Abstract
To evaluate the potential role of mitochondrial lactate dehydrogenase (LDH) in tissue lactate clearance and oxidation in vivo, isolated rat liver, cardiac, and skeletal muscle mitochondria were incubated with lactate, pyruvate, glutamate, and succinate. As well, α-cyano-4-hydroxycinnamate (CINN), a known monocarboxylate transport inhibitor, and oxamate, a known LDH inhibitor were used. Mitochondria readily oxidized pyruvate and lactate, with similar state 3 and 4 respiratory rates, respiratory control (state 3/state 4), and ADP/O ratios. With lactate or pyruvate as substrates, α-cyano-4-hydroxycinnamate blocked the respiratory response to added ADP, but the block was bypassed by addition of glutamate (complex I-linked) and succinate (complex II-linked) substrates. Oxamate increased pyruvate (≈10–40%), but blocked lactate oxidation. Gel electrophoresis and electron microscopy indicated LDH isoenzyme distribution patterns to display tissue specificity, but the LDH isoenzyme patterns in isolated mitochondria were distinct from those in surrounding cell compartments. In heart, LDH-1 (H4) was concentrated in mitochondria whereas LDH-5 (M4) was present in both mitochondria and surrounding cytosol and organelles. LDH-5 predominated in liver but was more abundant in mitochondria than elsewhere. Because lactate exceeds cytosolic pyruvate concentration by an order of magnitude, we conclude that lactate is the predominant monocarboxylate oxidized by mitochondria in vivo. Mammalian liver and striated muscle mitochondria can oxidize exogenous lactate because of an internal LDH pool that facilitates lactate oxidation.
Arterial lactate concentration is low and stable in resting humans (1, 2) and other mammals, such as rats (3) and dogs (4), giving no indication of relatively high flux rates that range from 0.5 to 1.0 mg/kg per min in resting men (1, 2). In resting mammals oxidation accounts for approximately half lactate disposal and gluconeogenesis approximately 20% (1–5). During sustained submaximal exercise, blood lactate rate of appearance increases as a direct function of metabolic rate (1–3); however, arterial lactate concentration increases little during sustained moderate intensity exercise because disposal through oxidation (≈80%) and gluconeogenesis (≈20%) matches appearance (1, 2, 6). During both rest and exercise, skeletal muscle is a major site of lactate oxidation as well as production (1, 2). Differences in circulating lactate concentration between individuals with similar rates of appearance are attributable to variations in clearance rate (3, 5). At exercise onset, net lactate release from working muscle contributes to the elevation of circulating lactate concentration, but as exercise continues, working muscle consumes and oxidizes lactate on a net basis as production continues (1, 7). Studies on mammalian muscles in situ demonstrate that in working muscle lactate uptake and oxidation are concentration dependent (8, 9), and similar data are available on human skeletal muscle (1, 2, 7, 10) and heart (11).
The cytosol of heart, skeletal muscle, and other cells is abundant in lactate dehydrogenase (LDH) (12). The equilibrium constant for LDH is 3.6 × 104 M−1, and muscle LDH isoforms demonstrate characteristics of low Km and high Vmax, resulting in lactate production regardless of the state of oxygenation (13, 14). That working skeletal muscle tissue can simultaneously produce, consume, and oxidize lactate has been explained by the lactate shuttle mechanism (15), a model that assumes fiber heterogeneity with lactate production occurring in fast-glycolytic (type IIB) fibers and oxidation in slow-oxidative (type I) fibers. However, the model of a cell–cell lactate shuttle is less adequate for predicting lactate metabolism in resting muscle tissue, which appears fully oxygenated (14), but which releases lactate on a net basis (1, 7).
To evaluate the role of mitochondria in balancing lactate production and oxidation as part of an intracellular lactate shuttle (16), we respired isolated rat cardiac, skeletal muscle, and liver mitochondria with lactate and pyruvate in the presence or absence of known inhibitors of metabolism. As well, we probed for the presence of LDH isoforms in mitochondria by electrophoresis and electron microscopy. Results support the conclusion of a mitochondrial role in cellular lactate oxidation (16) and provide an explanation of the high correlation between lactate clearance during exercise (3) and muscle mitochondrial respiratory capacity (17).
MATERIALS AND METHODS
Mitochondrial Isolation.
Rat hind limb skeletal muscle and heart mitochondria were isolated according to Makinen and Lee (18), as described (17, 19). The protocol involves: tissue isolation, mincing in cold isolation medium (IM), 2°C incubation with a proteolytic enzyme (trypsin), homogenization, and differential centrifugation. The initial mitochondrial pellet was resuspended and precipitated twice in IM. Liver mitochondria were prepared as described (20) and washed twice by resuspension and centrifugation.
Polarography.
Mitochondrial protein concentration was determined by bicinchoninic acid assay (Pierce), and incubations were carried out at a [mitochondrial] of 1 mg/ml (21). A Rank Bros. (Cambridge, U.K.) O2 electrode was used, and the respiration chamber was kept constant at 37°C. The respiratory medium consisted of: 15 mM KCl, 15 mM K2HPO4, 15 mM KH2PO4, 25 mM Tris, 45 mM sucrose, 12 mM mannitol, 5 mM MgCl2, 7 mM EDTA, and 0.2% BSA, pH 7.4 (18). Assays were performed in duplicate or triplicate on fresh mitochondria. To stimulate respiration, ADP was added to a concentration of 5 nM, and pyruvate and lactate were added to give a concentration of 10 mM (+2.5 mM malate). Glutamate and succinate were used at 10 mM. To block the mitochondrial monocarboxylate transporter (mMCT), 5 mM α-cyano-4-hydroxycinnamate (CINN) was used (22). To block mitochondrial LDH, 50 mM oxamate (OX) was used (23). NAD+ was not added to the reaction system.
Mitochondrial Fractionation.
Compartment fractions of liver, muscle, and cardiac mitochondria were prepared by digitonin fractionation as described by Schnaitman and Greenawalt (24). After digitonin treatment of intact mitochondria, the 10,000 × g pellet represents the inner membrane plus matrix constituent. The pellet from the 100,000 × g centrifugation represents outer membrane constituents, and the supernatant from the 100,000 × g spin represents the intermembrane (periplasmic) space.
LDH Separation and Analysis-Electrophoresis.
LDH isoenzymes present in isolated mitochondria or in mitochondrial fractions were separated by adding 1 μg protein to agarose (1%) gels (Reliant precast gels; FMC) and electrophoresing at 90 V for 30 min by using a Bio-Rad Sub-Cell system. An electrophoretic marker (LDH Isotrol; Sigma) containing LDH isoenzymes 1–5 was used as an aid in identification of isoenzymes. After electrophoresis, the areas of isoenzyme activity were visualized by using a colorimetric procedure (Sigma Procedure 705). The gels were fixed in 5% acetic acid. LDH isoenzyme bands were scanned and quantified by using a Bio-Rad GS-700 imaging densitometer.
Electron Microscopy.
Liver, heart, soleus, and extensor digitorum longus tissues were harvested from pentobarbital anesthetized rats and prepared for electron microscopy by high-pressure freezing and conventional tissue processing.
Conventional processing.
Dissected tissues were immediately placed into a fixative containing 2% paraformaldehyde and 0.1% glutaraldehyde in 0.1 M sodium cacodylate buffer, where they were minced into small (≈1 mm3) pieces. The samples then were dehydrated in a graded acetone series and embedded into LR White resin by using microwave processing. After polymerization, thin (60 nm) sections were cut by using an RMC MTX ultramicrotome with a diamond knife. Sections were immunolabeled and stained.
High-pressure freezing.
Tissues were removed by using a MANAN Pro-Mag 1.2 Biopsy System (Northbrook, IL) attached with an automatic cutting needle (25). Specimens then were loaded into brass planchettes, and the empty space was filled with 1-hexadecene (Sigma). Planchettes then were frozen in a BAL-TEC HPM 010 high-pressure freezing machine (Balzers, Liechtenstein) and immediately transferred under liquid nitrogen into a fixative containing 0.2% glutaraldehyde and 0.1% uranyl acetate in acetone (26). All specimens were stored in liquid nitrogen until used.
Freeze substitution.
Frozen samples were transferred to a Leica EM AFS where the temperature was gradually raised from −90°C to 20°C over a 4-day period. Specimens then were rinsed in pure acetone, embedded, sectioned, immunolabeled, and stained.
Immunolabeling.
Sections were placed on formvar and carbon-coated nickel grids and labeled (27). For high-pressure frozen and conventionally processed tissue samples, a 1:10 dilution of LDH-H4 and LDH-M4 mAbs (Anawa, Zurich) were used. For secondary antibody labeling, a 1:20 dilution of goat anti-mouse antibody conjugated to 15 nm gold particles was applied, with negative controls receiving only the secondary antibody. This form of control was used to demonstrate that the secondary antibody did not bind nonspecifically to the tissue. Immunolabeled sections then were stained by using 2% aqueous uranyl acetate and Reynold’s lead citrate solutions. Density of LDH labeling was determined by scanning micrographs into Adobe Photoshop 4.0 and quantitated by NIH Image 1.61.
Transmission electron microscopy.
Grids were viewed by using a JEOL 100CX TEM operating at 80 kV. For high-pressure frozen samples, three grids were examined for each tissue, and a total of 29 micrographs were developed and printed. Conventionally processed samples produced two grids per tissue, resulting in 17 micrographs. A calibration grid (Ted Pella, Redding, CA) was used to determine magnification of micrographs.
RESULTS AND DISCUSSION
Mitochondria isolated from liver, skeletal muscle, and heart (Table 1) readily oxidized lactate and pyruvate; the MCT-inhibitor, CINN, blocked respiration of both substrates. In the presence of CINN, state 3 respiration was restored with the addition of either succinate or glutamate. Restoration of respiration with succinate or glutamate indicates that CINN does not block mitochondrial respiratory complex I (glutamate, an NADH-linked substrate) or complex II (succinate, an FADH2-linked substrate). Upstream inhibition of mitochondrial lactate and pyruvate oxidation, such as a transport limitation, by CINN is indicated.
Table 1.
Respiratory rates and control parameters of isolated rat liver, skeletal muscle, and cardiac muscle mitochondria with either pyruvate, lactate, succinate, or glutamate as substrates in the presence or absence of OX or CINN
| Substrate | Tissue | State 3 | State 4 | RCI | ADP/O |
|---|---|---|---|---|---|
| Pyruvate | Liver | 120.3 ± 7.4 | 46.6 ± 3.7 | 5.2 ± 1.1 | 3.00 ± 0.01 |
| Muscle | 293.4 ± 22.6 | 72.4 ± 5.1 | 12.3 ± 0.9 | 3.04 ± 0.03 | |
| Heart | 168.9 ± 23.3 | 70.2 ± 7.8 | 5.7 ± 0.6 | 3.03 ± 0.01 | |
| Pyruvate + OX | Liver | 177.5 ± 21.5 | 80.1 ± 9.4 | 6.0 ± 0.4 | 3.00 ± 0.01 |
| Muscle | 413.0 ± 41.4 | 76.8 ± 5.4 | 9.7 ± 1.2 | 3.19 ± 0.16 | |
| Heart | 265.8 ± 42.4 | 80.1 ± 9.4 | 4.5 ± 0.5 | 3.25 ± 0.27 | |
| Pyruvate + CINN | Liver | 50.3 ± 3.1 | – | – | – |
| Muscle | 89.2 ± 2.0 | – | – | – | |
| Heart | 41.5 ± 6.0 | – | – | – | |
| Succinate | Liver | 177.5 ± 21.5 | – | 6.0 ± 1.0 | 2.23 ± 0.06 |
| Muscle | – | – | – | – | |
| Heart | – | – | – | – | |
| Pyruvate + CINN + succinate | Liver | 167.5 ± 5.0 | 57.8 ± 4.0 | 5.6 ± 1.0 | |
| Muscle | – | – | – | – | |
| Heart | – | – | – | – | |
| Lactate | Liver | 166.1 ± 17.9 | 66.5 ± 4.8 | 5.6 ± 0.9 | 3.00 ± 0.01 |
| Muscle | 314.4 ± 42.4 | 63.4 ± 11.8 | 21.1 ± 2.0 | 3.01 ± 0.03 | |
| Heart | 142.4 ± 14.7 | 59.3 ± 6.4 | 5.9 ± 0.9 | 3.16 ± 0.27 | |
| Lactate + OX | Liver | 85.2 ± 11.4 | – | – | – |
| Muscle | 38.0 ± 2.6 | – | – | – | |
| Heart | 46.7 ± 12.0 | – | – | – | |
| Lactate + CINN | Liver | 55.6 ± 1.0 | – | – | – |
| Muscle | 41.5 ± 6.0 | – | – | – | |
| Heart | 41.5 ± 6.0 | – | – | – | |
| Lactate + OX + glutamate | Liver | 258.7 ± 16.0 | 87.0 ± 13.0 | 8.7 ± 2.0 | – |
| Muscle | 322.8 ± 27.2 | 103.8 ± 8.6 | 3.5 ± 1.3 | – | |
| Heart | 258.7 ± 16.0 | 87.0 ± 13.0 | 8.9 ± 1.0 | – |
Respiratory rates in nmol O2/mg mitochondrial protein per min. Substrate concentrations: pyruvate and lactate (10 mM + 2.5 mM malate); glutamate (10 mM); succinate (10 mM); OX (50 mM); CINN (5 mM). RCI, Respiratory Control Index.
Previously, on rat white muscle and heart tissue homogenates made to respire with lactate or pyruvate, Molé et al. (28) demonstrated preferential lactate oxidation by the more oxidative tissue preparation. Subsequently, Baldwin et al. (29) showed that the ability of muscle homogenates to oxidize lactate was related to mitochondrial density. Our results extend those of previous workers and demonstrate a mitochondrial role in lactate oxidation. In our experiments, we normalized substrate concentration to 10 mM, but lactate concentration [L] exceeds that of pyruvate [P] by an order of magnitude or more, in vivo. Thus, it is likely that lactate represents a more important substrate than pyruvate for mitochondrial respiration in vivo, especially when the [lactate] and [L]/[P] are high such as during exercise.
The LDH inhibitor OX blocked oxidation of lactate by liver, muscle, and heart mitochondria (Table 1). In the absence of OX, lactate oxidation exceeded pyruvate oxidation by 10–40% in mitochondria isolated from the three tissues. However, in the presence of OX, pyruvate oxidation was similar to that of lactate in the absence of OX (Table 1) because pyruvate interacts directly with pyruvate dehydrogenase, bypassing the LDH step as required for lactate. Our results indicate the key role of LDH in the ability of isolated mitochondria to oxidize lactate. In mitochondria, the path of lactate oxidation involves conversion to pyruvate. Dependence of mitochondrial lactate oxidation on LDH is relevant to the issue of isolation artifact (see below). LDH in the mitochondrial intermembrane space and matrix adds functionality to the system, an effect unlikely to accrue from nonspecific binding during isolation.
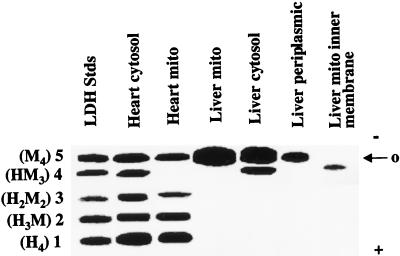
Agarose gel electrophoresis of LDH isoenzyme patterns in cytosolic fractions of different tissues (Fig. 1) are consistent with results of previous investigations showing tissue specificity (e.g., ref. 12). However, mitochondrial fractions also revealed the presence of LDH isoenzymes (Fig. 1). Further, LDH isoenzyme patterns differed among tissues, and between mitochondria and the surrounding cytosol in each tissue. Heart and muscle mitochondria were noted by the prevalence of both LDH-1 (H4) and LDH-5 (M4), whereas liver mitochondria were distinguished by the presence of LDH-5 (M4).
Figure 1.
Agarose gel electrophoresis of LDH in mitochondria from rat liver and heart. LDH isoenzyme patterns differ between cytosol and mitochondria in both tissues.
In agreement with Brandt et al. (30) and Kline et al. (31), our results indicate that mitochondrial LDH is present in the intermembrane space. However, we also found LDH present in the inner membrane + matrix fraction. In contrast, the mMCT is bound to the inner mitochondrial membrane (32–34).
Three conclusions with regard to electrophoretic separation of LDH isozymes in mitochondria and cytosol appear appropriate. First, differences in LDH patterns between mitochondria and surrounding cytosol suggest that LDH within mitochondrial preparations did not result from cytosolic contamination. Second, the greater abundance of LDH-1 (H4) in heart than liver is expected on the basis of the high oxidative capacity of cardiac tissue. However, like heart, liver is also a net lactate consumer. Thus, differences in mitochondrial LDH pattern in liver compared with heart may be more related to the gluconeogenic, as opposed to oxidative nature of liver. Third, the ability of mitochondria to convert lactate to pyruvate is governed by mitochondrial redox rather than LDH isoenzyme pattern. This latter conclusion is reached because of similar abilities of liver and cardiac mitochondria to oxidize exogenous lactate despite very different LDH isozyme patterns.
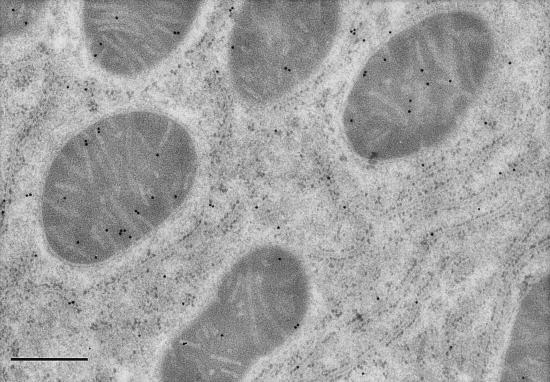
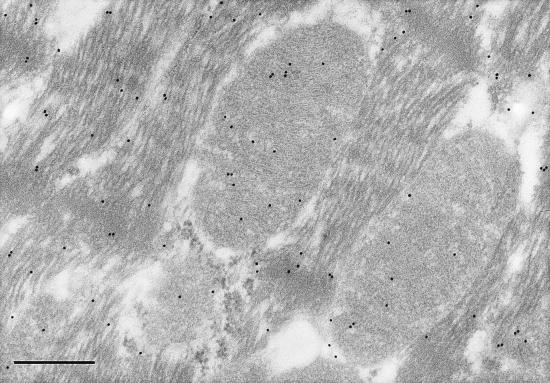
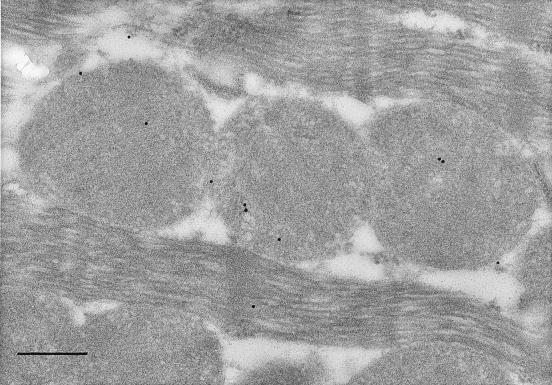
Electron micrographs of liver, skeletal, and cardiac muscle tissues probed with immunolabeled antibodies to LDH-1 and –5 produced labeling patterns consistent with those of gel electrophoresis. Negative control experiments did not show labeling of any tissues studied and preclude the possibility of accidental labeling. In liver, the LDH-5 (M4) antibody labeled sites in mitochondria, surrounding cytosol and organelles (Fig. 2). However, the density of labeling of liver mitochondria was significantly (2-fold, P < 0.05) greater than in surrounding compartments. In myocardium, the LDH-5 antibody labeled mitochondria and surroundings uniformly (Fig. 3). However, in myocardium the LDH-1 antibody labeled primarily mitochondria, with 2-fold greater (P < 0.05) labeling frequency than among myofibrils or extramitochondrial organelles (Fig. 4). In soleus (not shown), the frequency of mitochondrial LDH-1 labeling was 60% greater than for surrounding areas (P < 0.05). Also for soleus the LDH-1 antibody labeled mitochondria with a 2.6-fold greater mean frequency than in surrounding areas, but because of variation mitochondrial labeling was not greater than for surrounding areas (P ≈ 0.10) on a two-tailed test. Overall, results of immunolocalization of LDH in mitochondria are consistent with those of Baba and Sharma (32) who used electron microscope histochemistry and showed LDH to be associated with the inner membrane and matrix of rat pectoralis and cardiac muscle mitochondria. Importantly, visualization of LDH isoforms in mitochondria in situ supports the interpretation that results of polarographic and electrophoresis studies were not caused by isolation artifact (23).
Figure 2.
Electron micrograph of high-pressure frozen rat liver showing mitochondria, rough endoplasmic reticulum, and cytosol. Immunolocalization of anti-LDH-5 (M4) antibodies is indicated by the 15 nm gold particles. Note presence of LDH-5 in mitochondria and surrounding matrix and organelles. (Magnification, ×58,300; scale bar = 400 nm.)
Figure 3.
Electron micrograph of conventionally processed rat left ventricle. Immunolocalization of anti-LDH-5 (M4) antibodies is shown by the 15 nm gold particles. Note presence of LDH-5 in mitochondria and surrounding matrix and myofibrils and cytosol. (Final magnification, ×60,500; scale bar = 400 nm.)
Figure 4.
Electron micrograph of conventionally processed rat left ventricle. Immunolocalization of anti-LDH-1 (H4) antibodies is shown by the 15 nm gold particles. Note abundance of LDH-1 in mitochondria. (Final magnification, ×67,000; scale bar = 300 nm.)
Insertion of mitochondrial proteins, enzymes, and constituents is known to require N-terminal sequences permitting transmembrane movement. However, we examined the GenBank database and have been unable to identify LDH sequences with appropriate characteristics for transmembrane movement. Thus, at present we are unable to explain appearance of LDH as a constituent of either the mitochondrial inner membrane or matrix.
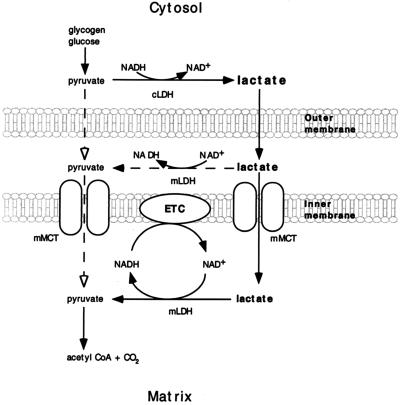
Probable locations of mitochondrial LDH and MCT make it possible to conceive a functional orientation of the two proteins (Fig. 5). Because the cytosolic concentration of lactate exceeds that of pyruvate by 10-fold or more, according to this model of an intracellular lactate shuttle, lactate is the monocarboxylate that enters the mitochondrial intermembrane space. The outer mitochondrial membrane is known to be more highly permeable to a variety of substances than is the inner membrane (36). Similarly, even though the mMCT has a higher affinity for pyruvate than lactate (33–35), by virtue of mass action lactate will be the predominant species entering the matrix. In the matrix, altered Redox during high tricarboxylic acid cycle and electron transport chain flux rates can effect the lactate-pyruvate conversion.
Figure 5.
Depiction of the functional relationship between mitochondrial LDH and mMCT in operation of the intracellular lactate shuttle. The predominant monocarboxylate entering the mitochondrial intermembrane space is lactate. Entry of lactate and pyruvate into the mitochondrial matrix is facilitated by mMCT. Thus, lactate enters mitochondria; lactate is oxidized to pyruvate via mitochondrial LDH when mitochondrial Redox decreases, and pyruvate is oxidized via the tricarboxylic acid cycle and electron transport chain (ETC).
The lactate shuttle hypothesis (15) posited a role of muscle fiber type heterogeneity in lactate exchange among tissues. This supposition was based on the known lactate concentration and mitochondrial density differences between types I and IIB muscle fibers (29, 37). Subsequently, studies on isolated sarcolemmal vesicles (38) showed that sarcolemmal transporters facilitate lactate flux according to concentration and pH gradients. More recently, candidates for seven putative cell membrane monocarboxylate transporters have been cloned and sequenced (39, 40). Based on current evidence, we believe that this scenario of lactate flux between glycolytic and oxidative fibers holds, even if no recruitment occurs. Given similar glycolytic rates in glycolytic and oxidative muscle fibers, glycolytic fibers will tend to accumulate and release lactate because of lesser mitochondrial density. In contrast, highly oxidative fibers will act as lactate sinks because of greater mitochondrial content.
Although the model of an intracellular lactate shuttle (16) is a unique proposal for mammalian liver and striated muscle metabolism, the paradigm is apparently not unique in nature. Mitochondrial matrix LDH is well characterized in sperm of boar (41), mouse, rat, and rabbit (42), and pyruvate-lactate shuttles have been proposed for these systems. Additionally, LDH has a predominant mitochondrial localization in placental trophoblast cells (43). Thus, parallel models of mitochondrial lactate oxidation involving mitochondrial LDH exist in cells of the same and related phylogenies. Further, in Saccharomyces cerevisiae Flavocytochrome b2 is a soluble l-lactate cytochrome c oxidoreductase found in the mitochondrial intermembrane space (44) that couples lactate dehydrogenation to reduction of cytochrome c (45). Hence, S. cerevisiae readily consume and oxidize lactate by a mechanism analogous to that which we propose for mammalian striated muscle and liver mitochondria.
In conclusion, results confirm the presence of mitochondrial LDH (30–32) and support a role for mitochondrial LDH in tissue lactate clearance and oxidation. By virtue of this role, long-standing problems of understanding why fully oxygenated cells readily form lactate are obviated. Glycolysis inevitably results in cytosolic lactate production because the equilibrium constant for LDH is far in the direction of product and the free energy change (ΔG°′ = −6 kcal/mol) is large. However, mitochondrial lactate oxidation can balance cytosolic production, with resultant being zero cellular net lactate release. Glycogenolysis and glycolysis in excess of the corresponding rate of mitochondrial PDH activity results in muscle lactate accumulation and net release regardless of absolute flux rates or state of tissue oxygenation (1, 7). For example, epinephrine stimulation of glycogenolysis in resting muscle results in lactate release during constant or elevated oxygen consumption (46). Moreover, in a respiratory steady state, elevation of arterial lactate concentration results in a switch from muscle lactate net release to uptake as well as suppression of glucose uptake because exogenous lactate substitutes for endogenous (8, 10). Further, under conditions of low glycolytic flux, resting mixed skeletal muscle will release lactate on a net basis (1, 2, 7), whereas working myocardium with a high glycolytic flux rate will consume and oxidize lactate on a net basis (11). Results are consistent with the presence of an intracellular lactate shuttle.
Acknowledgments
This work was supported by National Institutes of Health Grants DK19577 and AR42906. J.P.S. was supported by a Howard Hughes Medical Institute Summer Research Fellowship.
ABBREVIATIONS
- LDH
lactate dehydrogenase
- CINN
α-cyano-4-hydroxycinnamate
- MCT
monocarboxylate transport inhibitor
- mMCT
mitochondrial MCT
- OX
oxamate
References
- 1.Brooks G A, Butterfield G E, Wolfe R R, Groves B M, Mazzeo R S, Sutton J R, Wolfel E E, Reeves J T. J Appl Physiol. 1991;71:333–341. doi: 10.1152/jappl.1991.71.1.333. [DOI] [PubMed] [Google Scholar]
- 2.Stanley W, Gertz E, Wisneski J, Morris D, Neese R, Brooks G. J Appl Physiol. 1986;60:1116–1120. doi: 10.1152/jappl.1986.60.4.1116. [DOI] [PubMed] [Google Scholar]
- 3.Donovan C M, Brooks G A. Am J Physiol. 1983;244:E83–E92. doi: 10.1152/ajpendo.1983.244.1.E83. [DOI] [PubMed] [Google Scholar]
- 4.Depocas F, Minaire Y, Chatonnet J. Can J Physiol Pharmacol. 1969;47:603–610. doi: 10.1139/y69-106. [DOI] [PubMed] [Google Scholar]
- 5.Mazzeo R S, Brooks G A, Schoeller D A, Budinger T F. J Appl Physiol. 1986;60:232–241. doi: 10.1152/jappl.1986.60.1.232. [DOI] [PubMed] [Google Scholar]
- 6.Brooks G A, Wolfel E E, Groves B M, Bender P R, Butterfield G E, Cymerman A, Mazzeo R S, Sutton J R, Wolfe R R, Reeves J T. J Appl Physiol. 1992;72:2435–2445. doi: 10.1152/jappl.1992.72.6.2435. [DOI] [PubMed] [Google Scholar]
- 7.Brooks G A, Wolfel E E, Butterfield G E, Cymerman A, Roberts A C, Mazzeo R S, Reeves J T. Am J Physiol. 1998;275:R1192–R1201. doi: 10.1152/ajpregu.1998.275.4.R1192. [DOI] [PubMed] [Google Scholar]
- 8.Gladden L B, Crawford R E, Webster M J. Am J Physiol. 1994;266:R1095–R1101. doi: 10.1152/ajpregu.1994.266.4.R1095. [DOI] [PubMed] [Google Scholar]
- 9.Watt P W, MacLennan P A, Hundal H S, Kuret C M, Rennie M J. Biochim Biophys Acta. 1988;944:213–222. doi: 10.1016/0005-2736(88)90434-8. [DOI] [PubMed] [Google Scholar]
- 10.Richter E A, Kiens B, Saltin B, Christensen N J, Savard G. Am J Physiol. 1988;254:E555–E561. doi: 10.1152/ajpendo.1988.254.5.E555. [DOI] [PubMed] [Google Scholar]
- 11.Gertz E W, Wisneski J A, Neese R, Bristow J D, Searle G L, Hanlon J T. Circulation. 1981;63:1273–1279. doi: 10.1161/01.cir.63.6.1273. [DOI] [PubMed] [Google Scholar]
- 12.Kopperschlager G, Kirchberger J. J Chromatogr B. 1996;684:25–49. doi: 10.1016/0378-4347(96)00133-8. [DOI] [PubMed] [Google Scholar]
- 13.Connett R J, Gayeski T E, Honig C R. Am J Physiol. 1984;246:H120–H128. doi: 10.1152/ajpheart.1984.246.1.H120. [DOI] [PubMed] [Google Scholar]
- 14.Connett R J, Honig C R, Gayeski T E, Brooks G A. J Appl Physiol. 1990;68:833–842. doi: 10.1152/jappl.1990.68.3.833. [DOI] [PubMed] [Google Scholar]
- 15.Brooks G A. In: Proceedings of the First International Congress of Comparative Physiology and Biochemistry. Gilles R, editor. Berlin: Springer; 1985. pp. 208–218. [Google Scholar]
- 16.Brooks G A. Comp Biochem Physiol. 1998;120:89–107. doi: 10.1016/s0305-0491(98)00025-x. [DOI] [PubMed] [Google Scholar]
- 17.Davies K J, Packer L, Brooks G A. Arch Biochem Biophys. 1981;209:539–554. doi: 10.1016/0003-9861(81)90312-x. [DOI] [PubMed] [Google Scholar]
- 18.Makinen M W, Lee C P. Arch Biochem Biophys. 1968;126:75–82. doi: 10.1016/0003-9861(68)90561-4. [DOI] [PubMed] [Google Scholar]
- 19.Brooks G A, Hittelman K J, Faulkner J A, Beyer R E. Am J Physiol. 1971;220:1053–1059. doi: 10.1152/ajplegacy.1971.220.4.1053. [DOI] [PubMed] [Google Scholar]
- 20.Brooks G A, Hittelman K J, Faulkner J A, Beyer R E. Med Sci Sport. 1971;3:72–74. [PubMed] [Google Scholar]
- 21.Chance B, Williams G. Adv Enzymol. 1965;17:65–70. doi: 10.1002/9780470122624.ch2. [DOI] [PubMed] [Google Scholar]
- 22.Halestrap A P. Biochem J. 1974;138:313–316. doi: 10.1042/bj1380313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chretien D, Pourrier M, Bourgeron T, Sene M, Rotig A, Munnich A, Rustin P. Clin Chim Acta. 1995;240:129–136. doi: 10.1016/0009-8981(95)06145-6. [DOI] [PubMed] [Google Scholar]
- 24.Schnaitman C, Greenawalt J W. J Cell Biol. 1968;38:158–175. doi: 10.1083/jcb.38.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hohenberg H, Tobler M, Muller M. J Microsc. 1996;183:133–139. doi: 10.1046/j.1365-2818.1996.820642.x. [DOI] [PubMed] [Google Scholar]
- 26.McDonald K. In: Methods in Molecular Biology. Hajibagheri N, editor. Clifton, NJ: Humana; 1998. [Google Scholar]
- 27.McDonald K. Methods Cell Biol. 1994;41:411–444. doi: 10.1016/s0091-679x(08)60926-7. [DOI] [PubMed] [Google Scholar]
- 28.Molé P A, Van Handel P J, Sandel W R. Biochem Biophys Res Commun. 1978;85:1143–1149. doi: 10.1016/0006-291x(78)90661-7. [DOI] [PubMed] [Google Scholar]
- 29.Baldwin K M, Hooker A M, Herrick S E. Biochem Biophys Res Commun. 1978;83:151–157. doi: 10.1016/0006-291x(78)90410-2. [DOI] [PubMed] [Google Scholar]
- 30.Brandt R B, Laux J E, Spainhour S E, Kline E S. Arch Biochem Biophys. 1987;259:412–422. doi: 10.1016/0003-9861(87)90507-8. [DOI] [PubMed] [Google Scholar]
- 31.Kline E S, Brandt R B, Laux J E, Spainhour S E, Higgins E S, Rogers K S, Tinsley S B, Waters M G. Arch Biochem Biophys. 1986;246:673–680. doi: 10.1016/0003-9861(86)90323-1. [DOI] [PubMed] [Google Scholar]
- 32.Baba N, Sharma H M. J Cell Biol. 1971;51:621–635. doi: 10.1083/jcb.51.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolli R, Nalecz K A, Azzi A. J Biol Chem. 1989;264:18024–18030. [PubMed] [Google Scholar]
- 34.Halestrap A P. Biochem J. 1975;148:85–96. doi: 10.1042/bj1480085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paradies G, Papa S. FEBS Lett. 1975;52:149–152. doi: 10.1016/0014-5793(75)80659-4. [DOI] [PubMed] [Google Scholar]
- 36.Tzagoloff A. Mitochondria. New York: Plenum; 1982. [Google Scholar]
- 37.Baldwin K M, Campbell P J, Cooke D A. J Appl Physiol. 1977;43:288–291. doi: 10.1152/jappl.1977.43.2.288. [DOI] [PubMed] [Google Scholar]
- 38.Roth D A, Brooks G A. Arch Biochem Biophys. 1990;279:377–385. doi: 10.1016/0003-9861(90)90505-s. [DOI] [PubMed] [Google Scholar]
- 39.Garcia C K, Goldstein J L, Pathak R K, Anderson R G, Brown M S. Cell. 1994;76:865–873. doi: 10.1016/0092-8674(94)90361-1. [DOI] [PubMed] [Google Scholar]
- 40.Price N T, Jackson V N, Halestrap A P. Biochem J. 1998;3298:321–328. doi: 10.1042/bj3290321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calvin J, Tubbs P K. Eur J Biochem. 1978;89:315–320. doi: 10.1111/j.1432-1033.1978.tb20929.x. [DOI] [PubMed] [Google Scholar]
- 42.Gallina F G, Gerez de Burgos N M, Burgos C, Coronel C E, Blanco A. Arch Biochem Biophys. 1994;308:515–519. doi: 10.1006/abbi.1994.1072. [DOI] [PubMed] [Google Scholar]
- 43.Jones C J, Fox H. Invest Cell Pathol. 1978;1:217–225. [PubMed] [Google Scholar]
- 44.Daum G, Bohni P C, Schatz G. J Biol Chem. 1982;252:13028–13033. [PubMed] [Google Scholar]
- 45.Daff S, Ingledew W J, Reid G A, Chapman S K. Biochemistry. 1996;35:6351–6357. doi: 10.1021/bi9522561. [DOI] [PubMed] [Google Scholar]
- 46.Stainsby W, Sumners C, Eitzman P. J Appl Physiol. 1987;62:1845–1851. doi: 10.1152/jappl.1987.62.5.1845. [DOI] [PubMed] [Google Scholar]