Abstract
Whereas information is rapidly accumulating about the structure and position of genes encoded in the human genome, less is known about the complexity and relative abundance of their expression in individual human cells and tissues. Here, we describe the characteristics of the transcriptomes of two cultured cell lines, HB4a (normal breast epithelium) and HCT-116 (colon adenocarcinoma), using massively parallel signature sequencing (MPSS). We generated in excess of 107 short signature sequences per cell line, thus providing a comprehensive snapshot of gene expression, within the technical limitations of the method. The number of genes expressed at one copy per cell or more in either of the lines was estimated to be between 10,000 and 15,000. The vast majority of the transcripts found in these cells can be mapped to known genes and their polyadenylation variants. Among the genes that could be identified from their signature sequences, ≈8,500 were expressed by both cell lines, whereas 6,000 showed cellular specificity. Taking into account sequence tags that map uniquely to the genome but not to known transcripts, overall the data are consistent with an upper limit of 17,000 for the total number of genes expressed at more than one copy per cell in one or both of the two cell lines examined.
Determining the precise variations in gene expression patterns that drive embryogenesis, differentiation, cellular responses to environmental change, and pathological phenomena is a major challenge of the postgenomic era. Achieving this goal requires methods that are able to detect all transcripts present in a cell population in an unbiased manner, and to document significant differences in the abundance of even those derived from very poorly expressed genes. One approach to documenting the diversity of gene expression is through the generation of partial sequences such as ESTs or serial analysis of gene expression (SAGE) tags (1–5). Although we and others have generated hundreds of thousands of such sequences from individual human tissue types, truly redundant coverage of the estimated 200,000–300,000 transcripts (6) in a single cell population has not been previously reported. We have now undertaken such an experiment by using massively parallel signature sequencing (MPSS) (7). This technique can generate millions of signature tags proximal to the 3′ end of transcripts, representing expressed genes in cDNA libraries derived from cells or tissues. In the experiments described here, we have used in excess of 107 tags per cell line derived from libraries with 3.6 × 106 clones each. In principle, this number is sufficient to provide a 10-fold clone coverage of the transcripts present in a human cell.
Methods
Poly(A)+ RNA (1 μg each) was prepared from HB4a (8) and HCT-116 (9) cells, and used to prepare cDNA libraries comprising 21 bases adjacent to the poly(A) proximal DpnII site according to the Megaclone protocol (10) at Lynx Therapeutics (Hayward, CA). The resulting libraries were amplified and loaded onto microbeads. Approximately 1.6 × 106 microbeads were loaded into each flow cell, and the signature sequences were determined by serial enzymatic reactions. An average of 7 × 105 sequences were obtained from each sequencing run, and 16 runs were performed for each library. For technical reasons, the sequences in half of the runs were read in a phase offset by two nucleotides from the other half. All of the signature counts were normalized to 106, because some could be read in only one of the two phases.
A comprehensive reconstruction of human transcripts based on genome data were undertaken based on two datasets: (i) a set of alignments between transcripts and genome regions, thoroughly filtered to eliminate the effects of pseudogenes, highly conserved gene families, repetitive elements, and EST sequencing errors, and decomposed into graphs documenting the boundaries of exons and their connectivity (C.I., B.J.S., and C.V.J., unpublished results); (ii) a mapping onto the genome of all polyadenylation sites that could be extracted from the chromatograms of the EST sequencing projects, thus marking sites where polyadenylation has been experimentally documented (11). These two datasets were merged to produce fully connected graphs traversing all of a gene's exon boundaries and terminating at polyadenylation sites wherever possible; individual transcript sequences contributing to the definition of such a graph retain membership in the graph. Graphs were used to predict, starting from a confirmed polyadenylation site and following exon boundaries, the signature tags that would be produced by cleavage with DpnII and sequencing of the adjacent 13 nucleotides. The assignment of an annotation to a signature sequence was based on the best annotated member of the graph, in the order RefSeq, GenBank mRNA, and EST (RefSeq is the National Center for Biotechnology Information database that provides curated sequences for nucleic acids, including cDNAs, and proteins).
The primary output of our analysis was a tab-delimited file containing the following fields: signature tag sequence; tags per million (tpm) in HB4a library; tpm in HCT-116 library; accession number of the reference transcript used for the annotation; description of the gene (annotation); type of evidence supporting the annotation; ordinal position of signature sequence relative to poly(A); identifier of poly(A) site (internal use); and identifier of gene graph (internal use). The numbers provided in the paper were produced by parsing this file as well as the original data files by using ad hoc perl and awk scripts.
Results
DpnII-anchored MPSS was performed with poly(A)+ RNA preparations from the HCT-116 colon adenocarcinoma (9) and HB4a mammary luminal epithelium (8) cell lines. Tags of length 17 (including the sequence of the anchoring enzyme recognition site) were sequenced from each library. In the HB4a library, the signature sequences comprised 17,354 tags present at >3 tpm and 36,982 at <3 tpm. In the HCT-116 library, the numbers were 24,065 and 54,704, respectively. Tags detected by the MPSS protocol at a frequency of <3 tpm were not considered to be reliable, and were not analyzed further in the present study. It should be noted that a frequency of 3 tpm corresponds to roughly one transcript per cell. It is possible that there are rare transcripts of biological significance that give rise to some of these nonsignificant tags, but larger numbers of MPSS analyses will be required to ascertain whether they can be reproducibly obtained and mapped to experimentally verifiable transcripts.
Among the reliable tags observed in the HCT-116 library, 10,611 were not found in HB4a; conversely, 3,900 tags were unique to the HB4a cells, and 13,454 tags were common to the two libraries. Thus, the total number of tags is 27,965, of which 48% are common to both cell lines. Whereas it may be tempting to equate the number of tags with the number of genes expressed at one copy per cell or more in the cell populations under consideration, these numbers are not equivalent because (i) some of the tags are derived from contaminants, particularly of mitochondrial and ribosomal origin, (ii) many transcripts can be alternatively polyadenylated and generate more than one tag, (iii) cleavage at upstream DpnII sites can also generate more than one tag from a single transcript, (iv) many transcripts contain Alu family repetitive elements in their 3′ UTRs, giving rise to tags that cannot be distinguished from each other, (v) a tag may map to more than one transcript by chance alone, (vi) a transcript may not contain a DpnII site, and (vii) the DpnII site may map too close to the polyadenylation site to produce a meaningful tag. Nevertheless, the number of tags provides a rough first estimate of the complexity of gene expression in the cells from which the library was derived.
To establish as strong a correlation as possible between tags, transcripts, and genes, we have used a comprehensive map of the transcribed regions of the human genome, including experimentally defined polyadenylation sites (11) and the connectivity of exons. Using this map, we have reconstituted virtual transcripts whose sequence is derived from the genome and whose polyadenylation sites are known. These transcripts often extend the 3′ UTR of available mRNA sequences significantly, and the methodology to derive them is described in Methods. We were able to map to virtual transcripts 17,992 of 27,689 reliable tags (excluding those derived from mitochondrial contaminants and Alu elements) from the two combined datasets. Of these mapped tags, 12,234 could be associated with mRNAs from the annotated RefSeq database, 3,966 with mRNAs from GenBank/European Molecular Biology Laboratory (EMBL) not represented in RefSeq, 1,715 with spliced ESTs mapped to the genome, and 77 with anonymous cDNAs from high-throughput sequencing projects [GenBank/EMBL high throughput cDNA (HTC) section].
Using a similar tag to transcript matching procedure, we mapped the MPSS signatures to the current (October 2002) human reference transcript sequence collections from the National Center for Biotechnology Information (NCBI; RefSeq NM and XM sections) and the ensembl (ver. 8.30) project (Table 1). Only those signatures contained in the reference transcript collections were recorded, not those inferred from their extension on the genome. The results clearly show the advantages of fully exploiting combined genome and transcriptome data, because we were able to document the origin of ≈50% more tags than would be possible by using the reference databases alone (Table 1). In most cases, this improvement was due to the association of previously unannotated ESTs and EST clusters to defined genes. Meaningful annotation was available for 90% of the tags that could be mapped to genes. If we take a gene to be a transcribed region mapped to the genome, then the tags from the two libraries are derived from a total of 12,468 known genes (9,879 in the HB4a library and 11,353 in the HCT-116 library). Because known genes are usually more highly and commonly expressed than potentially novel ones, the tags unique to each library are expressed at lower levels and less likely to have been well-annotated: the average level of expression of mapped tags common to both libraries was 120 tpm, whereas that of tags unique to one or the other of the cell lines was almost 10-fold less at 15 tpm (8.3 and 22.2 for the HCT-116- and HB4a-specific tags, respectively).
Table 1.
Annotation of MPSS tags
| HB4a | HCT-116 | Combined | |
|---|---|---|---|
| Total tags | 17,354 | 24,065 | 27,965 |
| Contaminants* | 160 | 264 | 276 |
| Match virtual transcripts† | 12,109 (70%) | 14,699 (62%) | 17,992 (65%) |
| Match NCBI models‡ | 9,476 (55%) | 10,883 (46%) | 12,326 (44%) |
| Match ensembl transcripts | 8,561 (50%) | 9,842 (41%) | 11,105 (40%) |
Contaminants include mitochondrial and ribosomal RNAs and repetitive elements.
Percentages are calculated relative to total tags minus contaminants.
Combination of experimental and predicted transcripts (NM and XM identifiers) in RefSeq (November 8, 2002).
The origin of the remaining 10,471 reliable tags remains uncertain. One possibility is that sequencing errors and genetic polymorphisms account for a significant proportion of them (12). To test this hypothesis, we identified tags that had single nucleotide differences with annotated tags and that were present at less than half the abundance of the presumed “parent.” We thus found that 5,665 of the unmapped tags (53%) could have been generated by sequencing errors or polymorphisms. It is worth noting that, of the 5,665 tags marked as potential sequencing errors, 3,380 (60%) did not find matches anywhere within the human genome (version 30 of the NCBI/ensembl assembly, August 2002), whereas only 1,041 (22%) of the remaining 4,806 tags could not be mapped. This result is consistent with the majority of the tags flagged as potential sequencing errors being indeed generated by errors; tags that differ from abundant mapped tags by 2 nt or more are about three times as likely to be derived from novel transcripts or novel parts of known genes. Of the 4,806 tags that could not be mapped to transcripts nor attributed to potential sequencing errors, 3,765 matched the genome, and 2,645 did so in a unique position. Of these 2,645 tags, 958 (36%) mapped to introns of known genes, and 862 (42%) mapped within 5 kb of an exon, each time in the expected orientation relative to the direction of transcription. These numbers suggest that >70% of MPSS signature tags that can be matched to the genome but not to known transcripts could be derived from yet unmapped portions of known genes. Of the tags that could not be matched to the transcriptome, 1,120 occur in multiple locations on the genome; in fact, they collectively mark 23,870 genomic locations. It is unlikely that reliable tag-to-transcript assignments will ever be obtained for most of them.
Assuming that >90% of the human euchromatic genome sequence has been completed and that the procedure for identifying sequencing errors was mostly correct, it is very unlikely that >2,000 polyadenylated transcripts expressed at one copy per cell or more in either HB4a or HCT-116 cells remain to be discovered. This finding also puts an upper limit ≈17,000 to the total number of genes expressed in one or both of the two cell lines that we examined.
The most abundant signatures in both datasets were derived from mitochondrial contaminants, which make up ≈10% of the total (124,206 and 102,079 tpm). Alu family repetitive elements, presumably embedded within bona fide mRNA 3′ UTRs, also contributed significantly (15,622 and 35,332 tpm). Only 5 signatures among the 154 occurring at >1,000 tpm on the average could not be annotated; 3 of these did not map to the genome and their origin could not be ascertained. Whereas the 2 others could be mapped on the genome (one uniquely, and the other in 17 distinct locations), we have no explanation for their abundance. The unique tag mapped to an intron of the densin-180 (KIAA1365) gene, but in an antisense orientation, making it unlikely that it could be derived from an alternatively spliced form of this gene; EST and SAGE data offer no indication that this region could be highly expressed in the cell lines examined here. This finding underlines the importance of experimental verification in any attempts to use MPSS or SAGE data in a gene discovery pipeline (13). More generally, and as expected, the quality of the annotation decreased with the abundance of the tags, as illustrated in Table 2. It should be noted that >80% of the tags present at more than three copies per cell can be mapped to genes, and therefore that the quality of biological information that can be extracted from MPSS experiments is high.
Table 2.
Distribution of tag abundance and identification
| Abundance, tpm* | No. of tags | No. identified† | No. of genes |
|---|---|---|---|
| >10,000 | 7 | 7 | 3 |
| >5,000 | 25 | 24 | 14 |
| >1,000 | 154 | 149 | 120 |
| >500 | 298 | 280 | 229 |
| >100 | 1,719 | 1,600 | 1,397 |
| >50 | 3,261 | 3,060 | 2,631 |
| >10 | 10,519 | 9,608 | 8,018 |
| >5 | 15,145 | 13,517 | 10,876 |
| >1 | 27,965 | 25,779 | 17,992 |
Mean between the HB4a and HCT-116 libraries.
Identification includes a match to a gene, to a known contaminant, or to a potential sequencing error or polymorphism; it does not include a match to the genome.
Among the most highly represented transcripts in both cell lines were those encoding ribosomal proteins (e.g., L13, S2, P0, and P1), β and γ actins, EF-1α, and glyceraldehyde-3-phosphate dehydrogenase. This result is entirely consistent with a large corpus of published data. An examination of the genes that are differentially expressed between the two cell lines is also revealing. The HB4a breast epithelial cells abundantly express keratins and components of the extracellular matrix such as fibronectin and fibulin-like extracellular matrix protein. Consistent with their secretory role, they also express kallikrein, annexin, and complement components. All of the corresponding transcripts are found at one copy per cell or less in the colon carcinoma line. The biological significance of specific gene expression in the HCT-116 cells is much less clear; many of the transcripts expressed preferentially in these cells have not been functionally characterized. It is interesting to note that expression of MAGE-A family antigens (14) is easily detectable in the colon cell line (63 tpm) while being absent from the HB4a cells. More generally, the relative differences in levels of gene expression detected by MPSS seem to be significantly greater than those found by using other methods. For example, the signature tags for keratins 5 and 6 were found at 3,843 tpm in the HB4a library while being totally absent from the HCT-116 library.
A class of mRNAs of great biological importance but difficult to detect because of their low abundance are those encoding transcription factors. Detectable from our data were 308 mRNAs encoding transcription factors (as deduced from their annotation). Eighty-three percent (257) of these transcripts were expressed at <10 copies per cell (30 tpm) in one line or the other, and over one third (128) were specifically expressed in one cell line and not the other, as would be expected from their very different phenotypes. This result demonstrates the power of the technique in detecting specifically even transcripts of very low abundance.
Similarly to SAGE, MPSS relies on the production of sequence tags proximal to mRNA polyadenylation sites. Occasionally, because of incomplete cleavage of the cDNA, a tag is produced from an upstream site. The frequency of such tags has been reported to be ≈1% in SAGE (15). In the MPSS data, tags derived from the second DpnII site were present at essentially the same frequency as in SAGE (44,044/4,436,630, or 1%), relative to those that could be mapped unambiguously to the first site upstream of a polyadenylation event.
Because of the depth of their coverage, MPSS data can give us our first quantitative glimpse into the relative utilization of alternative polyadenylation sites (11, 16) and its potential regulation across cell types and environmental conditions. First, for each of the genes for which we had been able to document multiple polyadenylation sites based on EST data (11), we examined whether all of the sites were used in the two cell lines selected for this study. In most genes, only a subset of polyadenylation sites seemed to have been used at a frequency sufficient to be detectable at a level exceeding 3 tpm. In a total of 11,522 genes where we had EST-based evidence for the existence of multiple polyadenylation sites, we found 15,209 sites in 5,273 genes confirmed by MPSS tags, and 14,064 sites for which a tag was predicted but not found in our libraries. The average number of EST-documented polyadenylation sites per gene was 2.54, of which about half (1.32) were confirmed by the tags found in the HB4a and HCT-116 cell lines. These data strongly suggest that polyadenylation site selection is cell- and tissue-specific, and that further MPSS analyses will be required to sample the full repertoire of polyadenylation site selection.
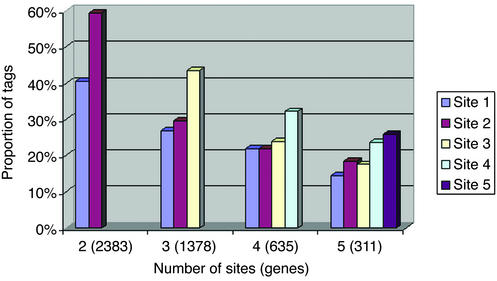
The remaining analyses focused on the genes where we could confirm the utilization of multiple polyadenylation sites from the MPSS signature tags. We investigated whether there was a systematic bias in the usage of polyadenylation sites as a function of their position relative to the transcript 5′ end. For each gene, we tabulated the proportion of tags from both libraries associated with each polyadenylation site. The results are shown in Fig. 1 for genes with two to five sites. It shows very clearly that, if there is a bias at all, it is for the 3′-most site, and not for the one closest to the mRNA coding region. This fact further emphasizes the need to fully document the extent of 3′UTRs, whether for the proper identification of SAGE or MPSS signature sequences or for the design of appropriate probes for microarrays.
Figure 1.
Utilization of polyadenylation sites as a function of their position relative to the transcript 5′ end (site 1 is the 5′ most). Only transcripts with five polyadenylation sites or fewer were taken into account, because the numbers obtained from of those with six or more (244 genes total) are insufficient to yield significant results.
We then looked for cases where the relative utilization of polyadenylation sites differed significantly between the two lines by flagging interlibrary differences between the relative MPSS signature tag representations within the same gene. We found 2,613 cases out of 5,273, or about half of all genes alternatively polyadenylated in HB4a and HCT-116 cells, where relative polyadenylation site usage differed by 3-fold or more between the two lines. It would be premature to speculate on the biological significance of such differences, but the data show conclusively that there are significant differences between cell types in selecting polyadenylation sites. Further research is needed to establish the extent to which site selection is tied to cellular differentiation, to the cellular response to changing environmental conditions, or to global patterns of gene expression.
Discussion
Our data present a comprehensive picture of gene expression in two human cell lines. Technical limitations inherent to the MPSS protocol and to the structure of the human transcriptome (highly conserved gene families, repetitive elements, pseudogenes, etc.) make it impossible to match all signature tags to genes in an unequivocal fashion. However, over the last 2 yr, the sequence of the human genome has been essentially completed, and the quantity and quality of transcriptome data has drastically increased through large-scale EST and full-length insert-sequencing projects (17). The combination of these efforts has made it possible to reliably assign the majority of signature tags to transcripts (15) and to interpret the results of MPSS experiments in a meaningful fashion. The highlights of the present analysis can be summarized as follows. (i) The complexity of the transcriptomes of individual human cell lines is rather limited. A normal mammary epithelial cell line expresses ≈10,000 genes, whereas a colon adenocarcinoma line expresses ≈15,000. We cannot be more precise at this point because we do not know how many signature tags actually document novel transcripts. (ii) The number of “housekeeping” genes expressed in all cell types is still a matter of conjecture; however, the data presented here put an upper limit to this number at ≈8,500, and the analysis of further cell types will undoubtedly reduce it further. (iii) The depth of transcript coverage generated by publicly available EST and SAGE sequences is sufficient to have documented all but a small number of poorly expressed genes. (iv) Long-range alternative polyadenylation can be documented in detail from MPSS data, and it is now clear that polyadenylation site selection is a cell type-specific phenomenon. (v). Finally, MPSS has been validated as a technique that is able to produce a comprehensive view of the transcriptional program of a cell.
It is informative to compare the numbers we obtained with those of a very careful cDNA-driven mRNA hybridization study performed by Hastie and Bishop (18) on RNA extracted from mouse kidneys, brains, and livers. They concluded that there were ≈12,000 different expressed mRNA sequences, distributed in three major abundance classes, and that ≈10,000 mRNAs were expressed in common in the three tissues, although at sometimes very different abundances. If we assume that the limit of detection in the Hastie and Bishop study was ≈6 tpm or two copies per cell, the numbers obtained in the present study are remarkably similar to theirs (Table 2), and, although we are now in a position to document the abundance of each individual RNA, our overall estimate of the complexity of gene expression has changed very little with the introduction of highly sophisticated techniques such as MPSS.
Clearly, there is still enormous and largely untapped value in producing MPSS data for many more cell types and experimental conditions. Whereas the cost and complexity of the technique make it unsuitable for high-throughput analysis or for diagnostic applications, its adoption by a growing number of laboratories working on a variety of experimental systems would greatly increase the value of the data, especially because they are produced in a format that is easy to document and to share. Therefore, we strongly support the creation of an openly accessible repository of MPSS data, to complement and extend what has already been achieved for sequence-based transcript-documenting technologies such as ESTs and SAGE (15).
Acknowledgments
We gratefully acknowledge the support of the Ludwig Institute for Cancer Research and of the National Cancer Institute for making this collaborative study possible.
Abbreviations
- MPSS
massively parallel signature sequencing
- tpm
tags per million
- SAGE
serial analysis of gene expression
References
- 1.Adams M D, Soares M B, Kerlavage A R, Fields C, Venter J C. Nat Genet. 1993;4:373–380. doi: 10.1038/ng0893-373. [DOI] [PubMed] [Google Scholar]
- 2.Strausberg R L, Riggins G J. Proc Natl Acad Sci USA. 2001;98:11837–11838. doi: 10.1073/pnas.221463598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camargo A A, Samaia H P B, Dias-Neto E, Simão D F, Migotto I A, Briones M R, Costa F F, Nagai M A, Verjovski-Almeida S, Zago M A, et al. Proc Natl Acad Sci USA. 2001;98:12103–12108. doi: 10.1073/pnas.201182798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Velculescu V E, Zhang L, Vogelstein B, Kinzler K W. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 5.Lal A, Lash A E, Altschul S F, Velculescu V, Zhang L, McLendon R E, Marra M A, Prange C, Morin P J, Polyak K, et al. Cancer Res. 1999;59:5403–5407. [PubMed] [Google Scholar]
- 6.Bishop J O, Morton J G, Rosbash M, Richardson M. Nature. 1974;250:199–204. doi: 10.1038/250199a0. [DOI] [PubMed] [Google Scholar]
- 7.Brenner S, Johnson M, Bridgham J, Golda G, Lloyd D H, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al. Nat Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 8.Harris R A, Hiles I D, Page M J, O'Hare M J. Br J Cancer. 1995;72:386–392. doi: 10.1038/bjc.1995.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brattain M G, Fine W D, Khaled F M, Thompson J, Brattain D E. Cancer Res. 1981;41:1751–1756. [PubMed] [Google Scholar]
- 10.Brenner S, Williams S R, Vermaas E H, Storck T, Moon K, McCollum C, Mao J I, Luo S, Kirchner J J, Eletr S, et al. Proc Natl Acad Sci USA. 2000;97:1665–1670. doi: 10.1073/pnas.97.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iseli C, Stevenson B J, De Souza S J, Samaia H B, Camargo A A, Buetow K H, Strausberg R L, Simpson A J, Bucher P, Jongeneel C V. Genome Res. 2002;12:1068–1074. doi: 10.1101/gr.62002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Velculescu V E, Madden S L, Zhang L, Lash A E, Yu J, Rago C, Lal A, Wang C J, Beaudry G A, Ciriello K M, et al. Nat Genet. 1999;23:387–388. doi: 10.1038/70487. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Sun M, Lee S, Zhou G, Rowley J D, Wang S M. Proc Natl Acad Sci USA. 2002;99:12257–12262. doi: 10.1073/pnas.192436499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S. Cancer Res. 2001;61:5544–5551. [PubMed] [Google Scholar]
- 15.Boon K, Osorio E C, Greenhut S F, Schaefer C F, Shoemaker J, Polyak K, Morin P J, Buetow K H, Strausberg R L, De Souza S J, Riggins G J. Proc Natl Acad Sci USA. 2002;99:11287–11292. doi: 10.1073/pnas.152324199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pauws E, van Kampen A H, van de Graaf S A, de Vijlder J J, Ris-Stalpers C. Nucleic Acids Res. 2001;29:1690–1694. doi: 10.1093/nar/29.8.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strausberg R L, Buetow K H, Greenhut S F, Grouse L H, Schaefer C F. Cancer Invest. 2002;20:1038–1050. doi: 10.1081/cnv-120005922. [DOI] [PubMed] [Google Scholar]
- 18.Hastie N D, Bishop J O. Cell. 1976;9:761–774. doi: 10.1016/0092-8674(76)90139-2. [DOI] [PubMed] [Google Scholar]