their full sibs, a situation that would arise when singly inseminated females lay multiple eggs on a food source, the expected response to selection would be 3,790βf. This reduction in the expected rate of evolution was predicted by Hamilton (see part c of section 3 in ref. 11) in his consideration of negative kin effects (see also ref. 46). Because the DGE–IGE covariance (GfS) is larger than the IGE variance (GSS), social performance cannot evolve in an “altruistic” direction, because there is no level of relatedness that can satisfy the conditions expressed in Eq. 5.
The diminished response to selection caused by the antagonistic counterevolution of IGEs (Eq. 7) and the further diminution expected when interactions are among relatives (Eq. 9), can be viewed as a constraint on phenotypic evolution. The constraint arises because evolution of body size is determined by the sum of changes in the mean contribution of both DGEs and IGEs (i.e., we can view the response to selection as Δāf + ΔāS), which show antagonistic correlated evolution (i.e., Δāf and ΔāS are of opposite sign) due to the negative IGE–DGE covariance. These antagonistic effects partially cancel each other out and, as a result, body size is expected to show much less of a response to selection than predicted by traditional QG theory (44). This type of constraint is fundamentally different from the usually considered genetic constraint on evolution arising from antagonistic pleiotropy (where genetic multicollinearity restricts multivariate evolution). The constraint arises from “socially antagonistic pleiotropy,” where genes that positively affect a character (via their effect on competitiveness) also negatively impact the social environment experienced by conspecifics (see ref. 47). This form of pleiotropy does not manifest at the individual level. Rather, it is a population-level phenomenon where the same genes that make individuals larger through direct mapping also make them smaller via indirect mapping. Because these two effects add together to determine the effects of genes on the evolution of the mean phenotype in a population, the IGE–DGE antagonism constrains the overall response to selection. Thus, even though a single individual does not manifest these pleiotropic effects (i.e., effects of genes are on the same trait expressed in two different individuals), they still affect how a trait is expected to evolve.
In the case of body size in Drosophila, we predict that selection for increased size would be constrained, because selection for size leads to an increase in the average competitiveness of the population. The change in average competitiveness would appear as a degradation of the social environment, which would partially counter the positive changes in DGEs affecting the trait. Although the phenotype is not expected to evolve at the rate predicted from the level of genetic variation and strength of selection, evolution at the genetic level (i.e., change in allele frequencies as measured by Δāf) continues. However, this genetic evolution is not matched by an equivalent magnitude of change at the phenotypic level. For example, if we were to select the largest individuals in a generation, they would on average also be the most competitive individuals. These individuals would have a set of genotypes that, under the current social conditions, make individuals large, and selection would therefore produce a genetic change in the population. However, the progeny of these individuals would find themselves in a more competitive environment, because they all inherited genes from the most competitive individuals in the previous generation. Thus, this new generation would not be as large as we would have expected based on the size of their parents, because they are experiencing a different social environment.
As an analogy, we can view body size evolving on a treadmill, where every step forward is accompanied by movement backward due to the associated negative changes in the environment. The result is that, depending on the speed of the treadmill, the trait either remains where it started or does not move as far as expected [Dickerson (48) referred to this as “slippage” on the treadmill]. When describing his fundamental theorem of natural selection, Fisher (47) recognized this process as the critical reason why populations do not continue to evolve to higher states of fitness (or character values) despite widespread recurrent directional selection. However, his intuition that these effects would exist and be potentially important had not been previously demonstrated. In addition, recurrent directional selection has been predicted to be a consequence of social selection, where the treadmill of social competition leads to a relentless force of selection (32).
This type of antagonistic evolution of the social environment has also been considered important in cases of evolution by sexual selection, where each sex provides an environment for the other. Antagonistic interactions of males and females (sexual conflict) create a situation where the adaptation of one sex is perceived as a degradation of the environment experienced by the other. Empirical evidence of this process has come from the evolution of seminal proteins in Drosophila, where males and females continually adapt to the environment provided by the other sex (49). This process has been referred to as the intraspecific Red Queen to reflect the fact that the evolving environment provides a constant force of selection (7).
The results of this study, and the general phenomenon of antagonism between IGEs and DGEs, may also help explain a number of interesting but unexplained patterns of variation in heritability found in other studies. For example, Scheiner and Lyman (50) found that the realized heritability of body size (measured as thorax size) was consistently smaller than the heritability value estimated by using full- or half-sib correlations. They estimated narrow-sense heritabilities both before and after artificial selection on body size and found that the two heritability estimates were very similar, indicating that selection did not significantly diminish genetic variance. However, realized heritabilities, estimated from the replicated responses to selection over 16 generations, were considerably smaller than the narrow–sense heritability estimates from the sib analysis (the realized heritabilities averaged ≈64% of the narrow-sense heritability values). This result would be expected if their population has a genetic architecture similar to the population analyzed here. Antagonistic evolution of IGEs would be expected to diminish the response to selection (as in Eq. 9), making the realized heritability smaller than the narrow-sense heritability. Given that they used a standard group-rearing protocol, this is likely to be the case. Another example comes from a study of realized heritabilities of body size (fresh weight) in both rich and poor larval environments (51). In this study, Hillesheim and Stearns found that nearly all realized heritability estimates were larger when larvae were reared in the richer environment. Assuming that the poor environment is also more competitive, we would expect the realized heritabilities to be smaller in that environment due to stronger antagonistic IGEs. In other words, because competition enforces the negative covariance between IGEs and DGEs, one would expect that the covariance would be larger (i.e., more negative) when competition is stronger. This would result in the smaller response to selection in the competitive environment, even if the direct narrow-sense heritabilities were similar in the two environments.
Because interactions between individuals are ubiquitous, the opportunity for phenotypic effects of these interactions, and thus IGEs, is considerable. This is particularly true for phenotypes like social behaviors, whose expression often depends on the social environment [or the “behavioral environment” (4, 52)], or for traits affected by social competition or other interactions during trait development. In addition, in many populations and in most laboratory analyses of quantitative genetics, interacting/competing individuals are related (e.g., ref. 53), providing the opportunity for kin effects and selection (54). Thus, the data presented here from Drosophila are expected to have significant implications for genetic analysis of a variety of traits in a diversity of taxa. These data suggest that the traditional paradigm, focused exclusively on direct effects of genes, is inadequate. To develop an accurate picture of genetic architecture that describes how genetic variation leads to phenotypic variation in a population, information on both direct and indirect effects of genes will be required whenever individuals interact.
Acknowledgments
I thank E. D. Brodie III, J. M. Cheverud, P. X. Kover, A. J. Moore, and M. J. Wade for insightful discussions during the development this work; A. J. Moore, M. Pigliucci, M. J. West-Eberhard, and an anonymous reviewer for thoughtful comments on the manuscript; A. Epps and P. X. Kover for help with the conducting of the experiments; and A. K. Chippindale for generously sharing the LHM population. This work was supported by a grant from Sigma Xi, a Postdoctoral Fellowship in Bioinformatics from the National Science Foundation (NSF), and NSF IBN-9896116 to E. D. Brodie III.
Abbreviations
- QG
quantitative genetic
- IGE
indirect genetic effect
- DGE
direct genetic effect
- sib
sibling
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 4357.
References
- 1.Roff D A. Evolutionary Quantitative Genetics. New York: Chapman & Hall; 1997. [Google Scholar]
- 2.Fisher R A. Trans R Soc Edinburgh. 1918;52:399–433. [Google Scholar]
- 3.Barton N H, Turelli M. Annu Rev Genet. 1989;23:337–370. doi: 10.1146/annurev.ge.23.120189.002005. [DOI] [PubMed] [Google Scholar]
- 4.Wcislo W T. Annu Rev Ecol Syst. 1989;20:137–169. [Google Scholar]
- 5.Wcislo W T. In: The Evolution of Social Behavior in Insects and Arachnids. Choe J C, Crespi B J, editors. New York: Cambridge Univ. Press; 1997. [Google Scholar]
- 6.Moore A J, Brodie E D, III, Wolf J B. Evolution (Lawrence, KS) 1997;51:1352–1362. doi: 10.1111/j.1558-5646.1997.tb01458.x. [DOI] [PubMed] [Google Scholar]
- 7.Rice W R, Holland B. Behav Ecol Sociobiol. 1997;41:1–10. [Google Scholar]
- 8.Wolf J B, Brodie E D, III, Cheverud J M, Moore A J, Wade M J. Trends Ecol Evol. 1998;13:64–69. doi: 10.1016/s0169-5347(97)01233-0. [DOI] [PubMed] [Google Scholar]
- 9.Griffing B. In: Proceedings of the International Conference on Quantitative Genetics. Pollak E, Kempthorne O, Bailey T B, editors. Ames: Iowa State Univ. Press; 1977. pp. 413–434. [Google Scholar]
- 10. Kirkpatrick, M. & Lande, R. (1989) Evolution (Lawrence, KS) 43.
- 11.Hamilton W D. J Theor Biol. 1964;7:1–16. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- 12.Cheverud J M. Evolution (Lawrence, KS) 1984;38:766–777. doi: 10.1111/j.1558-5646.1984.tb00349.x. [DOI] [PubMed] [Google Scholar]
- 13.Cheverud J M. Behav Ecol Sociobiol. 1985;16:239–243. [Google Scholar]
- 14.Lynch M. Proc Natl Acad Sci USA. 1987;84:8507–8511. doi: 10.1073/pnas.84.23.8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Queller D C. Evolution (Lawrence, KS) 1992;46:376–380. doi: 10.1111/j.1558-5646.1992.tb02045.x. [DOI] [PubMed] [Google Scholar]
- 16.Wolf J B, Brodie E D, III, Moore A J. Am Nat. 1999;153:254–266. doi: 10.1086/303168. [DOI] [PubMed] [Google Scholar]
- 17.Craig D M. Evolution (Lawrence, KS) 1982;36:271–282. doi: 10.1111/j.1558-5646.1982.tb05041.x. [DOI] [PubMed] [Google Scholar]
- 18.Goodnight C J. Evolution (Lawrence, KS) 1985;39:545–558. doi: 10.1111/j.1558-5646.1985.tb00394.x. [DOI] [PubMed] [Google Scholar]
- 19.Griffing B. Genetics. 1989;122:943–956. doi: 10.1093/genetics/122.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meffert L M. Genetics. 1995;139:365–374. doi: 10.1093/genetics/139.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bate M, Arias A M. The Development of Drosophila melanogaster. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. [Google Scholar]
- 22.Lewontin R C. Evolution (Lawrence, KS) 1955;9:27–41. [Google Scholar]
- 23.Lewontin R C, Matsuo Y. Proc Natl Acad Sci USA. 1963;49:270–278. doi: 10.1073/pnas.49.2.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weisbrot D R. Genetics. 1966;53:427–435. doi: 10.1093/genetics/53.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miranda J R, Hemmat M, Eggleston P. Heredity. 1991;66:333–342. doi: 10.1038/hdy.1991.42. [DOI] [PubMed] [Google Scholar]
- 26.Haj-Ahmad Y, Hickey D A. Nature. 1982;299:350–352. doi: 10.1038/299350a0. [DOI] [PubMed] [Google Scholar]
- 27.Cheverud J M, Moore A J. In: Quantitative Genetic Studies of Behavioral Evolution. Boake C R B, editor. Chicago: Univ. of Chicago Press; 1994. pp. 67–100. [Google Scholar]
- 28.Lande R, Arnold S J. Evolution (Lawrence, KS) 1983;37:1210–1226. doi: 10.1111/j.1558-5646.1983.tb00236.x. [DOI] [PubMed] [Google Scholar]
- 29.Lande R. Evolution (Lawrence, KS) 1976;30:314–334. doi: 10.1111/j.1558-5646.1976.tb00911.x. [DOI] [PubMed] [Google Scholar]
- 30.Frank S A. Foundations of Social Evolution. Princeton: Princeton Univ. Press; 1998. [Google Scholar]
- 31.Crook J H. In: Sexual Selection and the Descent of Man, 1871–1971. Campbell B, editor. Chicago: Aldine; 1972. pp. 231–281. [Google Scholar]
- 32.West-Eberhard M J. Proc Am Philos Soc. 1979;123:222–234. [Google Scholar]
- 33.West-Eberhard M J. Q Rev Biol. 1983;58:155–183. [Google Scholar]
- 34.West-Eberhard M J. Q Rev Biol. 1975;50:1–33. [Google Scholar]
- 35.Wade M J. Proc Natl Acad Sci USA. 1982;79:3575–3578. doi: 10.1073/pnas.79.11.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frank S A. Proc R Soc London B. 1994;258:153–161. doi: 10.1098/rspb.1994.0156. [DOI] [PubMed] [Google Scholar]
- 37.Chippindale A K, Rice W R. Proc Natl Acad Sci USA. 2001;98:5677–5682. doi: 10.1073/pnas.101456898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaw R G. Evolution (Lawrence, KS) 1987;41:812–826. doi: 10.1111/j.1558-5646.1987.tb05855.x. [DOI] [PubMed] [Google Scholar]
- 39.Fry J D. Evolution (Lawrence, KS) 1992;46:540–550. doi: 10.1111/j.1558-5646.1992.tb02057.x. [DOI] [PubMed] [Google Scholar]
- 40.Becker W A. Manual of Quantitative Genetics. Pullman, WA: Academic Enterprises; 1992. [Google Scholar]
- 41.Riska B J, Rutledge J J, Atchley W R. Genet Res. 1985;42:287–297. doi: 10.1017/s0016672300022278. [DOI] [PubMed] [Google Scholar]
- 42.Hoffmann A A. In: Adaptive Genetic Variation in the Wild. Mousseau T A, Sinervo B, Endler J, editors. Oxford: Oxford Univ. Press; 2000. pp. 200–218. [Google Scholar]
- 43.Roff D A, Mousseau T A. Heredity. 1987;58:103–118. doi: 10.1038/hdy.1987.15. [DOI] [PubMed] [Google Scholar]
- 44.Wright A J. Theor Appl Genet. 1986;72:256–263. doi: 10.1007/BF00267001. [DOI] [PubMed] [Google Scholar]
- 45.Paterson A H. Molecular Dissection of Complex Traits. New York: CRC; 1998. [Google Scholar]
- 46.Haldane J B S. Trans Camb Philos Soc. 1924;23:19–41. [Google Scholar]
- 47.Fisher R A. The Genetical Theory of Natural Selection. London: Oxford Univ. Press; 1930. [Google Scholar]
- 48.Dickerson G E. Cold Spring Harbor Symp Quant Biol. 1955;20:213–223. doi: 10.1101/sqb.1955.020.01.020. [DOI] [PubMed] [Google Scholar]
- 49.Rice W R. Nature. 1996;381:232–234. doi: 10.1038/381232a0. [DOI] [PubMed] [Google Scholar]
- 50.Scheiner S M, Lyman R F. J Evol Biol. 1991;4:23–50. [Google Scholar]
- 51.Hillesheim E, Stearns S C. Evolution (Lawrence, KS) 1991;45:1909–1923. doi: 10.1111/j.1558-5646.1991.tb02696.x. [DOI] [PubMed] [Google Scholar]
- 52.Wcislo W T. In: The Evolution of Social Behavior in Insects and Arachnids. Choe J C, Crespi B J, editors. Cambridge, U.K.: Cambridge Univ. Press; 1997. pp. 316–332. [Google Scholar]
- 53.Gebhardt M D, Stearns S C. Genet Res. 1992;60:87–101. doi: 10.1017/s0016672300030780. [DOI] [PubMed] [Google Scholar]
- 54.Alexander R D. Annu Rev Ecol Syst. 1974;4:325–383. [Google Scholar]


