Abstract
We report here a cancer drug therapy use of a gene involved in Down's syndrome. Using bioinformatics approaches, we recently predicted Single Minded 2 gene (SIM2) from Down's syndrome critical region to be specific to certain solid tumors. Involvement of SIM2 in solid tumors has not previously been reported. Intrigued by a possible association between a Down's syndrome gene and solid tumors, we monitored SIM2 expression in solid tumors. Isoform-specific expression of SIM2 short-form (SIM2-s) was seen selectively in colon, prostate, and pancreatic carcinomas but not in breast, lung, or ovarian carcinomas nor in most normal tissues. In colon tumors, SIM2-s expression was seen in early stages. Antisense inhibition of SIM2-s expression in a colon cancer cell line caused inhibition of gene expression, growth inhibition, and apoptosis. The administration of the antisense, but not the control, oligonucleotides caused a pronounced inhibition of tumor growth in nude mice with no major toxicity. Our findings provide a strong rationale for the genes-to-drugs paradigm, establish SIM2-s as a molecular target for cancer therapeutics, and may further understanding of the cancer risk of Down's syndrome patients.
With the completion of human genome sequencing efforts, a large number of new genes are likely to be discovered (1, 2). From these vast numbers of new genes, novel diagnostic and therapeutic targets for diseases like cancer are predicted to emerge (3, 4). For analyzing expression of a large number of sequences in diseased and normal tissues, high-throughput gene expression analysis is becoming increasingly useful with the availability of microarray and GeneChip technology (5, 6). A parallel way to initiate a search for genes relevant to cancer is to data mine the sequence databases (3, 7, 8). A large number of expressed sequences from diverse organ-, species-, and disease-derived cDNA libraries are being deposited in various databases in the form of ESTs.
For cancer-specific gene discovery, the Cancer Genome Anatomy Project (CGAP) database of the National Cancer Institute provides a comprehensive collection of expressed sequences in the form of ESTs as well as various data-mining tools to analyze these ESTs (9, 10). Recently we demonstrated that the CGAP database can be harnessed for discovery of cancer-specific genes (11). Using the Digital Differential Display tool of the CGAP database, organ- and tumor-specific genes were discovered, and a unique database encompassing both known and novel ESTs was established (11). One gene from the colon-specific novel EST collection was recently shown by us to be a highly specific colon tumor-related secreted factor, consistent with the bioinformatics prediction (12).
Another gene from this database (UniGene Hs.146186) showed homology to Human Single Minded 2 (SIM2). The SIM2 gene is present in chromosome 21 at the Down's syndrome critical region, which in triplication is associated with diverse phenotypic characteristics of Down's syndrome (13). Patients with Down's syndrome show various neurological symptoms and a high incidence of leukemia (14, 15). Members of the SIM family include SIM1 and SIM2, which map to 6q16.3-q21 and 21q22.2, respectively (16), and belong to a family of transcription factors containing a basic helix-loop-helix motif, two PAS (PER/ARNT/SIM) domains, and the HST (HIF1α/SIM/TRH) domains (17–19). In Drosophila, SIM is a master regulator of fruit fly neurogenesis, regulating the midline gene expression (17, 20). The SIM2 gene exists in two distinct forms, long and short (SIM2-l and SIM2-s), due to alternative splicing (16).
We explored a possible association of the SIM2 gene with solid tumors. SIM2 is expressed in an isoform-specific manner (SIM2-s) in select solid tumors and was detected in early-stage and advanced-colon carcinomas but not in the normal colon. Furthermore, SIM2-s expression was not seen in most normal tissues. Inhibition of SIM2-s by antisense technology in colon cancer cells caused apoptosis in cell culture and inhibition of tumor growth in nude mice. These findings have important implications for the future diagnosis and treatment of specific solid tumors as well as for understanding the cancer risk in Down's syndrome patients.
Materials and Methods
RT-PCR Analysis.
Normal and fetal-tissue cDNAs were from CLONTECH and the Biochain Institute (San Leandro, CA). Leukemia cell line-derived cDNAs were obtained from Geneka Biotechnology, Quebec, Montreal. Tumor and normal tissues were from the Cooperative Human Tissue Network, National Cancer Institute, and random primed cDNAs were synthesized as described (11). Early passage primary kidney and prostate cells were obtained from Clonetics Biowhittaker (Walkersville, MD) and were cultured following the manufacturer's instructions. The exon-specific RT-PCR primers used were SIM2-s1: (sense) 5′-TGGAGGACCGCCTTGTCTACCT-3′, (antisense) 5′-GCCCAAAGCGTGAGGGTTCTGTCT-3′, 619-bp product; SIM2-s2: (sense) 5′-TGGAGGACCGCCTTGTCTACCT-3′, (antisense) 5′-CCGGTGGCTCTGGAGGATTT-3′, 472-bp product; SIM2-l: (sense) 5′-TGCCCTTCGTGCTGCTCAACTACC-3′, (antisense) 5′-AGGAAACCAAGCCCCCAGCA-3′, 484-bp product; SIM1: (sense) 5′-GCTGGTGGAAGAGAGGCATT-3′, (antisense) 5′-TGGAGAACTGACCACACTAT-3′, 246-bp product and actin: (sense) 5′-CACTGTGTTGGCGTACAGGT-3′ and (antisense) 5′-TCATCACCATTGGCAATGAG-3′, 150-bp product. The PCR parameters included 94°C for 7 min followed by a 25 to 40-cycle amplification at 94°C, 45 sec; 60–67°C, 45 sec; and 72°C, 90 sec, with a final extension at 72°C for 10 min. Two independent PCR primer sets were used for SIM2-s analysis. Independent cDNA preparations were tested, and the quality of cDNA preparations was authenticated with actin.
LightCycler PCR.
The RT-PCR primers (designed by using light cycler probe design software, Ver. 1.0, Roche Applied Science, Indianapolis) used were SIM2-s3: (sense) 5′-GCCAGCCAGCGGTGAATGC-3′, (antisense) 5′-GCAAGTTTCCCAAAGCTGAGG-3′, 244-bp product. Titration of the cDNA template concentration and MgCl2 was done in preliminary experiments, and optimum concentrations were used in the reactions. FastStart DNA Master SYBR green I mix (Roche Molecular Biochemicals) was used, which required a 10-min denaturation step, then 45 cycles of amplification at 95°C, 10 sec; 68°C, 10 sec; 72°C, 30 sec; and an acquisition temperature (91°C), 1°C below the melting temperature (Tm) of the product held for 2 sec. This was followed by a melting curve analysis, which consisted of one cycle at 95°C for 1 sec; 65°C, 15 sec; and continuous acquisition reading up to 95°C. The crossover points were determined to quantify the reaction products. Product authenticity was validated by Tm measurement of the different PCR products, crossover point determination, serial dilution of the cDNAs, and agarose gel electrophoresis.
Immunohistochemistry (IHC).
Paraffin sections of the human tumor and normal colon tissues, tumor tissues from the nude mice experiments, and the antisense-treated cells in culture were analyzed by IHC by using affinity-purified rabbit antiserum to a SIM2-s-specific hydrophobic peptide SHGGGWQMETEPSRF (Sigma Genosys, The Woodlands, TX) as described (21). The specificity of IHC was established by excess peptide competition.
Antisense Drug.
The antisense (AS), 5′-GAGAGCAAGAAAGCACAGCAAGCC-3′ and reverse antisense (C), 5′-CCGAACGACACGAAAGAACGAGAG-3′ drugs were synthesized as a second-generation chimera (phosphorothioate-2′-O-methyl RNA chimera) by Oligos Etc. (Wilsonville, OR). Additional control sequences included sense and scrambled oligomers. The antisense sequence was chosen corresponding to the 3′ end within the coding region of the SIM2-s gene, which encodes the short form-unique region. The sequence was blast verified and from the secondary structure, no stem–loop structure was predicted. The oligomers were HPLC purified and purity ascertained by analytical ion exchange HPLC and capillary electrophoresis (>90%). Two independent preparations were tested.
Other Methods.
Detection of fragmented DNA was performed by using the Apoptag Apoptosis Detection Kit (Serologicals, Norcross, GA) according to the manufacturer's specifications. In addition, genomic DNA from the treated cells was separated by agarose electrophoresis and hybridized with 32P-labeled EcoRI restriction-digested normal human DNA. Antisense transfections in vitro were done by using the Lipofectin protocol (Life Technologies, Gaithersburg, MD). Tumorigenicity assays were done by using the MetaMouse model (Anticancer, San Diego). Briefly, NCr nu/nu nonirradiated male and female 5- to 6-wk-old 22- to 24-g body weight mice (n = 10) were injected s.c. with RKO colon carcinoma cell-derived stock tumors (four fragments per mouse), and 48 h later the mice were treated either with vehicle (PBS), antisense (AS), or reverse antisense (C), 1 mg/kg twice weekly s.c. on the contralateral side for 4 wk. The mean body weight and tumor volume (W2X L/2) were calculated on a weekly basis. The tumor volume was assessed by Student's t test with an α = 0.05 (two-sided). Spleen weight was measured at death, and tumors were characterized by histology. One representative experiment is shown.
Results
Specificity of SIM2-s in Solid Tumors.
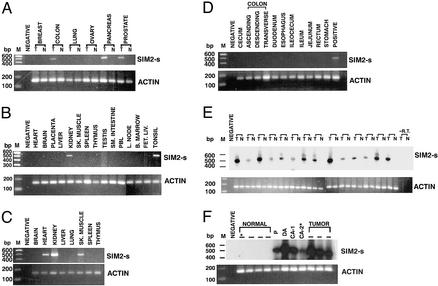
Using SIM2-s exon-specific primers, RT-PCR analysis of the cDNAs derived from a matched set of tumor and normal tissues from breast, colon, lung, ovary, pancreas, and prostate carcinomas was performed. The primer pair used was designed to discriminate between the SIM2-s and SIM2-l forms. Three independent primer pairs were used in the study (see Materials and Methods). The SIM2-s-specific PCR products were seen only in the colon, pancreas, and prostate tumor-derived cDNAs but not in the corresponding normal tissues (Fig. 1A). These results were confirmed by the use of independent sets of cDNAs from multiple patients (not shown). The SIM2–1 form was not detected in these samples (data not shown). Among many normal adult pooled tissues analyzed, SIM2-s expression was seen only in kidney- and tonsil-derived cDNAs (Fig. 1B). In the fetal tissues, SIM2-s was expressed in heart, kidney, and skeletal muscle, consistent with the developmental expression of SIM2-s (16) (Fig. 1C), whereas distinct parts of normal adult colon (pooled tissues) were negative (Fig. 1D). SIM2-s expression was seen in independent colon tumors (T) from 14 different patients (14/14) but not in the corresponding matched normal colon tissue (N) from the same patients. The authenticity of the SIM2-s-specific PCR products was verified by hybridization to a SIM2-s-specific internal oligonucleotide probe (Fig. 1E). Normal colon-derived cDNAs from unmatched pooled tissues were also negative for SIM2-s expression (data not shown). A stage-specific expression of SIM2-s was seen in early adenomas and carcinomas (Fig. 1F). The polyp was a retrospective sample from a colon cancer patient and was positive for SIM2-s expression (Fig. 1F). Another retrospective polyp-derived cDNA from an independent cancer patient was also positive for SIM2-s expression (data not shown). SIM2-s was expressed in diverse colon- (n = 4), prostate- (n = 3), and pancreas- (n = 4) derived cell lines, but not in the cell lines derived from breast (n = 4), ovary (n = 4), or lung (n = 4). These results suggested that SIM2-s is activated in select solid tumors, and that in colon tumors, it is activated at an early stage.
Figure 1.
Expression profile of SIM2-s. Random primed cDNAs were analyzed by RT-PCR. (A) Tumor (T) and normal (N) tissues (matched) from breast, colon, lung, ovary, pancreas, and prostate carcinomas; (B) CLONTECH pooled multiple normal tissue; (C) CLONTECH fetal tissue; (D) CLONTECH digestive tissue; (E) matched tumor and normal tissues from 14 independent colon carcinoma patients; and (F) cDNAs from normal colon, polyp (P), duodenal adenoma (DA) and colon adenoma (CA), and colon tumors were analyzed for SIM2-s and actin expression. PCR products in E and F were hybridized with an internal oligomer probe. *, Matched tissues from the same patient; M, 100-bp ladder; negative, template minus PCR control; positive, colon tumor cDNA; −RT, reverse transcriptase minus control. One representative experiment from three independent experiments is shown.
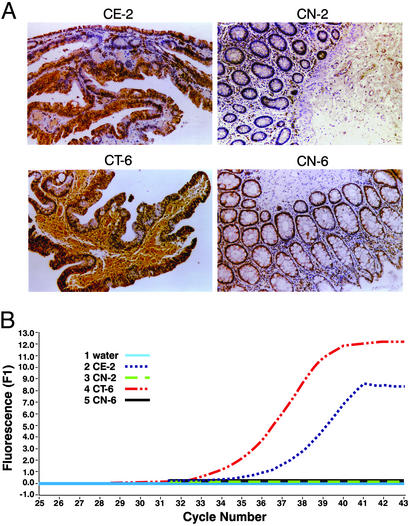
Elevated expression of SIM2-s was seen in the paraffin sections of colon adenoma (CE-2) and carcinoma (CT-6) by IHC, in comparison with the normal surrounding colon tissues (CN-2 and -6) from the same patients (Fig. 2A). Real-time RT-PCR of the RNAs from tumor and normal tissues of the same patients (Fig. 2B) showed quantitative changes (tumor>adenoma>normal) in the expression level of SIM2-s. The authenticity of the SIM2-s PCR product was confirmed by determining the Tm and the crossover point. No SIM2-s-specific PCR products were seen in the normal tissues, consistent with the results of the end point PCR (Fig. 1E). TaqMan quantification was not performed on these samples, because no PCR products were detected in the normal tissues.
Figure 2.
Correlation of SIM2-s expression (mRNA and protein) in colon tumors. Matched specimens of adenoma (CE-2) and corresponding normal (CN-2) and carcinoma (CT-6) and corresponding normal (CN-6) tissues were analyzed by IHC using SIM2-s-specific affinity-purified polyclonal antibodies (A). All sections were counterstained with hematoxylin (×360). Real-time quantitative RT-PCR of the same matched specimens are shown in B.
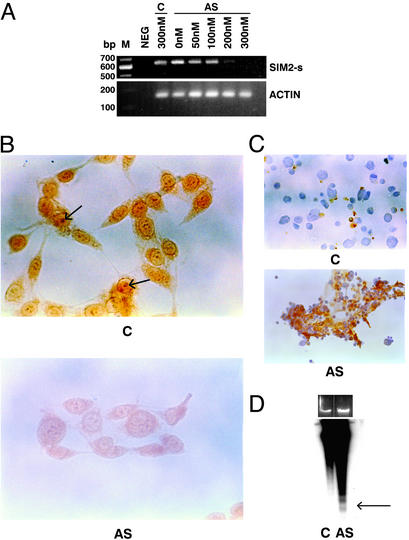
Antisense technology has been effective in blocking the expression of targeted genes and validating drug therapy use of targets (22, 23). To investigate whether the SIM2-s gene has a therapeutic potential, we have used antisense technology to inhibit the gene's function. The RKO colon carcinoma cells were treated with a second-generation (phosphorothioate-2′-O-methyl RNA chimera) SIM2-s antisense. In preliminary experiments, the antisense-treated cells showed rapid inhibition of growth (within 24–48 h), which was dose-dependent (50–300 nM), and the treated cells showed nuclear condensation, suggesting an apoptotic mechanism. Such an effect was not seen with various control sequences. The antisense-treated RKO cells showed inhibition of SIM2-s mRNA in a dose-dependent manner (Fig. 3A) and inhibition of the SIM2-s protein in the nucleus as monitored by IHC (Fig. 3B). The SIM2-s antibody detected a clear nuclear antigen stain (shown by arrow) and a diffused cytoplasmic stain. On antisense treatment, a pronounced inhibition of antigen stain was seen. The nuclear antigen stain was competed with 10-fold excess peptide (data not shown). In preliminary experiments, the SIM2-s antibody was not found to detect the SIM2-s protein in the cell lysates by Western blot. Similarly, a commercially available antibody to SIM2-s also failed to detect the SIM2-s protein in the cell lysates. Efforts are underway to generate a Western-compatible SIM2-s antibody to aid future studies. The antisense-treated cells were Apoptag positive, confirming DNA fragmentation in situ (Fig. 3C), and showed evidence of DNA laddering indicative of apoptosis (Fig. 3D). Slight differences in the amount of DNAs loaded (≈0.5-fold) were seen on the ethidium bromide-stained agarose gel between the control and the antisense-treated cells (Fig. 3D). The autoradiogram shown was deliberately overexposed, and ladder formation was not seen in the control DNA. The SIM2-s antisense also inhibited the growth of other colon cancer-derived cell lines, including HCT116 and SW480 as well as CAPAN-1 pancreatic carcinoma cells and induced apoptosis (data not shown). Treatment of MDA231 breast carcinoma cells or early-passage human colonic smooth muscle cells (both of which were negative for the target gene SIM2-s expression) with the same antisense drug did not cause growth inhibition, suggesting specificity of the antisense drug.
Figure 3.
Antisense efficacy in vitro. (A) RKO cells were treated with either 300 nM of control oligomers (C) or indicated concentrations of the antisense oligomers (AS) using the Lipofectin protocol, and 48 h later the RNA from the treated cells was analyzed by RT-PCR with SIM2-s and actin primers. Neg, template minus PCR control. (B) RKO cells were treated with 300 nM of control (C) or antisense (AS) oligomers for 48 h and the cells were analyzed by IHC by using the SIM2-s-specific antibody. The arrow indicates nuclear stain (×130). (C) The control (C)- or antisense (AS)-treated RKO cells were analyzed 48 h later by using an Apotag kit (×130). (D) Genomic DNA from control (C) or the antisense (AS)-treated RKO cells for 48 h was probed with 32P-labeled EcoR1-digested normal human DNA. Ethidium bromide-stained agarose gel is shown above the autoradiogram. An overexposed version of the autoradiogram is shown. Arrow indicates ladder representing oligosomes.
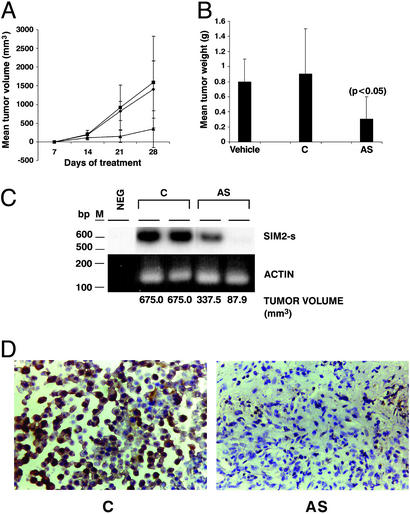
On the basis of these results, the antisense drug was tested by using RKO human colon cancer xenograft in a nude mouse tumorigenicity assay. The nude mice were injected with the RKO colon carcinoma-derived tumors s.c., and the tumor-bearing mice were treated with 1 mg/kg of the antisense drug (AS), control drug (C), or vehicle (PBS) twice weekly s.c. on the contralateral side for 1 mo. Significant tumor inhibition was seen in the antisense drug-treated mice (Fig. 4 A and B). Two independent tumors from the control drug- or the antisense drug-treated mice were analyzed for SIM2-s mRNA expression (Fig. 4C). Inhibition of SIM2-s mRNA was seen in the tumors from antisense-treated mice in comparison with the tumors from the control oligomer-treated mice. The tumor reduction in response to the antisense drug correlated with the reduced expression of the targeted gene. The tumor from the antisense-treated mice showed a pronounced inhibition of SIM2-s expression by IHC compared with the control-treated tumor (Fig. 4D). The confluency difference seen between the control and the antisense-treated tumors (Fig. 4D) is due to the paraffin sections chosen for the IHC analysis. The antisense-treated mice did not show loss of weight, changes in blood cell counts, or splenomegaly, an indicator of nonspecific effects of oligonucleotides (24), showing that the antisense drug was not toxic and was relatively specific. No histological changes in the liver, kidney, or spleen of the treated mice were seen. The antisense-treated tumors were Apoptag positive, consistent with the induction of apoptosis noted in the cell culture model.
Figure 4.
Antisense efficacy in a colon tumor model in vivo. Nude mice were injected with RKO-derived tumors s.c. and 48 h later were treated with the antisense (▴), control (♦), or vehicle (■) at 1 mg/kg, twice weekly s.c. on the contralateral side for 28 days, and the tumor volume was measured weekly (A). Mean tumor weight is shown in B. RNA from two independent tumors from the control (C) or antisense (AS)-injected mice were analyzed by RT-PCR for SIM2-s or actin expression (C). Neg, template minus PCR control; M, 100-bp ladder. The corresponding tumor volume is shown. Tumors from representative control (C) or antisense (AS)-treated mice were analyzed by IHC for SIM2-s expression (D).
Discussion
Human genome sequencing efforts have opened a new era for the discovery of highly specific cancer targets. Bioinformatics approaches and microarray technology are increasingly being used to move the gene sequences closer to the patient's bedside. Reasoning that the discovery of organ- and tumor-specific genes would provide a rationale for both novel diagnostic and therapy targets, we have used the Cancer Genome Anatomy Project database of the National Cancer Institute to create a solid tumor-specific EST database (11). Our recent identification of a colon cancer-specific gene (12) from this database provided a framework to test this database further. We chose another gene that was predicted to be specific to colon cancer. Contrary to the bioinformatics prediction of specificity to colon cancer, we observed that this gene (SIM2-s) was also expressed in pancreas and prostate cancers. This underscores the importance of validating computational predictions with relevant patient-derived materials. Nevertheless, the association of SIM2-s, a Down's syndrome-related gene with cancer, raised interesting possibilities of linking two very different diseases. Hence we investigated the status of SIM2-s in solid tumors.
A putative cancer-related role of the SIM family of genes is their ability to transcriptionally regulate key metabolic enzymes to inactivate carcinogens (25). Binding of carcinogens to the aryl hydrocarbon receptor (AhR), which is kept sequestered in the cytoplasm by HSP90 (26), dissociates HSP90. The ligand-bound AhR is then translocated into the nucleus, where it can dimerize with AhR Nuclear Translocator (ARNT) (27). This complex binds to the xenobiotic response element, present in the promoters of key oxidative enzymes and activates gene transcription (25, 28), thus causing inactivation of the carcinogen. The SIM proteins can inhibit the dimerization of the ligand bound AhR/ARNT complex (29) and hence prevent carcinogen metabolism. It is tempting to hypothesize that the SIM2-s gene undergoes activation (due to mutations, amplifications, or loss of repression) similar to that of oncogenes, tumor suppressor genes, and telomerase (30–32). Preliminary analysis of the Single Nucleotide Polymorphism (SNP) database revealed sim2-s-specific SNPs, supporting the hypothesis of activation of SIM2-s (data not shown).
The precise function and nature of the molecules the SIM2-s gene regulates in the normal and tumor tissues are not known. There is limited availability of reagents such as antibodies, protein, and specific inhibitors for distinct members of the SIM family. The putative transcription factor function of the SIM family of genes suggests transcriptional regulation by individual members of the family. The availability of this specific antisense drug to block the function of the SIM2-s gene opens up new avenues to decipher the putative targets of the SIM2-s gene.
The specificity of SIM2-s expression seen in colon tumors and early-stage colon adenomas and in a retrospective polyp sample from a colon cancer patient suggests activation at early stages of colon cancers, providing a diagnostic utility. Supporting this, in a recent report (21), using a large collection of colon, prostate, and pancreatic tumors and normal tissues, we have demonstrated an elevated expression of SIM2-s in tumors compared with the normal tissues, and a preliminary scoring system was established.
Progression of colon cancer involves activation and loss of expression of distinct genes (33). It is likely that the development of colonic adenomas and carcinomas involves multiple steps in which environmental or endogenous carcinogens induce or promote neoplasia through the accumulation of multiple, specific genetic mutations (34). Genetic predisposition to this process may take the form of inherited defects in control of cellular proliferation as in familial polyposis coli, or genetically determined polymorphism that affects enzyme activities relevant to the production or detoxification of carcinogens (35). Our results suggest SIM2-s as one of the determinants in this cascade.
Inhibition of SIM2-s expression by antisense technology, which induces apoptosis and inhibition of colon tumor growth in nude mice, offers a proof of concept for SIM2-s as a drug therapy target. The antisense-treated cells undergo apoptosis within 24–48 h. The induction of apoptosis we demonstrate in the colon tumor model could be due to a specific block at a distinct stage of the cell cycle or to an induction of differentiation. Efforts are underway to clarify the mechanism of antisense SIM2-s-induced cancer cell death. The half-life of SIM2-s has been shown to be 2 h (27), which can account for the rapidity of the antisense effects. The lack of expression of SIM2-s in most normal cells suggests that such a target would be low in toxicity. The toxic liability that can be anticipated by inhibition of SIM2-s is likely to be renal, because it is expressed in normal adult kidney. In preliminary experiments, treatment of early passage renal epithelial cells or prostate epithelial cells with the SIM2-s antisense drug did not show toxicity (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). We postulate that other members of the SIM family, which are also expressed in the normal kidney, may provide an alternative to SIM2-s function. However, this should be a matter of concern and further study is warranted.
A tantalizing link between Down's syndrome and cancer is the high incidence of leukemias (20-fold) seen in Down's children. Higher risks for acute lymphoblastic leukemia, myelodysplastic syndrome, and acute myeloid leukemia are seen in Down's children (36). Surprisingly, RT-PCR analysis of diverse leukemia-derived cell lines failed to reveal the presence of SIM2-s-specific transcript (see Fig. 6, which is published as supporting information on the PNAS web site). These results suggest that the observed epidemiological link between Down's patients and leukemia may not involve the function of the SIM2-s gene. Analysis of the bone marrow from Down's patients with or without leukemia for SIM2-s expression would clarify this issue. Increased transcription of the genes in the trisomic chromosome 21 may contribute to cancer in Down's patients (37, 38). Oncogenes and tumor suppressor genes present in the chromosome may further contribute to the cancers seen in these patients.
In summary, with the completion of the human genome sequencing efforts, discovery of genes with medicinal value has undergone a paradigm shift. Although in the past, cancer gene discovery followed conventional methods such as mapping the gene, loss of heterozygosity, and model organism studies, the future of cancer gene discovery will emanate from the human genome sequencing efforts. The prediction of SIM2-s specificity using bioinformatics of the human genome and the subsequent validation using tissue repository demonstrate the power of harnessing the human genome. The preclinical proof of concept we further demonstrate by using antisense technology provides one of the first few examples of the genes-to-drugs paradigm. A systematic search of other disease-associated genes for their possible role in cancer may provide additional novel cancer targets.
Supplementary Material
Acknowledgments
We thank G. Perry for critical evaluations, J. Narayanan for editorial assistance, and A. Spano for art work. Colon tumor and normal tissues were obtained from the Cooperative Human Tissue Network, which is funded by the National Cancer Institute. This work was supported by a grant from the Florida Atlantic Research Corporation (to R.N.). R.N. is an officer of Forseti Biosciences, Incorporated, and has an equity interest in the company.
Abbreviations
- IHC
immunohistochemistry
- SIM
Single Minded gene
References
- 1.Bentley D R. Med Res Rev. 2000;20:189–196. doi: 10.1002/(sici)1098-1128(200005)20:3<189::aid-med2>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 2.Waterston R H, Lander E S, Sulston J E. Proc Natl Acad Sci USA. 2002;99:3712–3716. doi: 10.1073/pnas.042692499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang L, Zhou W, Velculescu V E, Kern S E, Hruban R H, Hamilton S R, Vogelstein B, Kinzler K W. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 4.Subramanian G, Adams M D, Venter J C, Broder S. J Am Med Assoc. 2001;286:2296–2307. doi: 10.1001/jama.286.18.2296. [DOI] [PubMed] [Google Scholar]
- 5.Alon U, Barkai N, Notterman D A, Gish K, Ybarra S, Mack D, Levine A J. Proc Natl Acad Sci USA. 1999;96:6745–6750. doi: 10.1073/pnas.96.12.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper C S. Breast Cancer Res. 2001;3:158–175. doi: 10.1186/bcr291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lal A, Lash A E, Altschul S F, Velculescu V, Zhang L, McLendon R E, Marra M A, Prange C, Morin P J, Polyak K, et al. Cancer Res. 1999;59:5403–5407. [PubMed] [Google Scholar]
- 8.Wheeler D L, Chappey C, Lash A E, Leipe D D, Madden T L, Schuler G D, Tatusova T A, Rapp B A. Nucleic Acids Res. 2000;28:10–14. doi: 10.1093/nar/28.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strausberg R L, Dahl C A, Klausner R D. Nat Genet. 1997;15 Spec. No.:415–416. doi: 10.1038/ng0497supp-415. [DOI] [PubMed] [Google Scholar]
- 10.Riggins G J, Strausberg R L. Hum Mol Genet. 2001;10:663–667. doi: 10.1093/hmg/10.7.663. [DOI] [PubMed] [Google Scholar]
- 11.Scheurle D, DeYoung M P, Binninger D M, Page H, Jahanzeb M, Narayanan R. Cancer Res. 2000;60:4037–4043. [PubMed] [Google Scholar]
- 12.DeYoung M P, Damania H, Scheurle D, Zylberberg C, Narayanan R. In Vivo. 2002;16:239–248. [PubMed] [Google Scholar]
- 13.McCormick M K, Schinzel A, Petersen M B, Stetten G, Driscoll D J, Cantu E S, Tranebjaerg L, Mikkelsen M, Watkins P C, Antonarakis S E. Genomics. 1989;5:325–331. doi: 10.1016/0888-7543(89)90065-7. [DOI] [PubMed] [Google Scholar]
- 14.Kola I, Hertzog P J. Curr Opin Genet Dev. 1998;8:316–321. doi: 10.1016/s0959-437x(98)80088-9. [DOI] [PubMed] [Google Scholar]
- 15.Kola I, Hertzog P J. Hum Mol Genet. 1997;6:1713–1727. doi: 10.1093/hmg/6.10.1713. [DOI] [PubMed] [Google Scholar]
- 16.Chrast R, Scott H S, Chen H, Kudoh J, Rossier C, Minoshima S, Wang Y, Shimizu N, Antonarakis S E. Genome Res. 1997;7:615–624. doi: 10.1101/gr.7.6.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nambu J R, Lewis J O, Wharton K A, Jr, Crews S T. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- 18.Isaac D D, Andrew D J. Genes Dev. 1996;10:103–117. doi: 10.1101/gad.10.1.103. [DOI] [PubMed] [Google Scholar]
- 19.McGuire J, Coumailleau P, Whitelaw M L, Gustafsson J A, Poellinger L. J Biol Chem. 1995;270:31353–31357. doi: 10.1074/jbc.270.52.31353. [DOI] [PubMed] [Google Scholar]
- 20.Thomas J B, Crews S T, Goodman C S. Cell. 1988;52:133–141. doi: 10.1016/0092-8674(88)90537-5. [DOI] [PubMed] [Google Scholar]
- 21.De Young M P, Scheurle D, Damania H, Zylberberg C, Narayanan R. Anticancer Res. 2002;22:3149–3158. [PubMed] [Google Scholar]
- 22.Sharma H W, Narayanan R. BioEssays. 1995;17:1055–1063. doi: 10.1002/bies.950171210. [DOI] [PubMed] [Google Scholar]
- 23.Agrawal S, Kandimalla E R. Mol Med Today. 2000;6:72–81. doi: 10.1016/s1357-4310(99)01638-x. [DOI] [PubMed] [Google Scholar]
- 24.McIntyre K W, Lombard-Gillooly K, Perez J R, Kunsch C, Sarmiento U M, Larigan J D, Landreth K T, Narayanan R. Antisense Res Dev. 1993;3:309–322. doi: 10.1089/ard.1993.3.309. [DOI] [PubMed] [Google Scholar]
- 25.Hankinson O. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 26.Swanson H I, Bradfield C A. Pharmacogenetics. 1993;3:213–230. doi: 10.1097/00008571-199310000-00001. [DOI] [PubMed] [Google Scholar]
- 27.Swanson H I, Chan W K, Bradfield C A. J Biol Chem. 1995;270:26292–26302. doi: 10.1074/jbc.270.44.26292. [DOI] [PubMed] [Google Scholar]
- 28.Nebert D W, Roe A L, Dieter M Z, Solis W A, Yang Y, Dalton T P. Biochem Pharmacol. 2000;59:65–85. doi: 10.1016/s0006-2952(99)00310-x. [DOI] [PubMed] [Google Scholar]
- 29.Lees M J, Whitelaw M L. Mol Cell Biol. 1999;19:5811–5822. doi: 10.1128/mcb.19.8.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunter T. Cell. 1997;88:333–346. doi: 10.1016/s0092-8674(00)81872-3. [DOI] [PubMed] [Google Scholar]
- 31.Steele R J, Thompson A M, Hall P A, Lane D P. Br J Surg. 1998;85:1460–1467. doi: 10.1046/j.1365-2168.1998.00910.x. [DOI] [PubMed] [Google Scholar]
- 32.Helder M N, Wisman G B, van der Zee G J. Cancer Invest. 2002;20:82–101. doi: 10.1081/cnv-120000370. [DOI] [PubMed] [Google Scholar]
- 33.Fearon E R, Hamilton S R, Vogelstein B. Science. 1987;238:193–197. doi: 10.1126/science.2889267. [DOI] [PubMed] [Google Scholar]
- 34.Fettman M J, Butler R N, McMichael A J, Roberts-Thomson I C. J Gastroenterol Hepatol. 1991;6:81–89. doi: 10.1111/j.1440-1746.1991.tb01151.x. [DOI] [PubMed] [Google Scholar]
- 35.Deschner E E, De Cosse J J, Sherlock P. Clin Gastroenterol. 1981;10:755–771. [PubMed] [Google Scholar]
- 36.Hasle H, Clemmensen I H, Mikkelsen M. Lancet. 2000;355:165–169. doi: 10.1016/S0140-6736(99)05264-2. [DOI] [PubMed] [Google Scholar]
- 37.Chik K, Li C, Shing M M, Leung T, Yuen P M. J Pediatr Hematol Oncol. 1999;21:149–151. doi: 10.1097/00043426-199903000-00012. [DOI] [PubMed] [Google Scholar]
- 38.Satge D, Sasco A J, Cure H, Leduc B, Sommelet D, Vekemans M J. Cancer. 1997;80:929–935. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.