Abstract
When animal cells are exposed to stressful conditions, the tumor suppressor protein p53 restrains growth by promoting an arrested cell cycle or initiating a cell death program. How these distinct fates are specified through the action of a single protein is not known. To study its functions in vivo we produced a targeted mutation at the Drosophila p53 (Dmp53) locus. We show that Dmp53 is required for damage-induced apoptosis but not for cell-cycle arrest. Dmp53 function is also required for damage-induced transcription of two tightly linked cell death activators, reaper and sickle. When challenged by ionizing radiation, Dmp53 mutants exhibit radiosensitivity and genomic instability. Hence, elevated mutant loads were not caused by defective checkpoint functions but instead correlated with failures in p53-associated cell death. Our studies support the notion that core ancestral functions of the p53 gene family are intimately coupled to cell death as an adaptive response to maintain genomic stability.
The p53 tumor suppressor limits oncogenesis through activities that govern adaptive responses to stress. As such, p53 is thought to function as a “guardian of the genome” that becomes mutated or altered in most human cancers (1, 2). When cells are stressed by exposure to genotoxic agents, radiation, hypoxia, or inappropriate oncogene activation (1–3), p53 restrains growth through activities that reversibly arrest the cell cycle or promote apoptosis (4, 5), but how these alternative fates become specified through the action of this tumor suppressor is not understood. The primary mechanism of p53 action clearly involves transcriptional regulation (6–8), although other activities might contribute to accessory functions (9). The protein requires at least three functional domains to regulate downstream target genes but many mutations commonly found in human cancers map to the DNA-binding domain (10–13). A major effort in cancer biology is focused toward understanding downstream effectors of p53 function. The p53 target gene, p21, acts directly as a cyclin-dependent kinase inhibitor (14–16) and appears largely responsible for p53-dependent G1/S arrest. The in vivo downstream targets responsible for p53-mediated apoptosis, however, are less well defined. Important candidates for in vivo death effectors include members of the bcl2 family, bax (17), noxa (18), puma (19, 20), and the death receptor fas (21, 22). In many cases, regulated expression of these genes is context-specific and whether there exists a generic p53 apoptosis program for all cells is not known. Genome-wide searches for p53-responsive genes have also been reported, but it is not yet clear whether these loci represent direct or indirect targets (23, 24).
A Drosophila homolog of p53, Dmp53, was recently identified (25–27). Like mammalian counterparts (12, 13, 28–31), Dmp53 has a well conserved DNA-binding domain with a transcriptional activation domain at its N terminus, and an oligomerization domain at its C terminus. In previous studies, forced expression of wild-type and variant Dmp53 transgenes was used to establish that ectopic Dmp53 expression triggers apoptotic cell death and possibly affects the duration of M phase (27). Additional experiments with Dmp53 variants showed that radiation-induced apoptosis in the wing disc was suppressed by dominant negative transgenes. However, expression of these same transgenes had no effect on the damage induced cell-cycle-arrest response, even though many cell-cycle regulators, including p21, are conserved in the fly genome and expression of fly p21 alone can trigger G1 arrest (32). These observations supported a role for Dmp53 in apoptosis and, at the same time, suggest that Dmp53 may not engage checkpoint controls through the cell cycle like its mammalian counterparts.
In Drosophila, programmed cell death is governed by apoptosis activators mapping to a genomic interval referred to as the Reaper region (33). Several lines of evidence implicate one gene in this region, reaper (rpr), as a direct target of, and effector for, Dmp53 action in vivo. First, rpr transcription is acutely sensitive to damage signals, becoming induced within 90 min of γ-irradiation treatment (34). Second, this inductive response maps to a 20-bp radiation-responsive element that binds Dmp53 and resembles the consensus developed for mammalian counterparts (25). Third, recent loss-of-function analyses uncovered a partial requirement for rpr in a model of x-ray-induced cell death (35).
To directly examine the functions of Dmp53 and aid in the identification of direct in vivo targets, we isolated a mutation at the Dmp53 locus. Because there are no available P elements that map near the Dmp53 region, we adopted a gene-targeting method recently developed by Rong and Golic (36, 37). This method directs mutations at the desired locus through homologous recombination events that produce aberrant duplicates of the targeted gene. Although viable, Dmp53 mutants exhibit pronounced defects in damage-induced apoptosis. Radiation-induced expression of two proapoptotic genes, rpr and sickle (skl), and a p53-responsive reporter transgene failed to regulate normally in these mutants. However, under these same irradiation conditions, the damage-induced checkpoint response occurred normally. Dmp53 mutants are radiosensitive and also exhibit a pronounced mutator phenotype, manifested as high mutagenic loads in loss-of-heterozygosity (LOH) assays. Together, these results uncover important genomic consequences resulting from failures in p53-mediated cell death and establish the principle that preservation of genomic stability by p53 proteins can derive entirely from apoptotic determinants rather than cell-cycle checkpoint functions.
Materials and Methods
Materials and methods used for plasmid constructions, molecular verification of targeting, crosses and screen for targeting, oligonucleotide array analysis, and radiation sensitivity assay can be found in Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Crosses and Screens for Targeting.
Flies carrying the Dmp53 donor element on the second chromosome were crossed to flies carrying both 70FLP and 70I-SceI transgenes on the second chromosome to yield Dmp53 donor/70FLP, 70I-SceI;+/+ progeny. Induction of recombinations in the germ line as well as screens for targeting events were carried as described (36) and also can be found in Supporting Materials and Methods.
Histochemical Staining of rpr-lacZ Transgenic Embryos.
Transgenic flies containing the150-bp lacZ reporter construct (25) were crossed to Dmp53−ns mutant flies to generate 150-bp lacZ;p53−/− flies. Collection of 2- to 5-h embryos of the appropriate genotype and histochemical staining of those were carried out as described (25).
Cell Death Assay and Checkpoint Function Assay.
Cell death assay was carried out as described (25). Checkpoint function assays were done 90 min after γ-irradiation as described, with minor modifications described in Supporting Materials and Methods (27).
LOH Assay.
Three independent recombinant lines homozygous for Dmp53−ns and heterozygous for multiple wing hair (mwh) were generated. Each recombinant line was mock-treated or γ-irradiated with 250 rads during a third-instar larval stage. Wings were dissected and mounted in 1:1 methyl salicylate/Canada balsam (Sigma). Cells only with three or more hairs were scored as a mwh−/− phenotype.
Results
Targeted Mutation at the Dmp53 Locus.
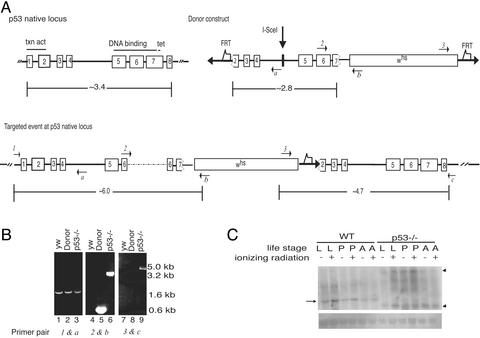
To obtain a Dmp53 mutant, we used a gene-targeting method developed by Rong and Golic for “ends-in” homologous recombination (36, 37). Like its mammalian counterpart, Dmp53 protein contains a putative transcriptional activation domain at the N terminus, a DNA-binding domain, and an oligomerization domain at the C terminus. A donor construct using an internal 2.8-kb Dmp53 fragment as a donor template from the target-gene sequence was prepared (Fig. 1A). This internal 2.8-kb fragment in the donor construct lacks two-thirds of the transcriptional activation domain, the entire oligomerization domain, and part of the DNA-binding domain (25–27). Note also that five of eight residues predicted to contact DNA (25–27) are absent in this donor template. Hence, an insertion of the 2.8-kb internal fragment at the Dmp53 native locus is predicted to generate loss-of-function mutations at this locus.
Figure 1.
Generation and verification of a targeted Dmp53 mutation. (A) Targeting scheme for the Dmp53 gene. The donor construct illustrated next to a schematic of the native Dmp53 locus was generated by the insertion of a 2.8-kb internal fragment of the Dmp53 gene into the targeting vector pTV2. The 2.8-kb fragment lacks part of an N-terminal transactivation domain (because of the elimination of the entire exon 1 and part of exon 2), and part of a central DNA-binding domain and an entire tetramerization domain (because of the elimination of part of exon 7 and the entire exon 8). A germ-line transformant donor strain bearing this construct on the second chromosome was used for targeted mutagenesis as described (36, 37). FRT, FLP recombination target. On heat-shock-mediated induction of FLP site-specific recombinase and I-SceI endonuclease, the donor construct is excised as an extrachromosomal molecule and may create a lesion at the native Dmp53 locus by homologous recombination. The targeted mutation at the native Dmp53 locus, referred to as Dmp53−ns, produced a tandem duplication of Dmp53 variants that sandwich the whs gene. The dotted line in exon 6 shown for the targeted event at the Dmp53 native locus indicates an unexpected insertion of partial pTV2 vector sequences (confirmed by PCR and sequencing). (B) PCR analysis of a Dmp53-targeting event. Genomic DNA from flies was used as template DNA for PCR to verify the targeting event at the Dmp53 locus. Lanes 1–3 use a primer pair, 1 and a. Lanes 4–6 use a primer pair, 2 and b, to verify the disruption of the N-terminal Dmp53 template. Lanes 7–9 use a primer pair, 3 and c, to assess the disruption at the C-terminal Dmp53 template. All PCR fragments were partially or fully sequenced to determine the variations introduced at the native Dmp53 locus. Locations of each primer are listed in A in italics. Genotype of flies is as follows: yw refers to the parental wild-type strain at the native Dmp53 site on the third chromosome; Donor refers to the Dmp53-donor construct on the second chromosome, wild-type at the native Dmp53 site on the third chromosome; and p53−/− refers to the disrupted native Dmp53 site on the third chromosome. (C) Northern blot analysis of a Dmp53-targeting event. Total RNA from wild-type (yw) and Dmp53−ns strains was isolated, blotted to a membrane, and hybridized with a Dmp53 probe. The arrow indicates the expected 1.6-kb Dmp53 transcript. In wild-type larval and pupae individuals, this transcript is moderately responsive to ionizing radiation. Dmp53 mutants lack the1.6-kb Dmp53 transcript, and instead express aberrantly sized transcripts, which are indicated by arrowheads. (Lower) The membrane stained with methylene blue to show ribosomal RNA as a loading control. RNA from untreated (−) or γ-irradiated (+) individuals in the wandering third-instar larvae (L), pupae (P), and adult (A) stages were analyzed.
We obtained seven individuals who had lost white+ mosaicism from the secondary screen (see Supporting Materials and Methods). Of these, we validated one line, Dmp53−ns, as a targeted mutation. PCR-based assays (Fig. 1B), with several primers specific to the Dmp53 genomic region as well as primers specific to the marker gene whs, confirm this interpretation. For example, the primer pair 3 and c gives a product only when there is a targeting event at the Dmp53 locus (Fig. 1 A and B).
Flies disrupted at the Dmp53 locus are viable, fertile, and exhibit no overt visible defects. We used Northern blot analyses to confirm that expression of native Dmp53 transcripts is disrupted in the Dmp53−ns strain. Hybridization with a Dmp53-specific probe to RNAs from three different stages showed a mildly radiation-responsive 1.6-kb Dmp53 transcript in wild-type flies that was absent from Dmp53−ns homozygotes (Fig. 1C). Instead, two aberrantly sized transcripts are evident in the mutant strain, which are probably derived from one or both of the targeted variants.
Dmp53 Is Necessary for Radiation-Induced Transcription of the Reaper Region.
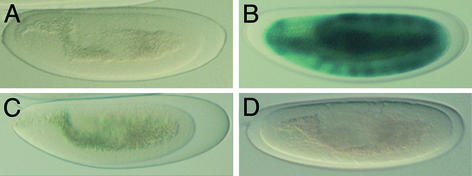
Programmed cell death in Drosophila requires three apoptosis activator genes; rpr, grim, and hid, which map to a 300-kb genomic interval (38–40). Of these, rpr is acutely sensitive to damage signals, becoming transcriptionally activated within a short period after exposure to ionizing radiation (34). The minimal radiation-responsive cis-element lies in a 150-bp fragment, 5 kb upstream of the rpr start codon, and within this radiation-responsive enhancer a Dmp53 binding site was identified (25). To determine whether Dmp53 is required for activation of this radiation-responsive enhancer in vivo, we used a lacZ reporter transgene referred to as rpr 150-bp-lacZ, which contains the minimal 150-bp radiation-responsive fragment. We examined β-galactosidase expression of the150-bp lacZ reporter gene in wild-type and Dmp53−ns mutant embryos with or without irradiation treatment. When mock-treated, neither wild-type nor Dmp53−ns mutant embryos showed β-galactosidase activity (Fig. 2 A and C). After irradiation treatment, wild-type embryos showed robust β-galactosidase activity (Fig. 2B). In contrast, Dmp53−ns mutant embryos showed no induction of β-galactosidase (Fig. 2D). These data show that Dmp53 is necessary for activation of the radiation-responsive enhancer upstream of the rpr locus.
Figure 2.
The radiation-responsive enhancer at the rpr locus is no longer radiation responsive in Dmp53−ns embryos. Transgenic lines carrying a 150-bp radiation-responsive enhancer from the rpr locus was tested in wild-type (A and B) or Dmp53−ns mutant embryos (C and D). Mock-treated controls (A and C) or γ-irradiated (B and D) samples were stained for β-galactosidase activity (25).
Like other genes in the Reaper region, skl encodes an apparent inhibitor of apoptosis protein (IAP) antagonist. This gene maps ≈41 kb proximal to rpr, and, like rpr, is acutely radiation responsive (41). We therefore compared radiation-responsive expression of rpr and skl in embryos from two parental wild-type strains and the Dmp53−ns mutants (Table 1). Microarray profiles of radiation-responsive genes confirmed that both rpr and skl are strongly induced after γ-irradiation (41). However, in the Dmp53−ns mutant strain, neither rpr nor skl was induced after γ-irradiation treatment. These results confirm that rpr is a transcriptional target of Dmp53 and also indicate that skl could be a Dmp53 target gene as well.
Table 1.
Radiation response: Fold-induction values
| Transcript | WTyw | WTw | Dmp53−ns |
|---|---|---|---|
| Reaper | 6.0 (↑) | 5.4 (↑) | No change |
| Sickle | 10.6 (↑) | 6.0 (↑) | No change |
Expression of the indicated transcripts was examined in irradiated and control samples from two wild-type strains (WTyw and WTw) and the Dmp53−ns mutant strain. Total RNAs isolated from staged embryos were analyzed by using the Affymetrix Drosophila GeneChip Array. Fold-induction values relative to basal levels are reported. In agreement with previous studies (41), reaper and sickle are induced from ≈6- to 10-fold, respectively, within 90 min of irradiation exposure. In Dmp53−ns, levels of reaper and sickle transcripts are unchanged.
Dmp53−ns Mutant Blocks Damage-Induced Apoptosis but Not Cell-Cycle Arrest.
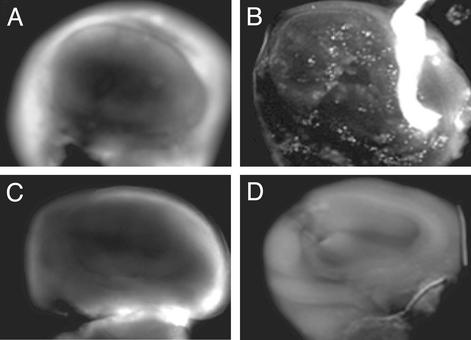
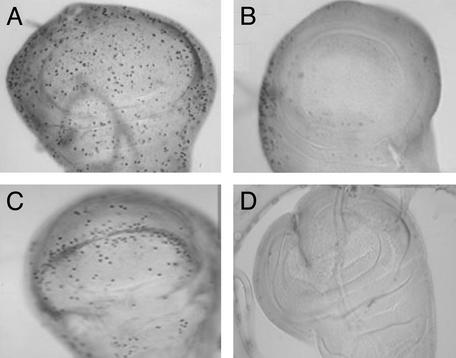
We examined requirements for Dmp53 in vivo in the context of damage-induced signaling. In the wing discs of wandering third instar larvae, apoptotic cells are rarely observed in wild-type flies (Fig. 3A; refs. 25 and 27) or in Dmp53−ns mutants (Fig. 3C). However, after irradiation, wild-type wing discs (and wing discs from Dmp53−ns heterozygotes) show notably increased levels of apoptotic cell death (Fig. 3B). This response was clearly absent from the Dmp53−ns strain because we observed no acridine orange-positive cells in irradiated wing discs from this strain (Fig. 3D). Under the same conditions and in the same tissue, we next tested the requirement for Dmp53 on damage-induced cell-cycle arrest. We used a phosphohistone H3 antibody to examine the presence of cells in mitosis as in previous studies (42, 43). After irradiation, both wild-type and Dmp53−ns mutant discs showed a complete absence of mitotic cells (Fig. 4 B and D) and, hence, without functional Dmp53, normal checkpoint functions were completely unaffected. Therefore, whereas Dmp53 status is an essential determinant of radiation-induced apoptosis, the gene plays no role in governing the cell-cycle-arrest response, at least in the developing wing discs. Together, these results demonstrate a requirement for Dmp53 in damage-induced apoptosis but not in cell-cycle arrest.
Figure 3.
Dmp53−ns mutants are defective in damage-induced apoptosis. A vital dye, acridine orange, specifically recognizes apoptotic cells. Wing discs from wild-type (A and B) or Dmp53−ns (C and D) were dissected from mock-treated (A and C) or γ-irradiation-treated (B and D) larvae, and apoptotic cells were visualized by acridine orange. For each genotype, at least five wing discs were dissected and stained. The Dmp53−ns phenotype was fully penetrant.
Figure 4.
Dmp53−ns mutants exhibit a normal cell-cycle arrest. Wing discs from wild-type (A and B) or Dmp53−ns (C and D) were dissected from mock (A and C) or γ-irradiated (B and D) third-instar wandering larvae. Histochemical staining with phosphohistone H3 antibody detects cells undergoing mitosis. For each genotype, approximately five wing discs were dissected and stained. Discs from wild-type or Dmp53−ns mutant were indistinguishable with respect to the incidence of immunoreactive cells, and staining seen for the Dmp53−ns control wing disc shown is within the normal range. The Dmp53−ns phenotype was fully penetrant.
Dmp53−ns Mutant Is Sensitive to Ionizing Radiation and Shows a Higher Rate of Genomic Instability.
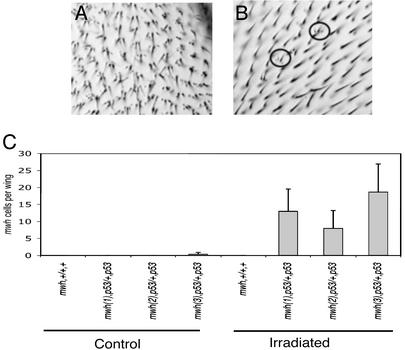
We assessed the Dmp53−ns line for potential mutator phenotypes by using a LOH assay (43, 44). Wandering third-instar larvae heterozygous for the mwh mutation and either wild-type or homozygous for the Dmp53−ns allele were collected and irradiated. LOH for the wild-type mwh allele (arising from chromosomal aberrations or point mutations) is readily assessed by scoring the recessive mwh phenotype in wing cells of adult flies (Fig. 5 A and B). The absence of Dmp53 alone did not significantly affect the basal incidence of mwh cells (Fig. 5C). However, if challenged by low doses of ionizing radiation (250 rads), each independent (mwh, Dmp53/+, and Dmp53) line showed substantially elevated levels of mwh cells under conditions that had no effect in wild-type flies for Dmp53. We also note that Dmp53−ns mutant larvae exhibited a radiosensitive phenotype, manifested as extensive lethality rates that were particularly noticeable at doses of 4,000 rads (Table 2). These results indicate that, in the context of genotoxic damage, Dmp53 functions to preserve organismal viability and genomic stability.
Figure 5.
Dmp53−ns adults carry high mutation loads after exposure to ionizing radiation. LOH is one measure of genomic instability. Here, the recessive mwh phenotype (A) was scored in flies heterozygous for mwh and either wild-type or homozygous for Dmp53−ns. (A) A wing from homozygous mwh fly. Each cell contains multiple hairs. (B) A wing from a fly heterozygous for mwh and homozygous for Dmp53−ns that has been treated with γ-irradiation. Cells within the circles have lost the wild-type copy of mwh, uncovering the recessive mwh phenotype. (C) Loss of Dmp53 leads to genomic instability after irradiation. Both mwh and Dmp53 genes are on the third chromosome, and three independent recombinant lines (mwh(1), p53; mwh(2), p53; and mwh(3), p53) were generated and tested. For each genotype, three wings were examined. The average number of mwh cells per wing in wild-type or Dmp53−ns mutant flies is indicated in flies that had or had not been exposed to moderate levels of ionizing radiation.
Table 2.
Radiation sensitivity: Percent survival to adult after exposure at 4,000 rads
| Strain | Survival, %
|
||
|---|---|---|---|
| Trial 1 | Trial 2 | Trial 3 | |
| WT | 35 | 67 | 78 |
| Dmp53−ns | 2 | 17 | 6 |
Wandering third-instar larvae (≈50 of each genotype per trial) were irradiated and eclosion was determined 5–6 days later. When tested at 1,000–3,000 rads, Dmp53−ns mutants exhibited moderate radiosensitivity. At doses of 500 rads or lower, eclosion rates for WT and Dmp53−ns mutants were between 80% and 90%.
Discussion
To infer in vivo functions for Dmp53, previous studies relied on forced expression of either wild-type or presumptive dominant-negative transgenes. Although these types of studies are informative, conclusions from them are confounded by considerations relating to highly expressed transgenes. Here we sought to determine the precise role or roles of Dmp53 through loss-of-function genetic analyses. Using the homologous recombination procedure devised by Golic and colleagues (36, 37), we isolated a targeted mutation at the native Dmp53 locus. As the product of ends-in recombination this allele, designated Dmp53−ns, generates two variant copies of Dmp53, both of which, if translated, would produce nonfunctional products. For example, the disrupted template mapping to the left of the whs gene lacks most of the oligomerization and DNA-binding domains, including five of eight residues thought to contact DNA (25–27). Likewise, the template mapping to the right of the whs gene lacks all contiguity with its native promoter and virtually all of the putative transactivation domain, including the initiating methionine and a critical serine residue (Ser-4) thought to be critical for regulation by fly Chk2 (45). Given the indispensable nature of these domains, Dmp53−ns can be considered a null allele with respect to transcriptional activity. It is formally possible that aberrant Dmp53−ns-derived activity could, in principle, engender neomorphic effects (e.g., if Dmp53 encodes significant functions beyond transcription) but, given that Dmp53−ns phenotypes are recessive and consistent with those previously documented for the transgenic dominant-negative alleles, we believe this is a remote possibility.
In agreement with Golic and colleagues (46), who briefly reported on a Dmp53 allele, flies homozygous for Dmp53−ns are viable and fertile and exhibit no overt visible defects as embryos, larvae, or adults. Knockout p53 mutations in mice are similarly viable and fertile and, hence, like its mammalian counterpart, Dmp53 function is not required for normal development. However, mice mutated at p53 also exhibit a strong predilection toward late-onset cancers and, although more subtle age-dependent effects might be uncovered in longevity studies not undertaken here, we observed no clear indications of late-onset overgrowth phenotypes in aged Dmp53−ns flies.
Studies on dominant-negative transgenes indicated important roles for Dmp53 in damage-responsive induction of rpr and the ensuing apoptotic reaction. Three compelling results with the Dmp53−ns strain validate these predictions for a loss-of-function Dmp53 allele, permitting us to account for cell killing by an external insult in this fly. First, whereas other aspects of rpr regulation appear normal, an upstream enhancer governing the acute radiation response at this locus is entirely unresponsive in a Dmp53−ns background (Fig. 2D). Second, direct examination of irradiated Dmp53−ns flies corroborates this defect, extending requirements for Dmp53 to regulation at the native rpr locus (Table 1). Third, individuals homozygous for Dmp53−ns exhibit profound failures in damage-induced apoptosis when directly tested under conditions of genotoxic stress. If placed under the same conditions, it is worth noting that discs from Dmp53−ns/+ heterozygous flies were indistinguishable from wild-type flies. Hence, in this context, we uncovered no evidence for dominant-negative effects exerted through the Dmp53−ns allele. Together, these data establish an absolute requirement for Dmp53 in radiation-induced apoptosis and also validate rpr as an authentic in vivo target of Dmp53.
Recently Peterson et al. (35) studied a synthetic deletion uncovering rpr and found a requirement for this gene in x-ray-induced cell death. Hence, pathways linking damage signals to rpr through Dmp53 are essential for optimal apoptotic cell death in this genetic model. At the same time, however, it should also be emphasized that evidence favoring the existence of additional, proapoptotic Dmp53 targets is strong. For example, flies that are singly mutated for rpr were only partially compromised for radiation-induced apoptotic cell death, and overexpression of Dmp53 in the eye was not significantly affected by the absence of rpr. Also, as seen here in the wing disc, removal of Dmp53 function completely abrogates radiation-induced apoptosis but removal of rpr only partially effects this same damage response. Together, these data argue that additional, proapoptotic targets of p53 exist in the Drosophila genome. One particularly attractive candidate in this regard is the inhibitor of apoptosis protein (IAP)-antagonist sickle (41, 47, 48), which, like rpr, also shows an acute, Dmp53-dependent induction provoked by damage signals (Table 1). This gene maps ≈40 kb to the 5′ side of rpr and so it is conceivable that the Dmp53-binding site regulating rpr might similarly govern the radiation-responsive induction of skl as well. Products encoded at rpr and skl provoke cell death, at least in part, by antagonizing native caspase inhibitors referred to as IAP proteins (41, 47, 48). IAP antagonists with orthologous activity (e.g., Smac/Diablo and Omi/Htra2) have also been reported in mammalian systems and it will therefore be of interest to determine whether any of these genes might represent p53 targets in mammals.
As in mammalian systems, Drosophila cells abruptly halt progression through the cell cycle in response to genotoxic stress. In vertebrate, and possibly invertebrate, models, the cell-cycle regulator p21 is essential for this response and, in stressed mammalian cells, p53 is the major regulator of p21 induction. Studies with dominant-negative transgenes suggested that Dmp53 may not similarly govern cell-cycle progression in flies, but, given the usual limitations on interpreting these types of studies, alternative technical explanations were also possible. Therefore, we directly examined the issue of damage-induced cell-cycle control in the Dmp53−ns strain. Using a mitosis marker to monitor this response, we found that irradiated wing discs from Dmp53−ns homozygotes were indistinguishable from wild-type samples. Hence, at least in the wing disc, Dmp53 is entirely dispensable for an arrested cell cycle and, together with earlier studies, the analyses uncover an important distinction between flies and mammals. Although p21 is possibly the arresting agent in both systems, our data suggest alternative p53-independent mechanisms for regulating cell-cycle arrest in the context of genotoxic damage.
Important implications arise from the fact that p53 functions to govern apoptosis, but not cell-cycle arrest, in Drosophila. From an evolutionary perspective, it can be inferred that ancestral functions of p53 were intimately coupled to the regulation of cell death in the face of genotoxic challenge. Hence, the core means for exerting negative growth control by this family relates to the effects of these proteins on cell death. An obvious corollary here is that checkpoint arrest provoked by p53 may reflect a more recently invented function, specific perhaps to the vertebrate or mammalian lineage. In agreement with this deduction, a recent study in the nematode similarly reported a role for Caenorhabditis elegans p53 in damage-induced cell death but found no role for the gene in the cell-cycle-arrest response (49). A second implication from these studies argues against the hypothesis that p53 launches a sequential damage response program. According to this widely held view, genotoxic stress first provokes an attempted repair program (manifested as cell-cycle arrest), which is then followed by apoptosis if repair efforts are unsuccessful. Inherent to this scenario is the assumption that p53 forces cells toward an arrested cell cycle before engaging a cell-death program. Because Dmp53 clearly elicits apoptosis without arresting the cell cycle, our findings strongly refute this notion. If there are universal effectors of this tumor suppressor, our data argue instead for a proximate connection between p53 signaling and cell death without the need for intervening cell-cycle signaling.
Perhaps the most substantive advance from these studies relates to the effect of Dmp53 on genomic stability. If exposed to moderate doses of radiation, Dmp53−ns homozygotes harbored significantly elevated mutagenic loads that were not detectable in their wild-type siblings. Interestingly, we uncovered no evidence that Dmp53−ns promotes a mutator effect in the absence of radiation and, hence, the effect of Dmp53−ns is clearly limited to conditions associated with genotoxic stress. These observations are clearly distinct from checkpoint/DNA replication/repair mutants (43, 50) where high mutagenic loads are detectable even in the absence of genotoxic stress. In mammalian systems, p53 is essential for the maintenance of genomic stability (51, 52), but the precise derivation of this activity has been difficult to resolve, given the multiple functions of this protein. For example, it is not clear whether failures in cell-cycle arrest or failures in cell death, or possibly a combination of both defective activities, is responsible for maintaining genomic stability. Because Dmp53−ns homozygotes exhibit a mutator phenotype (and yet are normal for cell-cycle arrest) we conclude that a failure in p53-associated cell death alone can account for genomic instability. Although defective cell-cycle control could contribute to many p53 phenotypes seen in mammalian settings, our data are consistent with Schmitt et al. (53), who argue from study of mouse models that mutator defects leading to robust transformation can be traced to p53-associated apoptosis rather than defective checkpoint functions.
Supplementary Material
Acknowledgments
We thank Dr. K. G. Golic for providing Drosophila strains, Joe Chapo and Anna Christich for assistance with the array analysis, and Yuichi Nishi for assistance with the LOH assay. This work was supported by National Institutes of Health/National Institute on Aging Grant R01 AG12466 and an American Cancer Society Research Scholar grant (to J.M.A.).
Abbreviations
- Dmp53
Drosophila p53
- LOH
loss of heterozygosity
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.May P, May E. Oncogene. 1999;18:7621–7636. doi: 10.1038/sj.onc.1203285. [DOI] [PubMed] [Google Scholar]
- 2.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 3.Vousden K H. Biochim Biophys Acta. 2002;1602:47–59. doi: 10.1016/s0304-419x(02)00035-5. [DOI] [PubMed] [Google Scholar]
- 4.Vousden K H. Cell. 2000;103:691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 5.Fisher D E. Apoptosis. 2001;6:7–15. doi: 10.1023/a:1009659708549. [DOI] [PubMed] [Google Scholar]
- 6.el-Deiry W S. Semin Cancer Biol. 1998;8:345–357. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 7.Jimenez G S, Nister M, Stommel J M, Beeche M, Barcarse E A, Zhang X Q, O'Gorman S, Wahl G M. Nat Genet. 2000;26:37–43. doi: 10.1038/79152. [DOI] [PubMed] [Google Scholar]
- 8.Chao C, Saito S, Kang J, Anderson C W, Appella E, Xu Y. EMBO J. 2000;19:4967–4975. doi: 10.1093/emboj/19.18.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caelles C, Helmberg A, Karin M. Nature. 1994;370:220–223. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- 10.Bargonetti J, Friedman P N, Kern S E, Vogelstein B, Prives C. Cell. 1991;65:1083–1091. doi: 10.1016/0092-8674(91)90560-l. [DOI] [PubMed] [Google Scholar]
- 11.Kern S E, Pietenpol J A, Thiagalingam S, Seymour A, Kinzler K W, Vogelstein B. Science. 1992;256:827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- 12.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 13.Pavletich N P, Chambers K A, Pabo C O. Genes Dev. 1993;7:2556–2564. doi: 10.1101/gad.7.12b.2556. [DOI] [PubMed] [Google Scholar]
- 14.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 15.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 16.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 17.Miyashita T, Reed J C. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 18.Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 19.Yu J, Zhang L, Hwang P M, Kinzler K W, Vogelstein B. Mol Cell. 2001;7:673–682. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- 20.Nakano K, Vousden K H. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 21.Owen-Schaub L B, Zhang W, Cusack J C, Angelo L S, Santee S M, Fujiwara T, Roth J A, Deisseroth A B, Zhang W W, Kruzel E, et al. Mol Cell Biol. 1995;15:3032–3040. doi: 10.1128/mcb.15.6.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munsch D, Watanabe-Fukunaga R, Bourdon J C, Nagata S, May E, Yonish-Rouach E, Reisdorf P. J Biol Chem. 2000;275:3867–3872. doi: 10.1074/jbc.275.6.3867. [DOI] [PubMed] [Google Scholar]
- 23.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 24.Yu J, Zhang L, Hwang P M, Rago C, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1999;96:14517–14522. doi: 10.1073/pnas.96.25.14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brodsky M H, Nordstrom W, Tsang G, Kwan E, Rubin G M, Abrams J M. Cell. 2000;101:103–113. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 26.Jin S, Martinek S, Joo W S, Wortman J R, Mirkovic N, Sali A, Yandell M D, Pavletich N P, Young M W, Levine A J. Proc Natl Acad Sci USA. 2000;97:7301–7306. doi: 10.1073/pnas.97.13.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ollmann M, Young L M, Di Como C J, Karim F, Belvin M, Robertson S, Whittaker K, Demsky M, Fisher W W, Buchman A, et al. Cell. 2000;101:91–101. doi: 10.1016/S0092-8674(00)80626-1. [DOI] [PubMed] [Google Scholar]
- 28.Unger T, Nau M M, Segal S, Minna J D. EMBO J. 1992;11:1383–1390. doi: 10.1002/j.1460-2075.1992.tb05183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fields S, Jang S K. Science. 1990;249:1046–1049. doi: 10.1126/science.2144363. [DOI] [PubMed] [Google Scholar]
- 30.Jeffrey P D, Gorina S, Pavletich N P. Science. 1995;267:1498–1502. doi: 10.1126/science.7878469. [DOI] [PubMed] [Google Scholar]
- 31.Kraiss S, Quaiser A, Oren M, Montenarh M. J Virol. 1988;62:4737–4744. doi: 10.1128/jvi.62.12.4737-4744.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denooij J C, Hariharan I K. Science. 1995;270:983–985. doi: 10.1126/science.270.5238.983. [DOI] [PubMed] [Google Scholar]
- 33.Abrams J M. Trends Cell Biol. 1999;9:435–440. doi: 10.1016/s0962-8924(99)01646-3. [DOI] [PubMed] [Google Scholar]
- 34.Nordstrom W, Chen P, Steller H, Abrams J M. Dev Biol. 1996;180:213–226. doi: 10.1006/dbio.1996.0296. [DOI] [PubMed] [Google Scholar]
- 35.Peterson C, Carney G E, Taylor B J, White K. Development (Cambridge, UK) 2002;129:1467–1476. doi: 10.1242/dev.129.6.1467. [DOI] [PubMed] [Google Scholar]
- 36.Rong Y S, Golic K G. Genetics. 2001;157:1307–1312. doi: 10.1093/genetics/157.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rong Y S, Golic K G. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- 38.Grether M E, Abrams J M, Agapite J, White K, Steller H. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- 39.Chen P, Nordstrom W, Gish B, Abrams J M. Genes Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- 40.White K, Grether M, Abrams J M, Young L, Farrell K, Steller H. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 41.Christich A, Kauppila S, Chen P, Sogame N, Ho S I, Abrams J M. Curr Biol. 2002;12:137–140. doi: 10.1016/s0960-9822(01)00658-3. [DOI] [PubMed] [Google Scholar]
- 42.Hendzel M J, Wei Y, Mancini M A, Van Hooser A, Ranalli T, Brinkley B R, Bazett-Jones D P, Allis C D. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 43.Brodsky M H, Sekelsky J J, Tsang G, Hawley R S, Rubin G M. Genes Dev. 2000;14:666–678. [PMC free article] [PubMed] [Google Scholar]
- 44.Baker B S, Carpenter A T, Ripoll P. Genetics. 1978;90:531–578. doi: 10.1093/genetics/90.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peters M, DeLuca C, Hirao A, Stambolic V, Potter J, Zhou L, Liepa J, Snow B, Arya S, Wong J, et al. Proc Natl Acad Sci USA. 2002;99:11305–11310. doi: 10.1073/pnas.172382899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rong Y S, Titen S W, Xie H B, Golic M M, Bastiani M, Bandyopadhyay P, Olivera B M, Brodsky M, Rubin G M, Golic K G. Genes Dev. 2002;16:1568–1581. doi: 10.1101/gad.986602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wing J P, Karres J S, Ogdahl J L, Zhou L, Schwartz L M, Nambu J R. Curr Biol. 2002;12:131–135. doi: 10.1016/s0960-9822(01)00664-9. [DOI] [PubMed] [Google Scholar]
- 48.Srinivasula S M, Datta P, Kobayashi M, Wu J W, Fujioka M, Hegde R, Zhang Z, Mukattash R, Fernandes-Alnemri T, Shi Y, Jaynes J B, Alnemri E S. Curr Biol. 2002;12:125–130. doi: 10.1016/s0960-9822(01)00657-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Derry W B, Putzke A P, Rothman J H. Science. 2001;294:591–595. doi: 10.1126/science.1065486. [DOI] [PubMed] [Google Scholar]
- 50.Xu J, Xin S, Du W. FEBS Lett. 2001;508:394–398. doi: 10.1016/s0014-5793(01)03103-9. [DOI] [PubMed] [Google Scholar]
- 51.Wahl G M, Linke S P, Paulson T G, Huang L C. Cancer Surv. 1997;29:183–219. [PubMed] [Google Scholar]
- 52.Robles A I, Harris C C. Acta Oncol. 2001;40:696–701. doi: 10.1080/02841860152619106. [DOI] [PubMed] [Google Scholar]
- 53.Schmitt C A, Fridman J S, Yang M, Baranov E, Hoffman R M, Lowe S W. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.