Abstract
Gilles de la Tourette syndrome (GTS) is a potentially debilitating neuropsychiatric disorder defined by the presence of both vocal and motor tics. Despite evidence that this and a related phenotypic spectrum, including chronic tics (CT) and Obsessive Compulsive Disorder (OCD), are genetically mediated, no gene involved in disease etiology has been identified. Chromosomal abnormalities have long been proposed to play a causative role in isolated cases of GTS spectrum phenomena, but confirmation of this hypothesis has yet to be forthcoming. We describe an i(18q21.1-q22.2) inversion in a patient with CT and OCD. We have fine mapped the telomeric aspect of the rearrangement to within 1 Mb of a previously reported 18q22 breakpoint that cosegregated in a family with GTS and related phenotypes. A comprehensive characterization of this genomic interval led to the identification of two transcripts, neither of which was found to be structurally disrupted. Analysis of the epigenetic characteristics of the region demonstrated a significant increase in replication asynchrony in the patient compared to controls, with the inverted chromosome showing delayed replication timing across at least a 500-kb interval. These findings are consistent with long-range functional dysregulation of one or more genes in the region. Our data support a link between chromosomal aberrations and epigenetic mechanisms in GTS and suggest that the study of the functional consequences of balanced chromosomal rearrangements is warranted in patients with phenotypes of interest, irrespective of the findings regarding structurally disrupted transcripts.
Gilles de la Tourette syndrome (GTS) [Online Mendelian Inheritance in Man (OMIM, www.ncbi.nlm.nih.gov/Omim/) 137580] is a developmental neuropsychiatric syndrome characterized by the presence of chronic motor and vocal tics. Several decades of evidence suggest that GTS and a spectrum of tic-related phenomena are heritable. Estimates of population prevalence vary with ascertainment methodology (1–3). Nonetheless, there is general agreement that first-degree relatives of GTS probands have a 10- to 100-fold greater risk of developing the disorder than do individuals in the general population (4). Monozygotic concordance rates have been found to be between 53% and 56%, versus a concordance of <10% in dizygotic twins (5, 6). When twin studies have used direct patient examination and included the diagnosis of chronic tics (CT), monozygotic concordance has been noted to be as high as 100% (7).‖‖
Several studies have suggested that GTS and a spectrum of related disorders, including CT and Obsessive Compulsive Disorder (OCD), are transmitted in an autosomal dominant fashion with partial penetrance (8–11). Despite this evidence, linkage analyses have not yet led to the identification of a gene involved in disease etiology. It is likely that the combination of genetic and clinical heterogeneity has hindered population genetic approaches to disease gene identification (12, 13).
In addition to linkage and association strategies, multiple investigators have studied chromosomal abnormalities in individuals and families with GTS in the hopes of identifying a gene or genes of major effect disrupted by the rearrangement (14–16). This strategy is predicated on the notion that such patients, although unusual, may help to identify genes that are of consequence for a subgroup of patients with GTS, OCD, and CT, and provide important insights into physiologic pathways that more commonly contribute to trait development.
A review of all published cases of chromosomal translocations or inversions identified in patients with GTS reveals that three segments of the genome, on chromosomes 18q, 7q, and 8q, have been reported to be rearranged in more than one unrelated individual (14–17). Nonetheless, only one report to date has identified a structurally disrupted transcript (16), and its relevance to GTS has yet to be confirmed.
In this paper, we report on a young man with CT and OCD who was found to carry a paracentric inversion involving chromosome 18q22. We mapped the telomeric end of the inversion to a genomic location that is within 1 Mb of a previously described translocation that cosegregated in a family with the range of clinical phenomena encompassing GTS, CT, and OCD (14). Our detailed characterization of this rearrangement breakpoint revealed a relatively gene-poor region with two nearby transcripts, neither of which was structurally altered by the chromosomal abnormality. Multiple reports have confirmed that balanced chromosomal abnormalities many hundreds of kilobases from disease-related genes may lead to the expected disease phenotypes (18). These findings raise the possibility that long-range position effects may be playing a role in the identified case. We undertook experiments assessing replication synchrony versus asynchrony in the patient and controls to evaluate this hypothesis and characterize the epigenetic phenomena in this genomic interval.
Materials and Methods
Case Material.
The patient is a 12-year-old boy of Korean descent who was adopted by an American couple at the age of 3 months. The patient developed eye-blinking tics at age 6, shoulder jerking at age 7, and teeth grinding and unwanted mouth movements at age 8. The tics were present on a daily basis over several years. He reported feelings of physical tension in his joints before having tics and noted “just-right” phenomena. There was no history of vocal tics.
The patient was assessed with a Yale–Brown Obsessive Compulsive Scale (Y-BOCS), a standard diagnostic instrument that includes a symptom checklist of obsessions and compulsions (19). His total Y-BOCS score for current obsessions and compulsions was found to be 30 of a maximum of 40 points. His worst-ever score was 34 of a maximum of 40 points. On the basis of the clinical history and presentation and scores on standardized instruments, the patient met Diagnostic and Statistical Manual of Mental Disorders IV (DSM-IV) diagnostic criteria for OCD and chronic motor tics.
No data were available regarding prenatal or family history due to the adoption from a foreign country. The patient had a normal developmental history, an above-average full-scale IQ, and a normal physical examination. Chromosomal testing revealed an inversion of chromosome inv(18)(q21;q22). Fragile X testing was negative. Routine laboratory tests including complete blood count, electrolytes and thyroid function studies were unremarkable.
Fluorescence in Situ Hybridization (FISH).
FISH studies were carried out as previously described (20). Bacterial artificial chromosome (BAC) or yeast artificial chromosome (YAC) DNA was labeled by nick translation. Biotin-labeled probes were detected with FITC-conjugated avidin (1:400 dilution), and digoxigenin-labeled probes were detected with an antidigoxigenin antibody conjugated to rhodamine (1:150 dilution).
Identification of Putative Coding Sequence.
The identities of BAC clones spanning the 18q22 inversion breakpoint were determined before the availability of relevant draft sequence from the human genome project. As a result, one spanning BAC was fully sequenced through collaboration with the Lawrence Livermore Laboratory of the Department of Energy. Sequence fragments were analyzed at Yale University by dividing larger contigs into 10-kb increments. These were “masked” for repetitive sequence by using the repeatmasker software package and were then examined for expressed sequences by using the blast search engine at the National Center for Biotechnology Information (NCBI, www.ncbi.nlm.nih.gov/blast/html/blastcgihelp.html#nucleotide_databases). Human, mouse and other EST databases were queried.
A sequence contig of the full 194-kb BAC clone (NCBI; accession no. AC096865) was generated by using sequencher (Gene Codes, Ann Arbor, MI) software, masked for repetitive elements, and evaluated by using the GENSCAN (http://genes.mit.edu/GENSCAN.html), FGENES, and GRAIL gene prediction algorithms. Translations of putative genes were evaluated by protein–protein homology searches by using the blast program and nonredundant and SWISS-PROT databases available at the NCBI.
Expression of putative coding regions was assessed first by attempting amplification using PCR with various cDNAs as template including Human Fetal, Adult and Immune System MultiTissue cDNA panels (CLONTECH). For predicted multiexon transcripts or spliced ESTs, attempts were made to amplify across each intron boundary. Predicted genes were evaluated by PCR by using primers from every combination of putative exons within a single predicted transcript.
Additional evaluation of putative coding regions was undertaken using RT-PCR. Tissue sources included adult human brain-derived neuronal stem cells, adult peripheral lymphocytes, SKN-B5 neuroblastoma, U118 glioblastoma multiform, donor patient glioma, American Type Culture Collection (ATCC) CCL-2 HeLa, ATCC CRL-2159 Primitive Neuroectodermal Tumor, ATCC HTB-169 human retinoblastoma, ATCC CCL-105 human small-cell carcinoma, donor patient human aorta, bovine aortic endothelial cells, human vascular endothelial cells (Cambrex, East Rutherford, NJ), human microvascular endothelial cells (Cambrex), human umbilical vein smooth muscle cells (Cambrex), human umbilical vein endothelial cells (Cambrex), and donor human T lymphocytes. RNA expression assays were performed on whole cellular RNA extracted from cell cultures according to the Trizol reagent manufacturer's protocol (GIBCO/BRL). First-strand cDNA was generated with either 1 μl of Oligo-dT20 primer (50 μM) or 1 μl of a 10 μM forward or reverse complement gene-specific primer. PCR was then performed on 2 μl of the RT reaction volume by using gene-specific primers from each exon and between all exons of the putative transcript.
PCR primers from the 5′ region of EST xb65d06.x1 (NCBI accession no. AWO82319) were provided to Incyte Genomics (Palo Alto, CA) for a human superpool library screen that evaluated 11 oligo-dT primed libraries from multiple human tissues including adult colon, heart, kidney, lung, liver, pancreas, prostate, spleen, and bone marrow as well as fetal brain and infant thymus.
Heteroduplex Analysis.
PCR primers were designed to parse the coding regions of cytokine inducible SH-2 containing protein 4 (CIS4) and Gilles de la Tourette syndrome chromosomal region 1 (GTSCR-1) into amplicons ranging in size from 200 to 350 bp. Amplified fragments were analyzed on a Transgenomic (Omaha, NE) WAVE DHPLC instrument per the manufacturer's recommendations by using the integrated software package to determine optimal column temperatures and solvent gradients for each amplicon. Fragments yielding chromatograms that deviated from the homozygous waveform were sequenced on both strands and evaluated for sequence variation by using sequencher software.
Replication Timing Studies.
FISH was carried out by using BAC clones as noted above with the following modifications: (i) Cell cultures were pulsed with BrdUrd (10 μM) for 90 min before harvesting to allow for incorporation into newly synthesizing DNA. (ii) Three-color fluorescence imaging was performed: one BAC probe was labeled by using dUTP-11-digoxigenin, and a second was direct-labeled with dUTP-DEAC. The digoxigenin-labeled probe was detected with antidigoxigenin rhodamine (1:150 dilution), and the pulsed S phase cells were detected with a 1:10 dilution of anti-BrdUrd-FITC (Pharmingen). (iii) Interphase nuclei were examined for the number of signals present at each locus. For all experiments, only those interphase nuclei showing BrdUrd incorporation as a marker for S phase were counted. (iv) Two colocalizing BAC probes were used in each experiment to assist in identifying signals that were representative of true hybridization as well as to distinguish the normal versus inverted chromosome. (v) Each slide was counted independently by two individuals in the lab. Whenever possible, the identity of the probe and/or the source of the cell material in a given experiment were not known to the raters. Certain experiments, for example those involving probes spanning the inversion breakpoint, precluded blinding. (vi) Several slides containing cells from a single control were hybridized at different times with the identical probe to ensure the reproducibility of hybridizations and counting procedures. In addition, a statistical analysis of inter-rater reliability was undertaken by using a simple κ test. The analysis of singlet–doublet (SD) signal percentages from the replication timing experiments was undertaken by using a χ2 test with 1° of freedom. All reported significance levels are two-tailed.
Results
Molecular Mapping of Breakpoints in an 18q21-22 Paracentric Inversion.
Mapping of the inversion was undertaken before the availability of the relevant draft sequence from the human genome project. Databases at NCBI were used to identify sequence-tagged sites (STSs) mapping to the 18q21 and 18q22–23 regions and YAC clones corresponding to the STSs and mapping to the region of interest were identified via database searches at the Whitehead Institute (www.genome.wi.mit.edu). These clones were used in FISH hybridizations to lymphocytes from the affected individual. YAC clone CEPH-B 846A2 was found to span the 18q22 breakpoint, and clone CEPH-B 804 B10 was found to span the 18q21.1 breakpoint (data not shown).
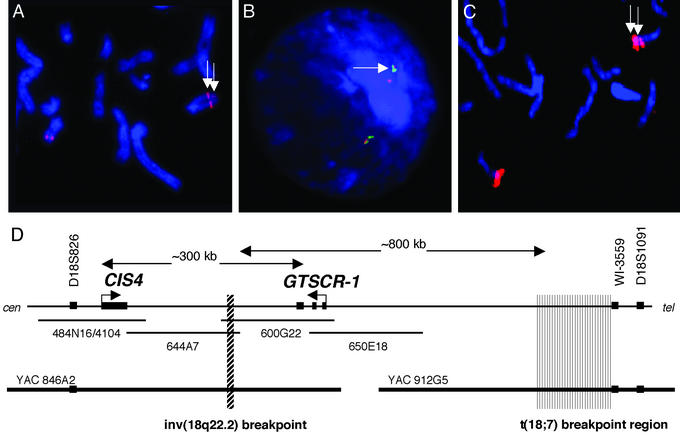
The 18q22 aspect of the inversion breakpoint was pursued intensively due to its close proximity to the 18q22 breakpoint mapped in a previously reported family with GTS, OCD, and CT (14). Once spanning YACs were identified, corresponding BAC clones from an RPCI-11 library were chosen by screening with radio-labeled EST and STS markers. Clone walking was then undertaken by using thermal asymmetric interlaced (TAIL) PCR (21) to identify end-sequences from the mapped BAC clones. Nonrepetitive end sequences were radio-labeled and used for successive library screenings. The overlapping BAC clones RPCI-11 600G22 (NCBI accession no. AC096865.2) and RPCI-11 644A7 (NCBI accession no. AC091305) were both found by FISH to span the 18q22 inversion breakpoint (see Fig. 1).
Figure 1.
FISH mapping of the telomeric aspect of a inv(18q21;q22) paracentric inversion. (A) Metaphase FISH using BAC 644A7 as a probe. The signal is split on the chromosome bearing the inversion (white arrows). The majority of the hybridization is seen in a centromeric position, whereas a small portion of the BAC probe remaining in the normal telomeric position is visible on metaphase spreads. (B) A control probe (green), 650E18, that maps telomeric to the breakpoint shows colocalization with the telomeric portion of probe 644A7. (C) FISH results from BAC 600G22 that also spans the inversion breakpoint. The majority of the hybridization signal remains in the telomeric position and the smaller signal is seen on the centromeric side of the inversion (white arrows) (D) The results of FISH mapping of both YAC and BAC clones in the region around the 18q22 inversion breakpoint. BACs are represented by thin horizontal lines, and YAC clones are represented by heavy black lines at the bottom of the diagram. The approximate position of the 18q22 inversion breakpoint is noted in the region of overlap between BAC clones 644A7 and 600G22. The approximate position of the previously noted t(18, 7) translocation breakpoint (14) is marked by the gray box on the right of the diagram. The breakpoint was noted to be centromeric to the flanking marker WI-3559. Two transcripts were identified in the vicinity of the 18q22 breakpoint. CIS4 was mapped to BAC 4104, and GTSCR-1 was mapped to BAC clones 600G22 and 650E18. Approximate distances separating the transcripts and the two rearrangement breakpoints are noted in the diagram and were derived from the NCBI database.
BAC clone 600G22 was fully sequenced. TAIL PCR was used to identify the end sequence from the overlapping BAC 644A7. Sequence comparisons demonstrated that this region of overlap between BACs was 47 kb. Given that both clones spanned the inversion, the 18q22 breakpoint was narrowed to this interval. The Bhogosian-Sell et al. (14) breakpoint mapped to YAC 964F4. Thus, the two independently identified 18q22 rearrangements in patients with GTS phenotypes localized to the interval flanked on the centromeric side by marker D18S826 and on the telomeric aspect by marker WI-3559.
Identification of Coding Sequences in the Vicinity of the 18q22 Breakpoint.
The regions surrounding the inversion breakpoint including that between the 18q22 inversion and the 18q22 translocation (14) breakpoints were evaluated for coding sequence by using blast EST homology searches, comparison with mouse sequence, and GENSCAN, GRAIL, and FGENEs prediction algorithms.
A single EST xb65d06.x1 (NCBI accession no. AW082319.1) was identified on BAC 600G22, mapping ≈100 kb telomeric to the inversion. The source of the EST was the Soares_NFL_T_GBC_S1 library derived from a mixture of three normalized cDNA libraries: fetal lung, testis, and B cell. The clone contained two intron–exon splice sites, and we subsequently identified it in a human fetal thymus library (Incyte Genomics) and named it GTSCR-1. Sequencing of this clone and comparison to the genomic sequence in the region demonstrated a putative initiator methionine with an in-frame stop codon 5′ to this ATG, as well as canonical splice sites (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). The gene encodes a putative product of 138 aa in three exons; the protein shows no significant homology to other proteins, ESTs, or protein motifs.
Two GENSCAN gene predictions were identified on clone 600G22 (data not shown). No coding sequence could be confirmed by using PCR reactions against multitissue cDNA libraries, Northern blotting of predicted exons, or RT-PCR.
FISH analyses confirmed the positions of BAC clones 41O4 (NCBI accession no. AC013558), 644A7, and 650E18 (NCBI accession no. AC037476) near the breakpoint (see Fig. 1). These BACs were also evaluated for coding sequence. Several overlapping ESTs were mapped to clone 4104, ≈150 kb centromeric to the inversion breakpoint. A contig of ESTs was constructed by using blast searches and evaluated by alignment to the genomic DNA sequence. The translation of the longest ORF for this EST contig was found to be identical to the known gene CIS4 [Online Mendelian Inheritance in Man (OMIM) 605118; NCBI accession no.: NM_004232.1] (22). These results were subsequently confirmed by mapping data derived from the human genome project.
Mutation Screening.
DHPLC evaluation of coding regions and intron–exon splice junctions for the transcripts CIS4 and GTSCR-1 revealed no missense or nonsense mutations in a group of 96 patients with GTS, OCD, and CT (Tables 1 and 2, which are published as supporting information on the PNAS web site).
Assessment of Replication Synchrony.
FISH analyses of interphase nuclei were used to assess replication synchrony. Loci that are replicating synchronously show either a single hybridization on each homologue, a singlet–singlet pattern, or two hybridization signals on each homolog a doublet–doublet pattern, depending on whether the locus is prior to or post replication. Loci at which one homolog has replicated and the other has not are visualized as a SD pattern (Fig. 2).
Figure 2.
Replicating timing assay using FISH probes on interphase nuclei. Singlet–singlet (SS), doublet–doublet (DD), and SD patterns are visualized on interphase nuclei, respectively.
We first examined the degree of replication asynchrony in the patient by using a BAC probe (600G22) that spanned the inversion breakpoint and found that 45% of 129 cells showed the SD pattern (Fig. 3A). We compared this to the frequency of SD signals found by using the same BAC probe in a cytogenetically normal, unaffected control. Twenty-seven percent of 100 S phase cells showed asynchronous replication. The difference was statistically significant (χ2 = 7.04; P < 0.01). We further evaluated the status of replication timing in the patient by hybridizing BACs from genomic regions known either to be imprinted or to express from both chromosomes. A probe from the SNRPN-SNURF locus that normally replicates asynchronously showed a significantly higher rate of SD signals (43/100 cells) than did a probe from the CFTR region that is known to replicate synchronously (24/100 cells) (χ2 = 7.27; P < 0.01) when hybridized to patient cells (Fig. 3A). Additional analysis showed that the rate of asynchrony found at the 18q22 breakpoint in the patient was significantly higher than for the biallelic CFTR locus in the patient cells (χ2 = 9.88, P < 0.01) (Fig. 3A).
Figure 3.
(A) Percentages for SD signals are shown for four conditions. Lane 1, a BAC probe containing the gene SNURF-SNRPN known to replicate asynchronously is hybridized to patient cells. Lane 2, BAC probe 600G22 spanning the patient's inversion breakpoint is hybridized to patient cells. Lane 3, BAC 600G22 is hybridized to cytogenetically normal control cells. Lane 4, a BAC probe mapping to the CFTR gene known to replicate synchronously is hybridized to patient cells. Inter-rater reliability was calculated by using the κ statistic and showed excellent agreement (k = 0.9583). A χ2 test with 1° of freedom was carried out comparing the 18q probe hybridized to the patient cell line versus the same probe hybridized to control cells (lane 2 vs. 3). The difference was significant (†, P < 0.01). A difference between the SD percentages at the 18q breakpoint locus compared with the CFTR locus in the patient's cells (lane 2 vs. 4) was also shown to be statistically significant (*, P < 0.005). A comparison of the Prader–Willi syndrome locus and the 18q22 breakpoint locus in the patient (lane 1 vs. 2) was indistinguishable (#, P > 0.5). (B) Percentages of singlet signals on the normal versus inverted chromosome 18. BAC 600G22 was hybridized to patient cells. Eighty-one percent of the SD signals (47/58) were the result of the singlet on the inverted (inv) chromosome (lane 1), whereas only 19% of singlets were found on the normal chromosome (lane 2). The difference was significant (*, P < 0.0001).
The patient's two chromosomes 18 were distinguishable in interphase nuclei via the use of FISH probes lying very close to, but on opposite sides of, the inversion breakpoint. This probe selection results in the colocalization of hybridization signals on the normal chromosome, whereas the abnormal chromosome is identified by the marked separation of hybridization signals. An analysis of the BAC probe spanning the inversion breakpoint showed that 81% of cells found to replicate asynchronously in the patient had the singlet (representative of later replication) on the chromosome bearing the inversion compared with 19% on the normal homolog (P < 0.0001) (Fig. 3B).
The frequency of SD signals was also evaluated for BACs RPCI-11 644A7 and RPCI-11 650E18, which overlap RPCI 11 600G22 on the centromeric and telomeric flanks respectively. These clones were hybridized to interphase nuclei from the patient as well as to a cytogenetically normal control cell line. In each instance, the degree of replication asynchrony in the patient was found to be significantly higher than for the control cells (χ2 = 3.91, P < 0.05 and χ2 = 6.86, P < 0.01, respectively), defining at least a 500-kb region of abnormal replication due to this chromosomal rearrangement (Fig. 4).
Figure 4.
Replication timing in the region surrounding the 18q22 breakpoint. The frequency of SD signals for three BACs in the region surrounding the 18q22.2 inversion breakpoint is shown. The top line represents data from the patient cells. The bottom line shows data from controls. The total number of interphase nuclei counted for each slide was as follows: BAC 644A7, patient = 60, control = 60; BAC 600G22, patient 129, control = 88; and BAC 650E18, patient = 60, control = 60. All differences between control and experimental conditions were significant and are noted in the respective positions on the graph.
Discussion
A patient with a chromosome 18 inversion was identified with a GTS phenotype involving CT and OCD. The fine mapping of this rearrangement indicated that the 18q22 breakpoint is within 1 Mb of a previously reported translocation that segregated with GTS, OCD, and CT in the pedigree (14). A comprehensive assessment of the affected genomic interval did not reveal a structurally disrupted gene.
Studies of replication timing were undertaken to determine whether the chromosomal inversion might have altered the epigenetic properties of the region, potentially leading to a functional haploinsufficiency of one or more genes. A marked degree of asynchrony was identified at the inversion breakpoint in the patient that extended beyond the immediate abnormality to an interval of several hundred thousand base pairs. Control experiments demonstrated no evidence of preexisting asynchrony at the 18q22 locus in normal individuals. Moreover, the anticipated patterns of replication were found in the patient's cells at loci expected to demonstrate either asynchronous or synchronous timing (SNRPN-SNURF and CTFR, respectively). Of particular note, the inverted chromosome was found to be relatively delayed in replication timing compared with the normal homolog, a finding consistent with decreased or absent gene expression on the rearranged chromosome.
A variety of investigations have confirmed the association between the replication timing of a locus and its transcriptional activity (23–25). Expressed loci have been found to replicate early in S phase, whereas nonexpressing loci replicate late in S phase. Asynchronous replication has been identified in instances in which one homologue shows decreased or silent gene expression, such as in the case of genomic imprinting (26, 27), X-chromosome inactivation (28) olfactory receptor expression (29) allelic exclusion involving B cell antigen receptor loci (30) and epigenetic silencing of CD4 in developing thymocytes (31).
In the current case, the identification of asynchrony resulting from a relative delay in replication of the patient's rearranged chromosome 18 suggests that decreased or absent expression of one or more genes in the 18q22 interval on the affected homologue may be responsible for the phenotype observed in the cases noted. Given our negative findings on heteroduplex analysis of 96 patients, it is not likely that mutations in either CIS4 or GTSCR-1 are a common cause of GTS. Nonetheless, by mapping and comparing the extent of epigenetic modifications in cases with rearrangements in this region, a candidate interval could be defined in which to narrow the search for a gene or genes responsible for a subset of patients with related phenomena. Given the high likelihood of locus heterogeneity in GTS, the identification of a disease-related gene or genes, even a small group of individuals, could provide important clues into the genetic and physiological mechanisms that underlie the disorder.
The use of replication timing as an assay for epigenetic phenomena has important advantages in studying chromosomal abnormalities. The method allows for investigation of large regions of the genome when compared with alternatives such as methylation-sensitive restriction enzymes or bisulfite sequencing. The ability to readily screen regions of interest in 100- to 200-kb segments is particularly valuable in the identification and mapping of long-range position effects when the potentially relevant transcripts are not yet known. In addition, the approach offers a useful assay in peripheral lymphocytes, regardless of whether genes of interest are expressed in blood. Although the absolute timing of replication is determined in part by whether a gene is expressed in a particular cell type, the synchrony of replication may be evaluated independent of tissue-specific expression. This allows for the investigation of epigenetic phenomena involving genes that may be expressed in inaccessible tissue, an asset that my turn out to be particularly valuable in the study of neuropsychiatric disorders.
The identification of an 18q22 rearrangement in a patient with a GTS spectrum phenotype and the proximity of two well characterized rearrangements in this genomic interval suggest that the region is a good candidate for containing a gene or genes responsible for the observed phenotype and deserves continued scrutiny. The demonstration of long-range epigenetic dysregulation of this region provides a model for how rearrangement located ≈1 Mb apart could be causing similar phenotypes without physically disrupting a single gene. The possibility that epigenetic changes resulting from a chromosomal abnormality can lead to a GTS phenotype suggests a novel mechanism for neuropsychiatric pathogenesis as a result of a balanced autosomal rearrangement and, accordingly, may have relevance for the study of other chromosomal abnormalities.
Supplementary Material
Acknowledgments
This publication is dedicated to the memory of Donald J. Cohen, without whom this work would not have been possible. We thank Sherman Weissman for many thoughtful suggestions and Xiangyang Lu, Gretchen Graff, Laszlo Furu, Jonas Lai, Tyler Briggs, Tobin Abraham, Manjunath Nimmakayalu, Allyson Lachowicz, Tim Andries, and Astrid Terry for generous technical assistance. Finally, we express our deepest gratitude to the patients and families who participated in the studies described above. This work was funded by a Bovenizer Research Fellowship (to M.W.S.) and grants from the Smart Family Foundation (to M.W.S.), the National Institutes of Health (RR16118 and NS43520 to M.W.S., MH18268 to M.W.S. and J.F.L., MH49351 to J.F.L. and D.L.P., DK02467 and DK56786 to J.M.G., and HD35947 to D.C.W.), and the Howard Hughes Medical Institute Medical Student Research Fellowship (to A.C.).
Abbreviations
- BAC
bacterial artificial chromosome
- YAC
yeast artificial chromosome
- OCD
Obsessive Compulsive Disorder
- GTS
Gilles de la Tourette syndrome
- CT
chronic tics
- NCBI
National Center for Biotechnology Information
- STSs
sequence-tagged sites
- GTSCR-1
Gilles de la Tourette syndrome chromosomal region 1
- CIS4
cytokine inducible SH-2 containing protein 4
- SD
singlet-doublet
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY262164).
Electronic database information: Repeat Masker, http://repeatmasker.genome.washington.edu; Baylor College of Medicine Search Launcher, http://searchlauncher.bcm.tmc.edu/seq-search/gene-search.html; BLOCKS, http://searchlauncher.bcm.tmc.edu/seq-search/gene-search.html; Cognator, http://searchlauncher.bcm.tmc.edu/seq-search/gene-search.html; PipMaker, http://bio.cse.psu.edu/pipmaker; UniSTS, www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unists; ORF-Finder, www.ncbi.nlm.nih.gov/gorf/gorf.html.
References
- 1.Apter A, Pauls D L, Bleich A, Zohar A H, Kron S, Ratzoni G, Dycian A, Kotler M, Weizman A, Cohen D J. Adv Neurol. 1992;58:61–65. [PubMed] [Google Scholar]
- 2.Apter A, Pauls D L, Bleich A, Zohar A H, Kron S, Ratzoni G, Dycian A, Kotler M, Weizman A, Gadot N, et al. Arch Gen Psychiatry. 1993;50:734–738. doi: 10.1001/archpsyc.1993.01820210068008. [DOI] [PubMed] [Google Scholar]
- 3.Pauls D L, Alsobrook J P, Gelernter J, Leckman J F. In: Tourette's Syndrome: Tics, Obsessions, Compulsions. Leckman J F, Cohen D J, editors. New York: Wiley; 1999. pp. 194–212. [Google Scholar]
- 4.Pauls D L, Raymond C L, Stevenson J M, Leckman J F. Am J Hum Genet. 1991;48:154–163. [PMC free article] [PubMed] [Google Scholar]
- 5.Hyde T M, Aaronson B A, Randolph C, Rickler K C, Weinberger D R. Neurology. 1992;42:652–658. doi: 10.1212/wnl.42.3.652. [DOI] [PubMed] [Google Scholar]
- 6.Price R A, Kidd K K, Cohen D J, Pauls D L, Leckman J F. Arch Gen Psychiatry. 1985;42:815–820. doi: 10.1001/archpsyc.1985.01790310077011. [DOI] [PubMed] [Google Scholar]
- 7.Walkup J T, Leckman J F, Price R A, Hardin M, Ort S I, Cohen D J. Psychopharmacol Bull. 1988;24:375–379. [PubMed] [Google Scholar]
- 8.Pauls D L, Leckman J F. N Engl J Med. 1986;315:993–997. doi: 10.1056/NEJM198610163151604. [DOI] [PubMed] [Google Scholar]
- 9.Curtis D, Robertson M M, Gurling H M. Br J Psychiatry. 1992;160:845–849. doi: 10.1192/bjp.160.6.845. [DOI] [PubMed] [Google Scholar]
- 10.Price R A, Pauls D L, Kruger S D, Caine E D. Psychiatry Res. 1988;24:251–261. doi: 10.1016/0165-1781(88)90107-2. [DOI] [PubMed] [Google Scholar]
- 11.Baron M, Shapiro E, Shapiro A, Rainer J D. Am J Hum Genet. 1981;33:767–775. [PMC free article] [PubMed] [Google Scholar]
- 12.Walkup J T, LaBuda M C, Singer H S, Brown J, Riddle M A, Hurko O. Am J Hum Genet. 1996;59:684–693. [PMC free article] [PubMed] [Google Scholar]
- 13.Seuchter S A, Hebebrand J, Klug B, Knapp M, Lehmkuhl G, Poustka F, Schmidt M, Remschmidt H, Baur M P. Genet Epidemiol. 2000;18:33–47. doi: 10.1002/(SICI)1098-2272(200001)18:1<33::AID-GEPI3>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 14.Boghosian-Sell L, Comings D E, Overhauser J. Am J Hum Genet. 1996;59:999–1005. [PMC free article] [PubMed] [Google Scholar]
- 15.Matsumoto N, David D E, Johnson E W, Konecki D, Burmester J K, Ledbetter D H, Weber J L. Eur J Hum Genet. 2000;8:875–883. doi: 10.1038/sj.ejhg.5200549. [DOI] [PubMed] [Google Scholar]
- 16.Petek E, Windpassinger C, Vincent J B, Cheung J, Boright A P, Scherer S W, Kroisel P M, Wagner K. Am J Hum Genet. 2001;68:848–858. doi: 10.1086/319523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donnai Lancet. 1987;14:627. (lett.). [Google Scholar]
- 18.Kleinjan D J, van Heyningen V. Hum Mol Genet. 1998;7:1611–1618. doi: 10.1093/hmg/7.10.1611. [DOI] [PubMed] [Google Scholar]
- 19.Goodman W K, Price L H, Rasmussen S A, Delgado P L, Heninger G R, Charney D S. Arch Gen Psychiatry. 1989;46:36–44. doi: 10.1001/archpsyc.1989.01810010038006. [DOI] [PubMed] [Google Scholar]
- 20.Lichter P, Tang C J, Call K, Hermanson G, Evans G A, Housman D, Ward D C. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y G, Whittier R F. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- 22.Masuhara M, Sakamoto H, Matsumoto A, Suzuki R, Yasukawa H, Mitsui K, Wakioka T, Tanimura S, Sasaki A, Misawa H, et al. Biochem Biophys Res Commun. 1997;239:439–446. doi: 10.1006/bbrc.1997.7484. [DOI] [PubMed] [Google Scholar]
- 23.Karube T, Watanabe S. Cancer Res. 1988;48:219–222. [PubMed] [Google Scholar]
- 24.Hansen R S, Canfield T K, Lamb M M, Gartler S M, Laird C D. Cell. 1993;73:1403–1409. doi: 10.1016/0092-8674(93)90365-w. [DOI] [PubMed] [Google Scholar]
- 25.Gunaratne P H, Nakao M, Ledbetter D H, Sutcliffe J S, Chinault A C. Genes Dev. 1995;9:808–820. doi: 10.1101/gad.9.7.808. [DOI] [PubMed] [Google Scholar]
- 26.Kitsberg D, Selig S, Brandeis M, Simon I, Keshet I, Driscoll D J, Nicholls R D, Cedar H. Nature. 1993;364:459–463. doi: 10.1038/364459a0. [DOI] [PubMed] [Google Scholar]
- 27.Greally J M, Starr D J, Hwang S, Song L, Jaarola M, Zemel S. Hum Mol Genet. 1998;7:91–95. doi: 10.1093/hmg/7.1.91. [DOI] [PubMed] [Google Scholar]
- 28.White W M, Willard H F, Van Dyke D L, Wolff D J. Am J Hum Genet. 1998;63:20–28. doi: 10.1086/301922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chess A, Simon I, Cedar H, Axel R. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 30.Mostoslavsky R, Singh N, Tenzen T, Goldmit M, Gabay C, Elizur S, Qi P, Reubinoff B E, Chess A, Cedar H, Bergman Y. Nature. 2001;414:221–225. doi: 10.1038/35102606. [DOI] [PubMed] [Google Scholar]
- 31.Zou Y R, Sunshine M J, Taniuchi I, Hatam F, Killeen N, Littman D R. Nat Genet. 2001;29:332–336. doi: 10.1038/ng750. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.