Abstract
The discovery of circulating fetal nucleic acid in maternal plasma has opened up new possibilities for noninvasive prenatal diagnosis. Thus far, a gender- and polymorphism-independent fetal-specific target that can be used for prenatal screening and monitoring in all pregnant women has not been reported. In addition, the origin of such circulating nucleic acid has remained unclear. Here we provide direct evidence that the placenta is an important source of fetal nucleic acid release into maternal plasma by demonstrating that mRNA transcripts from placenta-expressed genes are readily detectable in maternal plasma. The surprising stability of such placental mRNA species in maternal plasma and their rapid clearance after delivery demonstrate that such circulating mRNA molecules are practical markers for clinical use. The measurement of such plasma mRNA markers has provided a gender-independent approach for noninvasive prenatal gene expression profiling and has opened up numerous research and diagnostic possibilities.
Noninvasive prenatal diagnosis is a long-sought goal in human genetics. Recent interest in cell-free DNA in plasma and serum (1, 2) has led to the discovery of fetal DNA in maternal plasma (3–5). This noninvasive source of fetal nucleic acid has already been shown to be clinically valuable in the prenatal investigation of many conditions, including fetal rhesus D status (6, 7), sex-linked diseases (8), and β thalassemia (9). In addition, quantitative aberrations of fetal DNA have been described in many pathological conditions, including preeclampsia (10, 11), fetal chromosomal aneuploidies (12, 13), and hyperemesis gravidarum (14).
Despite the promising clinical use of fetal DNA in maternal plasma for noninvasive prenatal diagnosis, a number of challenges remain and several fundamental biological issues about this phenomenon are unresolved. First, in studies reporting the quantitative abnormalities involving fetal DNA in maternal plasma, the Y chromosome is commonly used as a fetal-specific marker in women carrying male fetuses (10–14). The use of such Y-specific markers has limited the application of this technology to the 50% of pregnant women who are carrying male fetuses. The eventual routine clinical application of this technology, e.g., as a screening tool for fetal chromosomal aneuploidies (12, 13), will be catalyzed by the development of a gender- and polymorphism-independent fetal nucleic acid marker, which can be used in all pregnancies. Second, the source of fetal DNA in maternal plasma remains unclear. Although it has been suggested that such fetal DNA could have originated from the placenta (4), no empirical proof of this hypothesis has been put forward to date.
Recently, a number of investigators have shown that in addition to DNA, RNA is also present in the plasma of human subjects, particularly those with cancer (15–18). The inherent lability of RNA has made these observations rather surprising. It has been suggested that circulating RNA may be stabilized by being protected in apoptotic bodies (19–22). These observations have led to the demonstration of the presence of fetal RNA in the plasma of pregnant women (23). Thus far, no quantitative information has been reported on this phenomenon. The clearance kinetics of maternal plasma fetal RNA after delivery also is unexplored at present.
In this article, we aim to address each of the unresolved areas outlined above. We reason that if the placenta is a major source of fetal nucleic acids in maternal plasma, then mRNA transcripts from genes expressed in the placenta should be detectable in maternal plasma. We tested this strategy by developing real-time quantitative RT-PCR to mRNA transcripts of two placenta-expressed genes, namely the genes coding for human placental lactogen (hPL) (24) and the β subunit of human chorionic gonadotropin (βhCG) (25). If these transcripts were detectable in maternal plasma, we would also measure the concentrations of such circulating placenta-derived mRNA and their clearance dynamics after delivery. The potential achievement of these goals would allow us to make a significant step toward the development of a universal marker for noninvasive prenatal gene expression profiling of the fetus by maternal plasma analysis.
Materials and Methods
Subjects.
Blood samples (15 ml) were collected with informed consent and Research Ethics Committee approval from healthy women with singleton uncomplicated pregnancies who visited the Department of Obstetrics and Gynecology at the Prince of Wales Hospital.
Processing of Blood Samples.
The blood samples were collected in EDTA-containing and plain tubes, and centrifuged at 1,600 × g for 10 min at 4°C. Plasma and serum were then carefully transferred into plain polypropylene tubes. The serum samples were stored at −20°C for immunoassays for the hPL and βhCG proteins. The plasma samples were recentrifuged at 16,000 × g for 10 min at 4°C, and the supernatants were collected into fresh polypropylene tubes. All placental tissue samples were immediately stored in an RNAlater stabilizing solution (Ambion, Austin, TX) and kept at −80°C until RNA extraction. For the filtration study, plasma samples were divided into three portions: two were individually passed through filters (Millex-GV, Millipore) with pore sizes of either 0.45 or 5 μm, and the other was not subjected to filtration.
RNA Extraction.
For plasma samples, 1.6 ml of plasma was mixed with 2 ml of TRIzol LS reagent (Invitrogen) and 0.4 ml of chloroform (20). For placental tissues, samples were homogenized in Trizol reagent (Invitrogen), and chloroform was then added according to the manufacturer's recommendations. The mixture was centrifuged at 11,900 × g for 15 min at 4°C, and the aqueous layer was transferred into new tubes. One volume of 70% ethanol was added to 1 vol of the aqueous layer. The mixture was then applied to an RNeasy minicolumn (RNeasy mini kit, Qiagen) and was processed according to the manufacturer's recommendations. Total RNA was eluted with 30 μl of RNase-free water and stored at −80°C. DNase treatment was carried out to remove any contaminating DNA (RNase-Free DNase Set, Qiagen).
Real-Time Quantitative RT-PCR.
One-step real-time quantitative RT-PCR was used for all mRNA quantitation (20). The primers for all of the hPL, βhCG, and GAPDH RT-PCR assays were intron-spanning. The hPL primer sequences were 5′-CATGACTCCCAGACCTCCTTC-3′ (sense) and 5′-TGCGGAGCAGCTCTAGATTG-3′ (antisense), and the dual-labeled fluorescent probe was 5′-(FAM)TTCTGTTGCGTTTCCTCCATGTTGG(TAMRA)-3′ (FAM, 6-carboxyfluorescein; TAMRA, 6-carboxy-N,N,N′,N′-tetramethylrhodamine). The βhCG-primer sequences were 5′-CTACTGCCCCACCATGACCC-3′ (sense) and 5′-TGGACTCGAAGCGCACATC-3′ (antisense), and the dual-labeled fluorescent probe was 5′-(FAM)CCTGCCTCAGGTGGTGTGCAACTAC(TAMRA)-3′. Calibration curves for hPL and βhCG quantifications were prepared by serial dilutions of HPLC-purified single-stranded synthetic DNA oligonucleotides (Genset Oligos, Singapore) specifying the hPL and βhCG amplicons, respectively, with amounts ranging from 1 × 107 to 1 × 101 copies. These assays were able to detect 100 copies of the respective calibrator targets. Absolute concentrations of hPL and βhCG mRNA were expressed as copies per ml of plasma. Previous data have shown that such single-stranded oligonucleotides reliably mimic the products of the reverse transcription step and produce calibration curves that are identical to those obtained by using T7-transcribed RNA (26). The sequences of the synthetic DNA oligonucleotides for hPL and βhCG calibrations were 5′-TGCGGAGCAGCTCTAGATTGGATTTCTGTTGCGTTTCCTCCATGTTGGAGGGTGTCGGAATAGAGTCTGAGAAGCAGAAGGAGGTCTGGGAGTCATGC-3′ and 5′-GATGGACTCGAAGCGCACATCGCGGTAGTTGCACACCACCTGAGGCAGGGCCGGCAGGACCCCCTGCAGCACGCGGGTCATGGTGGGGCAGTAGCC-3′, respectively. A calibration curve for GAPDH quantification was prepared as described, with results expressed in picograms per milliliter of plasma (20).
The RT-PCRs were set up according to the manufacturer's instructions (EZ rTth RNA PCR reagent set, Applied Biosystems) in a reaction volume of 50 μl. The fluorescent probes (Genset Oligos) were used at concentrations of 100 nM. The PCR primers (Genset Oligos) were used at a concentration of 300 nM for hPL and 200 nM for both βhCG and GAPDH. Extracted plasma RNA (5 μl) and 0.1 ng of extracted total placental RNA were used for amplification. Each sample was analyzed in duplicate, and the corresponding calibration curve was run in parallel for each analysis. Samples were also tested to ensure they were negative for DNA by replacing the rTth polymerase with the AmpliTaq Gold enzyme (Applied Biosystems). No amplification was observed for this control analysis, indicating the specificity of the assays for the respective mRNA. Multiple negative water blanks were also included in every analysis.
The thermal profile used for the hPL, βhCG, and GAPDH analysis was as follows: The reaction was initiated at 50°C for 2 min for the included uracil N-glycosylase to act, followed by reverse transcription at 60°C for 30 min. After a 5-min denaturation at 95°C, 40 cycles of PCR were carried out by using denaturation at 94°C for 20 s and 1 min of annealing/extension at 56°C, 58°C, and 60°C for hPL, βhCG, and GAPDH, respectively.
Replicate RNA extraction and RT-PCR analysis indicated that the coefficients of variation of threshold cycle (Ct) values of the analytical systems for hPL and βhCG mRNA were 2.3% and 3.2%, respectively.
Protein Assays.
The hPL and hCG protein concentrations were determined in maternal serum by an RIA (Diagnostic Products, Los Angeles) and an electrochemiluminescence immunoassay (Roche modular E170), respectively.
Results
Detectability and Stability of Placental mRNA in Maternal Blood.
We obtained peripheral blood samples from 10 pregnant women (gestational age, 7–14 wk). In half of the blood sample from each subject, plasma harvesting and RNA extraction were performed immediately on arrival at the laboratory (within 1 h of venesection). To test whether hPL and βhCG mRNA transcripts were detectable in maternal plasma, we analyzed the extracted RNA by using the respective real-time RT-PCR assays. We observed hPL and βhCG mRNA signals in all 10 plasma samples. These results demonstrated that placental mRNA was indeed detectable in the plasma of pregnant women. As a positive control, GAPDH mRNA was also detectable in all of these plasma samples.
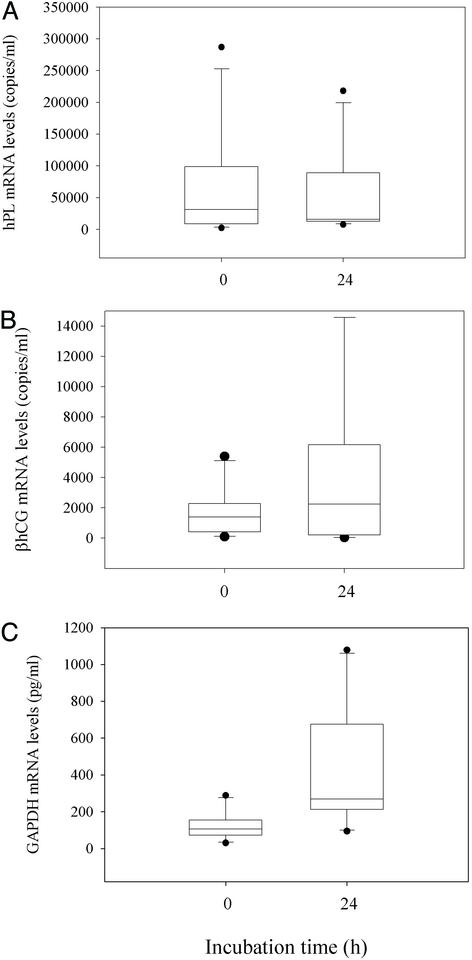
To investigate the stability of placental mRNA in maternal blood, we left the remaining aliquot from each of these maternal blood samples for 24 h at room temperature. We then extracted RNA from these samples and measured the levels of hPL, βhCG, and GAPDH transcripts. No significant difference was observed in the levels of hPL and βhCG mRNA transcripts, whereas the GAPDH mRNA was significantly higher in samples that had been left at room temperature for 24 h than those that had been processed immediately (Wilcoxon test, P = 0.770 for the hPL study, Fig. 1A; P = 0.275 for the βhCG study, Fig. 1B; P < 0.05 for the GAPDH study, Fig. 1C). These results indicate that hPL and βhCG mRNA in plasma was stable for up to 24 h at room temperature.
Figure 1.
Stability of placenta-derived mRNA in maternal blood. Box plots of mRNA levels in maternal plasma before and after incubation of the blood sample at room temperature for 24 h. (A) hPL. (B) βhCG. (C) GAPDH. The lines inside the boxes denote the medians. The boxes mark the interval between the 25th and 75th percentiles. The whiskers denote the interval between the 10th and 90th percentiles. The filled circles mark the data points outside the 10th and 90th percentiles.
Filtration Studies.
To exclude the possibility that the placental mRNA signals that we observed were coming from residual fetal cells left in the centrifuged plasma (27), e.g., trophoblasts, we filtered plasma samples from 14 pregnant women (gestational age, 7–14 wk) through 5-μm filters. We observed no significant change in maternal plasma hPL, βhCG, and GAPDH mRNA levels in the pre- and postfiltration samples (Fig. 2). The results suggest that these mRNA transcripts were not contained within intact cells, which would not have passed through the 5-μm filters.
Figure 2.
Particle-associated nature of placenta-derived mRNA in maternal plasma. Box plots show plasma mRNA levels of pregnant women after different pore-sized filtration treatments. (A) hPL. (B) βhCG. (C) GAPDH. The lines inside the boxes denote the medians. The boxes mark the interval between the 25th and 75th percentiles. The whiskers denote the interval between the 10th and 90th percentiles. The filled circles mark the data points outside the 10th and 90th percentiles.
The surprising stability of placental mRNA in maternal plasma prompted us to test whether such mRNA species might be contained inside subcellular particles. We thus passed these plasma samples through 0.45-μm filters. We observed a clearly observable reduction in hPL, βhCG, and GAPDH mRNA levels after this 0.45-μm filtration step (Fig. 2). Statistical analysis on data obtained before and after filtration through 5- and 0.45-μm filters shows a statistically significant difference (Friedman test, n = 14, P < 0.001, Fig. 2). Pairwise analysis confirms that statistically significant difference is detected between the 5-μm and 0.45-μm filtered groups (Dunn's test, P < 0.05 for hPL, βhCG, and GAPDH, Fig. 2). Overall, the plasma hPL and βhCG mRNA levels decreased by median values of 5.5-fold (interquartile range, 5.2–15) and 3.8-fold (interquartile range, 2.8–4.9), respectively, when comparing paired samples from the 5- and 0.45-μm filtered groups.
Variation of Circulating Placental mRNA with Gestational Age.
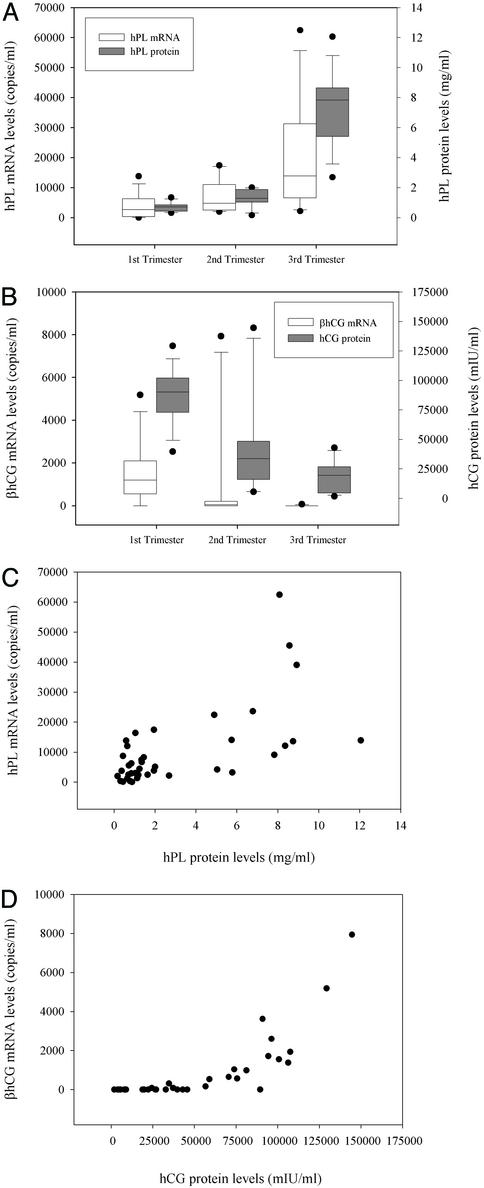
We obtained plasma samples from 39 pregnant women at various stages of gestation. We detected hPL mRNA in 100% (39 of 39) of all plasma tested in all three trimesters of pregnancy. For βhCG mRNA, the detection rates were 100% (14/14) for first- trimester (gestational age, 7–14 wk) samples, 42% (5/12) for second-trimester (gestational age, 15–23 wk) samples, and 7.7% (1/13) for third-trimester (gestational age, 35–41 wk) samples. In the first-trimester plasma samples, the median levels of hPL and βhCG mRNA were 2,671 (interquartile range, 375–6,217) and 1,205 copies per ml (interquartile range, 566–1,927), respectively (Fig. 3 A and B). The median levels of plasma hPL and βhCG mRNA from second-trimester pregnancies were 4,784 (interquartile range, 2,679–10,139) and 0 copies per ml (interquartile range, 0–125), respectively. In the third-trimester plasma samples, the median levels of plasma hPL and βhCG mRNA were 13,869 (interquartile range, 7,829–27,434) and 0 copies per ml (interquartile range, 0–0), respectively. Overall, circulating hPL and βhCG mRNA levels show an increasing and a decreasing trend, respectively, with gestational age. The corresponding levels of hPL and βhCG proteins were also determined (Fig. 3 A and B). The overall gestational variation of circulating hPL and βhCG mRNA shows a resemblance to the trends exhibited by the corresponding proteins (Pearson correlation analysis, r = 0.622 and P < 0.001 for hPL and r = 0.784 and P < 0.001 for βhCG, Fig. 3 C and D).
Figure 3.
Levels of placenta-derived mRNA in maternal plasma in normal pregnancy. (A) Box plot of hPL mRNA in maternal plasma and protein levels in maternal serum at various stages of gestation. (B) Box plot of βhCG mRNA in maternal plasma and hCG protein levels in maternal serum at various stages of gestation. The lines inside the boxes denote the medians. The boxes mark the interval between the 25th and 75th percentiles. The whiskers denote the interval between the 10th and 90th percentiles. The filled circles mark the data points outside the 10th and 90th percentiles. (C) Correlation between hPL mRNA in maternal plasma and protein levels in maternal serum. (D) Correlation between βhCG mRNA in maternal plasma and protein levels (IU, international units) in maternal serum.
Rapid Clearance of Placental mRNA from Maternal Plasma After Delivery.
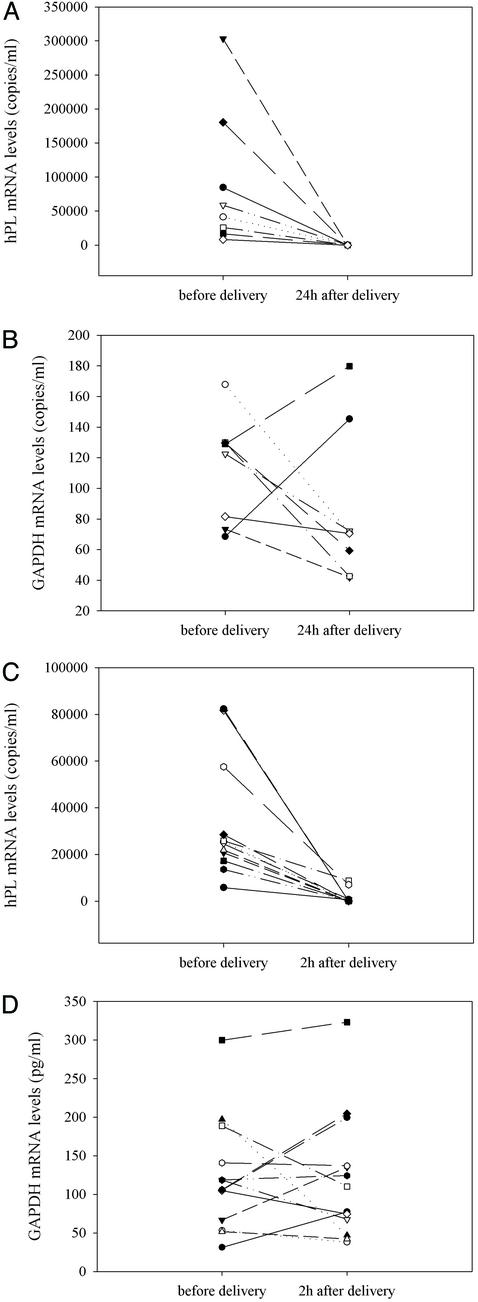
We next investigated whether delivery would result in the clearance of placental mRNA from maternal plasma. We chose hPL mRNA as our target because of the relative abundance of hPL mRNA in maternal plasma during the last trimester of pregnancy (gestational age of studied subjects, 38–42 wk). In eight predelivery plasma samples, the median level of hPL mRNA transcripts was 50,004 copies per ml. hPL mRNA was not detected in any of the postpartum samples 24 h after delivery (Fig. 4A). As a control, GAPDH mRNA was detected in all pre- and postdelivery plasma samples (Fig. 4B). No systematic alteration in maternal plasma GAPDH mRNA levels was observed (Wilcoxon test, P = 0.313). We next investigated whether clearance of circulating hPL mRNA might be observable if a shorter postpartum time point (2 h) were studied. In 13 subjects recruited for this second study, the median predelivery level of hPL mRNA was 24,499 copies per ml (Fig. 4C). Nine of the 13 women had no detectable plasma placental RNA by 2 h postpartum. The remaining subjects had ≈66–97% of maternal plasma fetal RNA cleared by 2 h after delivery. We detected GAPDH mRNA in all plasma samples, thus demonstrating the quality of the samples (Fig. 4D). No systematic alteration in maternal plasma GAPDH mRNA levels was observed (Wilcoxon test, P = 1.000).
Figure 4.
Clearance of fetus-derived mRNA from maternal plasma after delivery. (A and B) Maternal plasma hPL mRNA levels (A) and maternal plasma GAPDH mRNA levels (B) before delivery and 24 h after delivery. The results of eight plasma samples are shown, and each line represents one plasma sample obtained from one subject. (C and D) Maternal plasma hPL mRNA levels (C) and maternal plasma GAPDH mRNA levels (D) before delivery and 2 h after delivery. The results of 13 plasma samples are shown, and each line represents one plasma sample obtained from one subject.
Unidirectional Transfer of hPL mRNA Across the Placenta.
To investigate whether placental mRNA transfer was a bidirectional process (i.e., from the placenta to both the maternal and fetal circulations), we studied six women who delivered by cesarean section (gestational age, 37–39 wk). From each case, we collected maternal peripheral blood, placental tissues, and umbilical cord blood. The interval between maternal blood and cord blood sampling was ≈10 min. We measured hPL and GAPDH mRNA levels in mRNA extracted from maternal plasma, placental tissues, and cord plasma (Table 1). hPL was chosen because the data presented above demonstrate the relatively high level of hPL mRNA in maternal plasma during the third trimester of pregnancy. Conversely, βhCG mRNA was not chosen for this part of the study because its level in maternal plasma was the lowest for this stage of gestation. Interestingly, we could not detect any hPL mRNA in any cord plasma sample. In contrast, we detected hPL mRNA in every maternal plasma and placental RNA sample. As a positive control, GAPDH mRNA was detected in every sample analyzed. Taken together, these data indicate that the transfer of hPL mRNA was unidirectional, from the placenta into the maternal plasma, with no detectable reverse transfer from the placenta into cord plasma.
Table 1.
Quantitative analysis of hPL mRNA in the placenta, maternal plasma, and cord plasma
| Sample | Placental tissues, copies/pg
|
Maternal plasma, copies/ml
|
Cord plasma, copies/ml
|
|||
|---|---|---|---|---|---|---|
| hPL | GAPDH | hPL | GAPDH | hPL | GAPDH | |
| 1 | 62,026,500 | 290 | 288,470 | 171 | 0 | 413 |
| 2 | 14,739,140 | 100 | 119,850 | 58 | 0 | 450 |
| 3 | 78,966,310 | 310 | 152,080 | 143 | 0 | 420 |
| 4 | 61,374,600 | 220 | 24,050 | 83 | 0 | 300 |
| 5 | 59,118,610 | 340 | 83,440 | 56 | 0 | 356 |
| 6 | 90,956,480 | 260 | 104,600 | 139 | 0 | 619 |
Values are given as copies per ml of plasma, except for the values of hPL and GAPDH in placental tissues, which are given as copies per pg of total placental RNA.
Discussion
In this study we have demonstrated that mRNA transcripts derived from the placenta are detectable in maternal plasma during pregnancy and rapidly cleared after delivery. These results represent the direct demonstration that the placenta is an important organ for releasing fetal RNA into maternal plasma. The present quantitative study of fetal RNA in maternal plasma will form the model for future larger-scale studies aiming at establishing reference ranges. Such reference values can then be compared to data obtained in studies involving pathologies, e.g., preeclampsia and fetal chromosomal aneuploidies.
Our data offer a striking demonstration of the placental origin of maternal plasma hPL mRNA (Table 1). Thus, hPL mRNA was easily detectable in all placental and maternal plasma samples. Conversely, no hPL mRNA was detected in any of the cord plasma samples. This latter observation suggests that the placental barrier is relatively impermeable to the transfer of hPL mRNA from the placenta into the fetal circulation. These results contrast with fetomaternal cellular transfer, which has been shown to be a bidirectional process (28). However, an answer as to whether this apparent unidirectionality of mRNA transfer is a general phenomenon will have to await similar studies using other placental mRNA species.
The data presented in this article demonstrate the feasibility of noninvasive profiling of placental gene expression, just by analyzing a maternal sample. This development has opened numerous avenues for further studies. For example, previous data have indicated that the concentrations of fetal DNA in maternal plasma are increased in several pathological conditions, including preeclampsia (10, 29) and trisomy 21 (12, 13). However, because most investigators have used the Y chromosome, which is present only in male fetuses, as a marker for fetal DNA (10, 12, 13, 29), this approach is not applicable to the 50% of pregnant women carrying female fetuses. The development of placental mRNA markers that can be detected in maternal plasma represents a significant step toward the development of molecular markers that can be used for all pregnant women without being limited by fetal genetic polymorphisms, such as gender. Insight into this possibility is provided by our demonstration of the presence of placental mRNA in all first-trimester samples analyzed in this study. The next important step for plasma RNA research is the systematic evaluation of genes that are known to be expressed in the placenta and testing of their detectability in maternal plasma.
The detection of placental mRNA in maternal plasma is perhaps surprising in view of the inherent instability of RNA. Our data indicate that hPL and βhCG mRNA molecules are very stable in maternal plasma (Fig. 1). This stability suggests that placental mRNA markers are practical molecular markers for clinical use. The data for GAPDH, on the other hand, suggest that GAPDH mRNA is released from the blood cells when blood samples are left for 24 h at room temperature (Fig. 1C). This latter observation is consistent with data from our group, which implicate hematopoietic cells as a predominant source of nucleic acid in plasma (30).
Previously published data have shown that most of the plasma GAPDH mRNA molecules in healthy subjects and cancer patients are associated with subcellular particulate matter (20). Data presented in this article demonstrate that this “particality” extends to placental mRNA species in the plasma of pregnant women. Thus, most of the hPL and βhCG mRNA species in maternal plasma appeared to be associated with subcellular particles. This association may be responsible for the unexpected stability of placental mRNA in maternal plasma. The precise nature of such particles requires further elucidation. It is also possible that the molecular characteristics of such particles might change in relation with certain physiological or pathological parameters.
Our data indicate a correlation between the plasma mRNA levels of hPL and βhCG and the corresponding protein levels at various gestational ages (Fig. 3). However, because the mechanisms of release of protein and their corresponding mRNA are likely to be very different for many proteins, we believe that the availability of plasma mRNA-based markers will open many new investigative and diagnostic opportunities. Although conventional protein markers have the advantage of widespread availability and are relatively inexpensive and widely validated, plasma RNA markers do have a number of theoretical advantages: First, one method, involving mRNA extraction and RT-PCR analysis, can be used for many, if not all, plasma RNA markers. For protein-marker detection, on the other hand, many technical parameters may have to be specifically considered for a particular marker and potential technical problems, such as crossreactivity of the Abs used in the respective immunoassays (31), have to be considered on a case-by-case basis. Second, the use of plasma RNA markers may increase the number of markers that one can use for prenatal monitoring, because certain mRNA species may not be coding for a protein product. One example is the H19 gene, expressed in the placenta (32), for which no protein product has been identified (33). Third, the advent of microarray technology (34) will also allow the very rapid development of new plasma RNA markers, which can be used for prenatal monitoring. We thus envision that numerous exciting diagnostic applications will be forthcoming over the next few years based on plasma RNA analysis, allowing noninvasive monitoring of fetal gene expression and ultimately making prenatal testing safer and more acceptable to pregnant women.
Finally, because hPL and βhCG are both hormones, our data may have broader implications in the field of endocrinology. If our results can be generalized to other hormone systems, then a radically new approach for studying and assessing endocrinological disorders may be possible.
Acknowledgments
This project was supported by the Innovation and Technology Fund (AF/90/99).
Abbreviations
- hPL
human placental lactogen
- βhCG
β subunit of human chorionic gonadotropin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 4360.
References
- 1.Nawroz H, Koch W, Anker P, Stroun M, Sidransky D. Nat Med. 1996;2:1035–1037. doi: 10.1038/nm0996-1035. [DOI] [PubMed] [Google Scholar]
- 2.Chen X Q, Stroun M, Magnenat J L, Nicod L P, Kurt A M, Lyautey J, Lederrey C, Anker P. Nat Med. 1996;2:1033–1035. doi: 10.1038/nm0996-1033. [DOI] [PubMed] [Google Scholar]
- 3.Lo Y M D, Corbetta N, Chamberlain P F, Rai V, Sargent I L, Redman C W, Wainscoat J S. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi D W. Am J Hum Genet. 1998;62:763–764. doi: 10.1086/301809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pertl B, Bianchi D W. Obstet Gynecol. 2001;98:483–490. doi: 10.1016/s0029-7844(01)01195-4. [DOI] [PubMed] [Google Scholar]
- 6.Lo Y M D, Hjelm N M, Fidler C, Sargent I L, Murphy M F, Chamberlain P F, Poon P M, Redman C W, Wainscoat J S. N Engl J Med. 1998;339:1734–1738. doi: 10.1056/NEJM199812103392402. [DOI] [PubMed] [Google Scholar]
- 7.Faas B H, Beuling E A, Christiaens G C, von dem Borne A E, van der Schoot C E. Lancet. 1998;352:1196. doi: 10.1016/s0140-6736(05)60534-x. (lett.). [DOI] [PubMed] [Google Scholar]
- 8.Costa J M, Benachi A, Gautier E. N Engl J Med. 2002;346:1502. doi: 10.1056/NEJM200205093461918. (lett.). [DOI] [PubMed] [Google Scholar]
- 9.Chiu R W K, Lau T K, Leung T N, Chow K C K, Chui D H K, Lo Y M D. Lancet. 2002;360:998–1000. doi: 10.1016/s0140-6736(02)11086-5. [DOI] [PubMed] [Google Scholar]
- 10.Lo Y M D, Leung T N, Tein M S, Sargent I L, Zhang J, Lau T K, Haines C J, Redman C W. Clin Chem. 1999;45:184–188. [PubMed] [Google Scholar]
- 11.Zhong X Y, Laivuori H, Livingston J C, Ylikorkala O, Sibai B M, Holzgreve W, Hahn S. Am J Obstet Gynecol. 2001;184:414–419. doi: 10.1067/mob.2001.109594. [DOI] [PubMed] [Google Scholar]
- 12.Lo Y M D, Lau T K, Zhang J, Leung T N, Chang A M, Hjelm N M, Elmes R S, Bianchi D W. Clin Chem. 1999;45:1747–1751. [PubMed] [Google Scholar]
- 13.Zhong X Y, Burk M R, Troeger C, Jackson L R, Holzgreve W, Hahn S. Prenatal Diagn. 2000;20:795–798. doi: 10.1002/1097-0223(200010)20:10<795::aid-pd897>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 14.Sekizawa A, Sugito Y, Iwasaki M, Watanabe A, Jimbo M, Hoshi S, Saito H, Okai T. Clin Chem. 2001;47:2164–2165. [PubMed] [Google Scholar]
- 15.Lo K W, Lo Y M D, Leung S F, Tsang Y S, Chan L Y, Johnson P J, Hjelm N M, Lee J C, Huang D P. Clin Chem. 1999;45:1292–1294. [PubMed] [Google Scholar]
- 16.Kopreski M, Benko F A, Kwak L W, Gocke C D. Clin Cancer Res. 1999;5:1961–1965. [PubMed] [Google Scholar]
- 17.Chen X Q, Bonnefoi H, Pelte M F, Lyautey J, Lederrey C, Movarekhi S, Schaeffer P, Mulcahy H E, Meyer P, Stroun M, et al. Clin Cancer Res. 2000;6:3823–3826. [PubMed] [Google Scholar]
- 18.Silva J M, Dominguez G, Silva J, Garcia J M, Sanchez A, Rodriguez O, Provencio M, Espana P, Bonilla F. Clin Cancer Res. 2001;7:2821–2825. [PubMed] [Google Scholar]
- 19.Hasselmann D O, Rappl G, Tilgen W, Reinhold U. Clin Chem. 2001;47:1488–1489. [PubMed] [Google Scholar]
- 20.Ng E K, Tsui N B, Lam N Y, Chiu R W, Yu S C, Wong S C, Lo E S, Rainer T H, Johnson P J, Lo Y M D. Clin Chem. 2002;48:1212–1217. [PubMed] [Google Scholar]
- 21.Tsui N B, Ng E K, Lo Y M D. Clin Chem. 2002;48:1647–1653. [PubMed] [Google Scholar]
- 22.Anker P, Stroun M. Clin Chem. 2002;48:1210–1211. [PubMed] [Google Scholar]
- 23.Poon L L, Leung T N, Lau T K, Lo Y M D. Clin Chem. 2000;46:1832–1834. [PubMed] [Google Scholar]
- 24.Barrera-Saldana H A. Gene. 1998;211:11–18. doi: 10.1016/s0378-1119(98)00092-4. [DOI] [PubMed] [Google Scholar]
- 25.Reimer T, Koczan D, Briese V, Friese K, Richter D, Thiesen H J, Jeschke U. Mol Biotechnol. 2000;14:47–57. doi: 10.1385/mb:14:1:47. [DOI] [PubMed] [Google Scholar]
- 26.Bustin S A. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 27.Van Wijk I J, De Hoon A C, Jurhawan R, Tjoa M L, Griffioen S, Mulders M A M, Van Vugt J M G, Oudejans C B M. Clin Chem. 2000;46:729–731. [PubMed] [Google Scholar]
- 28.Lo Y M D, Lo E S, Watson N, Noakes L, Sargent I L, Thilaganathan B, Wainscoat J S. Blood. 1996;88:4390–4395. [PubMed] [Google Scholar]
- 29.Swinkels D W, de Kok J B, Hendriks J C, Wiegerinck E, Zusterzeel P L, Steegers E A. Clin Chem. 2002;48:650–653. [PubMed] [Google Scholar]
- 30.Lui Y Y, Chik K W, Chiu R W, Ho C Y, Lam C W, Lo Y M D. Clin Chem. 2002;48:421–427. [PubMed] [Google Scholar]
- 31.Selby C. Ann Clin Biochem. 1999;36:704–721. doi: 10.1177/000456329903600603. [DOI] [PubMed] [Google Scholar]
- 32.Jinno Y, Ikeda Y, Yun K, Maw M, Masuzaki H, Fukuda H, Inuzuka K, Fujishita A, Ohtani Y, Okimoto T, et al. Nat Genet. 1995;10:318–324. doi: 10.1038/ng0795-318. [DOI] [PubMed] [Google Scholar]
- 33.Juan V, Crain C, Wilson C. Nucleic Acids Res. 2000;28:1221–1227. doi: 10.1093/nar/28.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipshutz R J, Fodor S P, Gingeras T R, Lockhart D J. Nat Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]