Abstract
Tumstatin and endostatin are two inhibitors of angiogenesis derived from precursor human collagen molecules known as α3 chain of type IV collagen and α1 chain of type XVIII collagen, respectively. Although both these inhibitors are noncollagenous (NC1) domain fragments of collagens, they only share a 14% amino acid homology. In the present study we evaluated the functional receptors, mechanism of action, and intracellular signaling induced by these two collagen-derived inhibitors. Human tumstatin prevents angiogenesis via inhibition of endothelial cell proliferation and promotion of apoptosis with no effect on migration, whereas human endostatin prevents endothelial cell migration with no effect on proliferation. We demonstrate that human tumstatin binds to αvβ3 integrin in a vitronectin/fibronectin/RGD cyclic peptide independent manner, whereas human endostatin competes with fibronectin/RGD cyclic peptide to bind α5β1 integrin. The activity of human tumstatin is mediated by αvβ3 integrin, whereas the activity of human endostatin is mediated by α5β1 integrin. Additionally, although human tumstatin binding to αvβ3 integrin leads to the inhibition of Cap-dependent translation (protein synthesis) mediated by focal adhesion kinase/phosphatidylinositol 3-kinase/Akt/mTOR/4E-BP1 pathway, human endostatin binding to α5β1 integrin leads to the inhibition of focal adhesion kinase/c-Raf/MEK1/2/p38/ERK1 mitogen-activated protein kinase pathway, with no effect on phosphatidylinositol 3-kinase/Akt/mTOR/4E-BP1 and Cap-dependent translation. Collectively, such distinct properties of human tumstatin and human endostatin provide the first insight into their diverse antiangiogenic actions and argue for combining them for targeting tumor angiogenesis.
Angiogenesis is the formation of new capillaries from preexisting capillaries (1). Pathogenic angiogenesis plays an important role in disease progression such as cancer, diabetic retinopathy, psoriasis, rheumatoid arthritis, etc. (2–4). Under stable conditions, vascular endothelium is in a quiescent state associated with a relatively slow turnover (4). The switch involving conversion of quiescent endothelial cells to an active proangiogenic phenotype requires both up-regulation of endogenous angiogenic stimulators and down-regulation of endogenous angiogenesis inhibitors (4, 5). Certain factors such as angiostatin, endostatin, tumstatin, canstatin, arresten, and thrombospondin-1 and -2 are regarded as endogenous inhibitors of angiogenesis in vivo (6–12).
Vascular basement membrane (VBM) constitutes an important component of a blood vessel/capillary (13). Along with providing structural support, VBM is also speculated to modulate capillary endothelial cell behavior, especially during the sprouting of new capillaries (13). Several fragments of VBM constituents, such as endostatin and tumstatin, have been identified as endogenous inhibitors of angiogenesis (7, 8). Human tumstatin (hTumstatin) is an inhibitor of tumor angiogenesis and growth in mice (8). It is the NC1 domain of the α3 chain of type IV collagen and a component of basement membranes (8). Within this 28-kDa protein, the antiangiogenic activity is localized to a 25 aa stretch in the middle of the molecule, called T7 peptide (14). hTumstatin inhibits proliferation of endothelial cells and induces apoptosis in an αvβ3 integrin dependent manner (15). Apoptosis of endothelial cells by recombinant hTumstatin is mediated by αvβ3 integrin and the phosphatidyl-3-kinase (PI3-kinase)/Akt/mTOR pathway leading to protein synthesis inhibition (15).
Human endostatin (hEndostatin) is the NC1 domain of the α1 chain of type XVIII collagen and an inhibitor of tumor angiogenesis and growth in mice (7, 16). hEndostatin is liberated from the collagenous domain by cleavage within a protease-sensitive hinge by enzymes, such as elastase and cathepsins. hEndostatin circulates in the blood at concentrations of 20–35 ng/ml (17, 18).
hEndostatin is an inhibitor of endothelial cell migration, whereas mouse endostatin inhibits migration and also and causes G1 cell cycle arrest (19–23). The reason for this difference is not clear yet. hEndostatin binds to many cell surface proteins, including heparan sulfate proteoglycans, glypicans, vascular endothelial growth factor (VEGF) KDR receptor and integrins (20, 24–27). Although many different intercellular pathways have been identified as mediators of hEndostatin action, a functional receptor mediating the action of hEndostatin via a defined signaling pathway is yet unidentified (23, 26, 28–30).
Here we report that hEndostatin and hTumstatin, two basement membrane collagen-derived inhibitors, use distinct integrin receptors and signaling pathways to induce diverse effects on vascular endothelial cells and angiogenic subprocesses.
Materials and Methods
For additional information regarding sources of materials see Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Cell Culture.
Primary human umbilical vein endothelial cells (HUVEC), were obtained from American Type Culture Collection. HUVEC were maintained in EGM-2 media (Clonetics, San Diego) at 37°C in a humidified 5% CO2 atmosphere. Passages 3–5 were used for experiments as described (15).
Production of Recombinant Tumstatin and Synthetic T7 Peptide.
Recombinant hTumstatin (rhTum) was expressed by using 293 human embryonic kidney cell as described (15). Synthetic peptide T7 (TMPFLFCNVNDCNFASRNDYSYWL) was synthesized as described (15).
Endothelial Tube Formation Assay.
The endothelial tube formation assay was performed as described with minor modification (8) (see Supporting Materials and Methods).
Migration Assay.
Migration assays with HUVEC were performed by using a Boyden chamber system (Neuroprobe, Cabin John, MD) as described (9) (see Supporting Materials and Methods).
Proliferation Assay.
The proliferation assay was performed by using [3H]thymidine incorporation as described (8) (see Supporting Materials and Methods).
Cell Attachment Assay.
The attachment assay was performed as previously described with minor modification (31) (see Supporting Materials and Methods).
Immunoprecipitation and Immunoblotting Analysis.
rhEndo treated HUVEC were lysed in RIPA-DOC buffer (27), and cell lysates were subjected to immunoprecipitation using antibodies to α5β1 as described with minor modifications (32). The samples were separated by SDS/10% PAGE under reducing conditions and transferred to nitrocellulose. The blots were immunolabeled with primary antibodies to hEndostatin or α5β1 and horseradish peroxidase-labeled secondary antibodies and product was visualized by chemiluminescence (enhanced chemiluminescence kit, Amersham Pharmacia).
Cell Signaling Experiments.
For cell signaling experiments, 1 × 106 HUVEC were seeded into 10-cm2 dishes coated with fibronectin or vitronectin. According to the experimental protocol the cells were preincubated in some experiments with rhTum, rhEndo, or Rapamycin. The cells were lysed and the cell extracts were subjected to SDS/PAGE and immunoblotting using antibodies to phosphorylated and unphosphorylated proteins as described (15).
In Vitro Kinase Assay for mTOR Activity.
Phosphorylation of mTOR and GST 4EBP-1 fusion protein (mTOR substrate) was evaluated in HUVEC transfected with HA-mTOR/FRAP expression plasmid as described (15). Briefly, HUVEC were serum-starved and transiently transfected with HA-mTOR/FRAP plasmid by using Lipofectamine Plus (Invitrogen). Transfected HUVEC (4 × 106 cells) were treated with rhTum, rhEndo (40 μg/ml), or rapamycin (100 ng/ml) for 24 h according to the experimental protocol. The cells were lysed as described, and the cell lysates (200 μg) were subjected to immunoprecipitation with anti-HA antibody (15). HA-mTOR/Anti-HA complexes were incubated with recombinant GST-4E-BP1 fusion protein in the presence of 10 μCi of [γ32P]-ATP (1 Ci = 37 GBq) in kinase buffer. The reactions were terminated by boiling before the samples were subjected to SDS/PAGE, and phosphorylated proteins (mTOR-P and GST-4EBP1-P) were detected by autoradiography (15).
Determination of eIF4E–4E-BP1 Complexes.
HUVEC were serum starved for 24 h and then treated with rhTum, T7 peptide, rhEndo (40 μg/ml), or rapamycin (100 ng/ml), in the presence of 10% FCS for 24 h. Cell lysates were incubated with 7-methyl-GTP-Sepharose (Amersham Pharmacia) for 30 min at 25°C. The bound proteins were eluted with 2× SDS sample buffer, and the samples were analyzed by SDS/PAGE and immunoblotting using antibodies to eIF4E and 4E-BP1 as described (15).
Results and Discussion
Distinct Antiangiogenic Activities of rhEndo and rhTum on HUVEC.
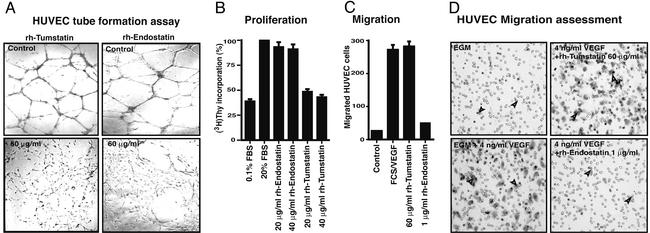
hEndostatin and hTumstatin both inhibit tumor angiogenesis in mice by targeting vascular/capillary endothelial cells in the tumors (7, 8). Here we attempted to delineate the antiangiogenic mechanisms of these two human molecules. We compared the anti-endothelial cell activity of recombinant human Endostatin (rhEndo) and recombinant human Tumstatin (rhTum) by using human umbilical vein endothelial cells (HUVEC) and cow pulmonary arterial endothelial cells (C-PAE cells). First, we used an antiangiogenic assay known as endothelial tube formation assay (33). Tube formation involving endothelial cells represents the net contribution of cell migration, proliferation and survival (33). Addition of rhTum to the culture media significantly inhibited HUVEC tube formation in a dose dependent manner with a maximal inhibition seen at 60 μg/ml (Fig. 1A Left). rhEndo inhibits the formation of tubes significantly at 60 μ/ml and higher (Fig. 1A Right) (21). Next, the antimigratory and antiproliferative effect of rhEndo and rhTum was assessed. In [3H]thymidine-incorporation assays, in vitro proliferation of HUVEC was significantly inhibited by rhTum (Fig. 1B), whereas hEndostatin demonstrated an insignificant effect on [3H]thymidine incorporation into HUVEC at equimolar concentration (Fig. 1B). Similar results were also obtained when C-PAE cells were used (data not shown). The capacity of HUVEC to migrate through a fibronectin-coated membrane toward FCS/VEGF gradients in a Boyden chamber is not inhibited by rhTum even at a concentration of 60 μg/ml (Fig. 1 C and D), whereas hEndostatin starting at a concentration of 1 μg/ml significantly inhibited migration of HUVEC in this same assay, as also shown by others (Fig. 1 C and D) (19–21). Similar results were also obtained when C-PAE cells were used (data not shown). Collectively, these results demonstrate that HUVEC tube formation is inhibited by both rhEndo and rhTum, because both migration and proliferation are essential for endothelial tube formation.
Figure 1.
In vitro antiangiogenic activities of rhTum and rhEndo. (A) Tube formation assay. HUVEC were plated on Matrigel-coated plates in presence of rhTum (Left) or rhEndo (Right). Tube formation was evaluated after 24 h by using a light microscope, and representative fields (×100 magnification) are pictured. (B) Proliferation assays. The graph summarizes the relative [3H]thymidine incorporation in HUVEC on treatment with rhEndo or rhTum compared with the 20% FCS control of three independent experiments. (C and D) Migration assays. (C) The graph displays the average results of three independent experiments. (D) Representative photographs of the underside of Boyden chamber membrane. FCS and VEGF in the lower chamber induce HUVEC migration (arrows, lower left).
Binding of rhTum and rhEndo to Integrins.
Next, we examined the binding characteristics and the role of integrins in mediating the distinct antiangiogenic properties of rhEndo and rhTum.
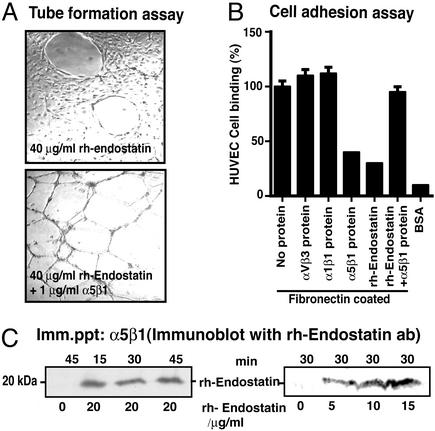
Attachment of HUVEC is significantly enhanced on plates coated with rhTum or rhEndo when compared with uncoated plates (data not shown). Binding of HUVEC to rhTum is inhibited by blocking antibodies specific for αvβ3 and the antiproliferative action of rhTum is mediated by αvβ3 integrin, suggesting that αvβ3 integrin is the functional receptor for rhTum (31). Preincubation of HUVEC with rhTum does not inhibit binding of endothelial cells to vitronectin, fibronectin, or RGD cyclic peptide (31). Binding of HUVEC to rhEndo-coated plates was significantly inhibited by blocking antibodies specific for α5β1 integrins, whereas blocking of αvβ3 or α1β1 integrins (two other prominent proangiogenic integrins) had no significant effect (data not shown). This observation supports recent studies, which demonstrate possible α5β1 integrin binding to rhEndo (20, 27). HUVEC tube formation is inhibited by rhEndo (Fig. 1A) and preincubation of HUVEC with equimolar amount of α5β1 integrin soluble protein reverses the antiangiogenic action of rhEndo (Fig. 2A). Such reversal is not seen with αvβ3 or α1β1 integrin soluble proteins (Fig. 2A). Similar results were also obtained when migration assays were used (data not shown). These experiments suggest that α5β1 integrin serves as a functional receptor for rhEndo.
Figure 2.
α5β1 integrin is a receptor for rhEndo. (A) Tube formation assay. HUVEC were plated on Matrigel-coated plates in media containing rhTum (40 μg/ml) without (Upper) or with (Lower) soluble α5β1 protein (1 μg/ml). Representative fields are shown (×100 magnification). Soluble αvβ3 and α1β1 integrins had no significant effect on rhEndo-induced inhibition of HUVEC tube formation (not shown). (B) Cell attachment assay. HUVEC were allowed to attach to plates that were coated with fibronectin in presence of soluble integrins α5β1, αvβ3, α1β1, rhEndo or without protein, BSA-coated plates were used as negative control. The graph summarizes the average results of three independent experiments. (C) Immunoprecipitation and immunoblot. HUVEC treated with rhEndo in a dose-dependent manner for 30 min (Right) or over a time course at a constant dose (20 μg/ml, Left) were lysed and α5β1 protein immunoprecipitated. Western analysis of the precipitate for bound rhEndo are shown. A specific band at 20 kDa was present after 15, 30, and 45 min of incubation with rhEndo. This band was absent in lysates of untreated cells.
In contrast to rhTum, preincubation of HUVEC with hEndo significantly decreased attachment of HUVEC to fibronectin, the major ligand for α5β1 integrin (Fig. 2B). In the same experimental setting, the addition of soluble α5β1 integrin captured rhEndo and reversed the impaired binding of HUVEC to fibronectin, further suggesting that rhEndo competes for fibronectin binding to α5β1 integrin (Fig. 2B). Immunocytochemistry experiments revealed that rhEndo colocalizes with α5β1 integrin on proliferating endothelial cells (see Supporting Materials and Methods and Fig. 7, which is published as supporting information on the PNAS web site). To further confirm binding of hEndostatin to α5β1 integrin, we performed immunoprecipitation and immunoblotting experiments to demonstrate rhEndo binding to α5β1 integrin (Fig. 2C).
RGD-Sequence Requirement for rhEndo/rhTum Binding to αvβ3 and α5β1 Integrins.
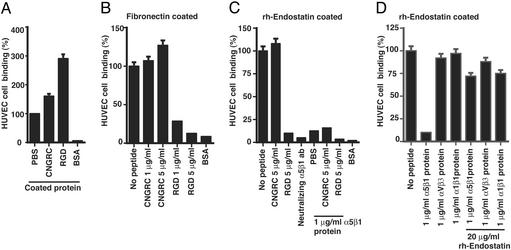
Integrins αvβ3 and α5β1 bind vitronectin/fibronectin by using their RGD-sequence motif (34). To further characterize the binding of rhTum and rhEndo to their receptors, we used synthetic RGD-4C cyclic peptide (competes for RGD-binding site on vitronectin and fibronectin) and CNGRC peptide (also binds to endothelial cells) in competition cell adhesion assays (31, 34). Attachment of endothelial cells to vitronectin and fibronectin is inhibited by blocking the RGD-site with RGD-4C cyclic peptide, whereas CNGRC peptide has no effect (31). Binding of endothelial cells to tumstatin-coated plates is not inhibited by RGD-4C cyclic peptide, CNGRC peptide, vitronectin, or fibronectin, suggesting that rhTum binds αvβ3 integrin in an RGD binding site-independent manner (31).
Blocking of the RGD-binding capacity within α5β1 integrin with soluble RGD-4C cyclic peptide inhibited attachment of HUVEC to fibronectin and also rhEndo-coated plates, whereas preincubation of HUVEC with CNGRC peptide had no significant effect (Fig. 3 B and C). We also demonstrated that soluble α5β1integrin protein could bind rhEndo that was coated onto cell culture plates and subsequently inhibited attachment of HUVEC to rhEndo (Fig. 3D). This effect was reversed by capturing the soluble α5β1 integrin with soluble rhEndo (Fig. 3D). These results suggest that rhEndo binds to α5β1 integrin in a RGD motif-dependent manner and competes for the RGD- binding sites within fibronectin. rhEndo does not contain a RGD sequence motif (20). Thus, the mechanism by which rhEndo inhibits RGD cyclic peptide or fibronectin binding to α5β1 integrin remains unclear.
Figure 3.
Cell attachment assays. The graphs display results of three independent experiments, presented as relative percentage compared with PBS control. (A) Attachment of HUVEC was increased to plates that were coated with synthetic peptides CNGRC (25 μg/ml) or RGD-4C (25 μg/ml) compared with uncoated control plates. (B) Attachment of HUVEC on fibronectin-coated plates was significantly inhibited by preincubation of the cells with RGD-4C peptide (73% at 1 mg/ml; 89% at a concentration of 5 μg/ml), whereas preincubation of with CNGRC-peptide had no effect on adhesion of HUVEC. (C) Attachment of HUVEC on rhEndo-coated plates was significantly inhibited by RGD cyclic peptide or α5β1 blocking antibodies, whereas CNGRC peptide had no significant effect. Addition of soluble α5β1 integrin protein into the wells significantly inhibited attachment of PBS or CNGRC peptide treated HUVEC, whereas it had no additional effect when the cells were pretreated with RGD cyclic peptide. (D) Attachment of HUVEC on rhEndo-coated plates was significantly inhibited by incubation with α5β1 soluble protein, whereas αvβ3 or α1β1 integrins had no significant effect. α5β1 integrin-induced inhibition was reversed by addition of soluble rhEndo to the culture media.
Signal Transduction Cascades Induced by rhTum and rhEndo.
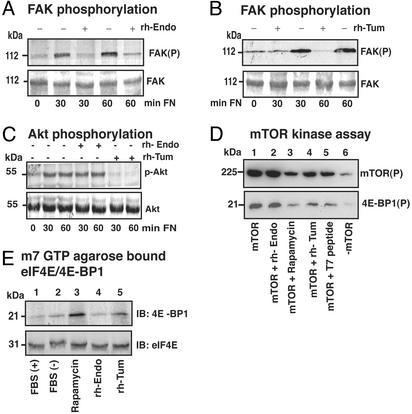
In endothelial cells, ligand binding to integrins induces phosphorylation of focal adhesion kinase (FAK), which serves as a platform for different downstream signals (35). rhTum and rhEndo both inhibit phosphorylation of FAK when endothelial cells are plated on fibronectin or vitronectin (Fig. 4A) (15, 26). Downstream of FAK, protein kinase B (PKB/Akt) plays an important role in mediating pathways that are involved in the regulation of endothelial cell survival, proliferation, and migration (36). Akt is a serine/threonine protein kinase that is activated by PI3-K (37). Akt is also known to regulate protein synthesis by mediating phosphorylation of eukaryotic initiation factor 4E-binding protein (4E-BP1) via mTOR kinase (15, 38, 39). Akt is also involved in regulation of cell cycle and cell senescence via E2F and p21 (36). Additionally, it has been suggested that Akt is involved in regulation of endothelial cell migration, possibly via an interaction with the Rho family of small G proteins (36).
Figure 4.
Effects of rhTum and rhEndo on the FAK/Akt/mTOR/4EBP1 Cap-dependent translation pathway. (A–C) Serum-starved HUVEC were plated on fibronectin-coated dishes in incomplete medium supplemented with rhEndo (20 μg/ml) or rhTum (20 μg/ml) for the indicated times and lysates analyzed by Western blot. (A and B) Phosphorylation of FAK. Immunoblots of phosphorylated FAK (Upper) and total FAK (Lower) demonstrate that both rhEndo (A) or rhTum (B) inhibit sustained phosphorylation. (C) Phosphorylation of Akt. Immunoblots of phosphorylated Akt (Upper) and total Akt (Lower) demonstrate no changes in phosphorylation on rhEndo treatment, whereas rhTum significantly inhibited the sustained phosphorylation of Akt. (D) mTOR kinase Assay. Autoradiography of autophosphorylated mTOR (Upper) and phosphorylated 4E-BP1 (Lower) isolated from hemagglutinin (HA)-mTOR transfected HUVEC. mTOR activity was not inhibited by rhEndo− (D, lane 2) compared with untreated transfected cells (D, lane 1). rhTum (D, lane 4) and T7 peptide (D, lane 5) significantly inhibited mTOR activity, similar to rapamycin (D, lane 3). Lane 6 is the nontransfected control. (E) eIF4E-4E-BP1 complex formation. Immunoblots for anti-4E-BP1 (Upper) and anti-eIF4E (Lower) of lysates precipitated with m7GTP-agarose beads. HUVEC were treated as indicated. Rapamycin (E, lane 3) and rhTum (E, lane 5) increased eIF4E-4E-BP1 complexes, whereas rhEndo (E, lane 4) had no significant effect compared with the untreated control (E, lane 2).
rhTum inhibited the sustained phosphorylation of Akt and activation of phosphatidyl-3-kinase in HUVEC that are plated on fibronectin or vitronectin (Fig. 4C) (15). Downstream of Akt, mTOR directly phosphorylates eukaryotic initiation factor 4E(eIF4E)-binding protein (4E-BP1) (Fig. 4D) (40). Unphosphorylated 4E-BP1 interacts with eIF4E and inhibits Cap-dependent protein synthesis (40). rhTum suppresses mTOR activity and thus inhibits phosphorylation of 4E-BP1 in HUVEC (Fig. 4 D and E) (15). Inhibition of 4E-BP-1 phosphorylation enhanced binding to eIF-4E, which in turn becomes unavailable to initiate cap-dependent translation (Fig. 4E) (15). rhEndo also inhibits phosphorylation of FAK, but phosphorylation (activation) of Akt was not inhibited (Fig. 4 A and C) (26). rhEndo had insignificant effect on the PI3-K-Akt-4E-BP-1 Cap-dependent translation pathway, in contrast to rhTum (Fig. 4 C and E). These results suggest that hEndostatin regulates migration of endothelial cells in an Akt-independent manner.
Collectively, our results suggest that despite different integrin binding characteristics, both rhEndo and rhTum inhibit phosphorylation of FAK. The mechanism by which rhEndo and rhTum mediate inhibition of FAK-phosphorylation is obviously quite different. Although Akt has been implicated as important for migration of cells, rhEndo does not inhibit activation of Akt (41). In contrast, rhTum inhibits Akt activation and leads to inhibition of Cap dependent protein synthesis with no effect on migration (15). Such differences in the signaling cascade by rhEndo and rhTum possibly explain their different effects on endothelial cell function. To investigate the molecular pathways leading to inhibition of endothelial cell migration by rhEndo, further experiments were carried out as described below.
rhEndo Signaling Downstream of FAK.
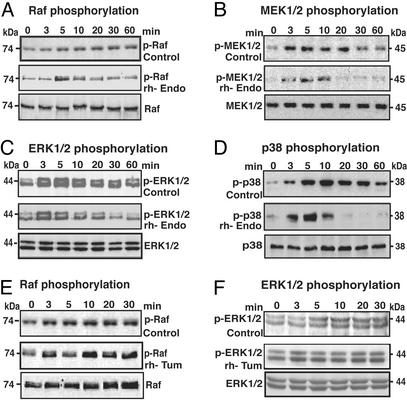
The mitogen-activated protein (MAP) kinase pathway is an important downstream target of FAK that is involved in the regulation of endothelial cell migration (42). Mice deficient in FAK or fibronectin are embryonic lethal and display severely impaired vasculogenesis and cell migration (43, 44). Mice deficient in the α5 integrin also suffer from embryonic lethality and exhibit defects in cell migration (45). Overexpression of FAK significantly increases the migratory capacity of cells via activation of the ERK pathway (46). These findings suggest that the fibronectin/α5β1 integrin/FAK/ERK pathway plays a major role in regulation of ECM-mediated migration. Attachment of endothelial cells to fibronectin via α5β1integrin activated the FAK/cRaf/MEK/ERK pathway in HUVEC (Fig. 5). Pretreatment of HUVEC with rhEndo inhibited sustained phosphorylation of Raf (Fig. 5A) and its downstream target MEK1/2 (Fig. 5B), which resulted in the attenuation of sustained phosphorylation of ERK1. Phosphorylation of ERK2 remained unchanged (Fig. 5C). ERK1 is involved in matrix-mediated migration as has been reported recently in experiments using the αvβ3 (47).
Figure 5.
Effects of rhEndo and rhTum on ERK and p38 MAP kinase signal transduction pathways. Serum-starved HUVEC were plated on fibronectin in incomplete medium containing 10 μg/ml rhEndo (A–D) or rhTum (E and F) and lysed at the indicated times (0–60 min). The levels of phosphorylated proteins after incubation with rhEndo or rhTum (Middle) were compared with untreated controls (Top) by immunoblot. (Bottom) The total signaling protein levels in rhEndo or rhTum-treated HUVEC. (A and E) Raf phosphorylation. Immunoblots for phopho-Raf indicate that sustained phosphorylation of Raf (p-Raf) (A and E, Upper) was inhibited by treatment with rhEndo (A Middle) but not with rhTum (E Middle). (B) MEK1/2 phosphorylation. The prolonged phosphorylation of MEK1/2 (B Top), downstream of Raf, was significantly inhibited by rhEndo (B Middle). (C and F) ERK1/2 phosphorylation. Immunoblots for phosphorylated ERK1/2 (p-ERK) indicate sustained phosphorylation of ERK1 (Upper) and ERK2 (Lower) after HUVEC were seeded on fibronectin (C and F, Upper). Treatment with rhEndo inhibited phosphorylation of ERK1 but had no significant effect on ERK2 phosphorylation (C Middle). Treatment with rhTum had no effect on phosphorylation of ERK1/2 (F). (D) p38 phosphorylation. Immunoblots for phosphorylated 38 MAP kinase (p-p38) indicate that prolonged phosphorylation of p38 (D Top) was decreased after treatment with rhEndo (D Middle).
Migration of endothelial cells requires cytoskeleton reorganization and cell adhesion (21). During VEGF-induced migration of endothelial cells, the ERK pathway mediates actin reorganization, whereas the MAP kinase homologue p38 pathway regulates the expression of integrins and proteases in endothelial cells, suggesting that these two pathways regulate endothelial cell migration in a coordinated manner (48). Our results demonstrate that rhEndo significantly decreases sustained phosphorylation of p38 (Fig. 5D), suggesting that rhEndo inhibits both ERK1 and p38 MAP kinase pathways. rhTum has no effect on phosphorylation of cRaf and ERK in HUVEC on fibronectin matrix, suggesting that rhTum has no effect on MAP kinase pathways downstream of FAK (Fig. 5).
These results demonstrate that although rhEndo and rhTum are both antiendothelial cell agents, rhEndo targets the migratory capacity of endothelial cells, whereas rhTum targets endothelial cell proliferation. These actions are mediated by α5β1 integrin in the case of rhEndo and by αvβ3 integrin in the case of rhTum. It is apparent that the functional receptors for these two inhibitors of angiogenesis are different, suggesting that the mechanism of receptor binding is also distinct. rhEndo competes with vitronectin/fibronectin/RGD cyclic peptide to bind α5β1 integrin on the endothelial cell surface. This finding suggests that rhEndo mediates its antimigratory effect by competing with ligands which provide proangiogenic/migratory cues, whereas rhTum does not compete with vitronectin/fibronectin/RGD cyclic peptide to bind αvβ3 integrin on endothelial cells. Thus, this finding suggests that rhTum binds to a site distinct from vitronectin/fibronectin/RGD cyclic peptide binding site. Because binding of rhTum to αvβ3 integrin does not displace proangiogenic ligands on αvβ3 integrin, the antiproliferative effect is likely caused by active reversal of pro-proliferative signals induced by vitronectin and fibronectin. Such action of rhTum therefore differs from the proposed action of rhEndo on endothelial cells.
Further insight into the diverse action of rhTum and rhEndo was realized when intracellular signaling cascades induced by these two inhibitors were examined. rhTum and rhEndo bind to different integrins by distinct mechanisms, but both induce inhibition of FAK phosphorylation. Inhibition of FAK activation by rhTum leads to inhibition of Cap-dependent translation (protein synthesis), mediated by PI3-K/Akt/mTOR/4EBP1 pathway. MAP kinase pathways are not affected by rhTum. In contrast, inhibition of FAK activation caused by rhEndo binding to α5β1 integrin leads to inhibition of ERK1/p38 MAP kinase pathways with no effect on PI3-K/Akt/mTOR/4EBP1 pathway (Fig. 6). Inhibition of FAK activation by rhEndo and rhTum, leading to such distinct downstream signaling, requires further investigation and might involve novel intracellular molecules downstream of FAK.
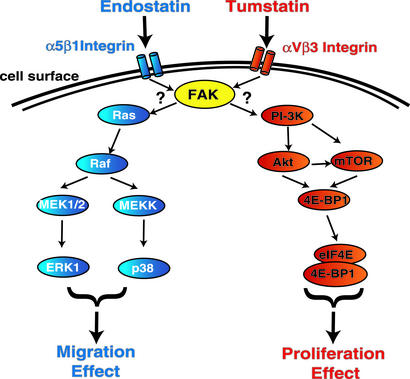
Figure 6.
Schematic illustration of distinct signaling pathways induced by rhTum and rhEndo. rhTum binds to αvβ3 integrin, whereas rhEndo binds to α5β1 integrin. Both rhEndo and rhTum inhibit phosphorylation of FAK (yellow). Downstream of FAK, rhTum inhibits the PI3-K/Akt/mTOR/4EBP1 pathway, resulting in inhibition of endothelial protein synthesis and proliferation. MAP kinase pathways are not affected by rhTum. In contrast, inhibition of FAK activation by rhEndo binding to α5β1 integrin leads to inhibition of ERK1/p38 MAP kinase pathways with no effect on PI3-K/Akt/mTOR/4EBP1 pathways, resulting in inhibition of endothelial cell migration.
Lastly, our results provide further support for the notion that basement membrane-derived angiogenesis inhibitors are not mutually exchangeable in their mode of action and offer hope that these inhibitors could be used in combination to realize effective synergy in the control of tumor angiogenesis and growth.
Supplementary Material
Acknowledgments
We thank Dr. N. Sonenberg and Dr. Judha Folkman for their reagent gifts and Nazia Akhtar for her technical assistance. This study was supported by National Institutes of Health Grants DK55001 and DK51711 and funds from the Program in Matrix Biology at Beth Israel Deaconess Medical Center. M.Z. was funded by Deutsche Forschungsgemeinschaft Grant ZE523/1-1. R.K. and Beth Israel Deaconess Medical Center are equity holders in ILEX Oncology, a company involved in the clinical development of rhTumstatin. Beth Israel Deaconess Medical Center also licensed patents related to rhEndostatin to ILEX Oncology in 1999.
Abbreviations
- MAP
mitogen-activated protein
- FAK
focal adhesion kinase
- PI3-K
phosphatidyl-3 kinase
- HUVEC
human umbilical vein endothelial cells
- VEGF
vascular endothelial growth factor
References
- 1.Folkman J. Ann Surg. 1972;175:409–416. doi: 10.1097/00000658-197203000-00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brooks P C, Montgomery A M, Rosenfeld M, Reisfeld R A, Hu T, Klier G, Cheresh D A. Cell. 1994;79:1157–1164. doi: 10.1016/0092-8674(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 3.Carmeliet P, Jain R K. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 4.Folkman J. Nat Med. 1995;1:27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Folkman J. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 6.O'Reilly M S, Holmgren L, Shing Y, Chen C, Rosenthal R A, Moses M, Lane W S, Cao Y, Sage E H, Folkman J. Cell. 1994;79:315–328. doi: 10.1016/0092-8674(94)90200-3. [DOI] [PubMed] [Google Scholar]
- 7.O'Reilly M S, Boehm T, Shing Y, Fukai N, Vasios G, Lane W S, Flynn E, Birkhead J R, Olsen B R, Folkman J. Cell. 1997;88:277–285. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 8.Maeshima Y, Colorado P C, Torre A, Holthaus K A, Grunkemeyer J A, Ericksen M D, Hopfer H, Xiao Y, Stillman I E, Kalluri R. J Biol Chem. 2000;275:21340–21348. doi: 10.1074/jbc.M001956200. [DOI] [PubMed] [Google Scholar]
- 9.Kamphaus G D, Colorado P C, Panka D J, Hopfer H, Ramchandran R, Torre A, Maeshima Y, Mier J W, Sukhatme V P, Kalluri R. J Biol Chem. 2000;275:1209–1215. doi: 10.1074/jbc.275.2.1209. [DOI] [PubMed] [Google Scholar]
- 10.Colorado P C, Torre A, Kamphaus G, Maeshima Y, Hopfer H, Takahashi K, Volk R, Zamborsky E D, Herman S, Ericksen M B, et al. Cancer Res. 2000;60:2520–2526. [PubMed] [Google Scholar]
- 11.Good D J, Polverini P J, Rastinejad F, Le Beau M M, Lemons R S, Frazier W A, Bouck N P. Proc Natl Acad Sci USA. 1990;87:6624–6628. doi: 10.1073/pnas.87.17.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volpert O V, Tolsma S S, Pellerin S, Feige J J, Chen H, Mosher D F, Bouck N. Biochem Biophys Res Commun. 1995;217:326–332. doi: 10.1006/bbrc.1995.2780. [DOI] [PubMed] [Google Scholar]
- 13.Darland D C, D'Amore P A. J Clin Invest. 1999;103:157–158. doi: 10.1172/JCI6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maeshima Y, Yerramalla U L, Dhanabal M, Holthaus K A, Barbashov S, Kharbanda S, Reimer C, Manfredi M, Dickerson W M, Kalluri R. J Biol Chem. 2001;276:31959–31968. doi: 10.1074/jbc.M103024200. [DOI] [PubMed] [Google Scholar]
- 15.Maeshima Y, Sudhakar A, Lively J C, Ueki K, Kharbanda S, Kahn C R, Sonenberg N, Hynes R O, Kalluri R. Science. 2002;295:140–143. doi: 10.1126/science.1065298. [DOI] [PubMed] [Google Scholar]
- 16.Fukai N, Eklund L, Marneros A G, Oh S P, Keene D R, Tamarkin L, Niemela M, Ilves M, Li E, Pihlajaniemi T, Olsen B R. EMBO J. 2002;21:1535–1544. doi: 10.1093/emboj/21.7.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sasaki T, Fukai N, Mann K, Gohring W, Olsen B R, Timpl R. EMBO J. 1998;17:4249–4256. doi: 10.1093/emboj/17.15.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feldman A L, Pak H, Yang J C, Alexander H R, Jr, Libutti S K. Cancer. 2001;91:1525–1529. doi: 10.1002/1097-0142(20010415)91:8<1525::aid-cncr1161>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 19.Yamaguchi N, Anand-Apte B, Lee M, Sasaki T, Fukai N, Shapiro R, Que I, Lowik C, Timpl R, Olsen B R. EMBO J. 1999;18:4414–4423. doi: 10.1093/emboj/18.16.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rehn M, Veikkola T, Kukk-Valdre E, Nakamura H, Ilmonen M, Lombardo C, Pihlajaniemi T, Alitalo K, Vuori K. Proc Natl Acad Sci USA. 2001;98:1024–1029. doi: 10.1073/pnas.031564998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo C J, LaMontagne K R, Jr, Garcia-Cardena G, Ackley B D, Kalman D, Park S, Christofferson R, Kamihara J, Ding Y H, Lo K M, et al. J Cell Biol. 2001;152:1233–1246. doi: 10.1083/jcb.152.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixelius J, Cross M, Matsumoto T, Sasaki T, Timpl R, Claesson-Welsh L. Cancer Res. 2002;62:1944–1947. [PubMed] [Google Scholar]
- 23.Hanai J, Gloy J, Karumanchi S A, Kale S, Tang J, Hu G, Chan B, Ramchandran R, Jha V, Sukhatme V P, Sokol S. J Cell Biol. 2002;158:529–539. doi: 10.1083/jcb.200203064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karumanchi S A, Jha V, Ramchandran R, Karihaloo A, Tsiokas L, Chan B, Dhanabal M, Hanai J I, Venkataraman G, Shriver Z, et al. Mol Cell. 2001;7:811–822. doi: 10.1016/s1097-2765(01)00225-8. [DOI] [PubMed] [Google Scholar]
- 25.Kreuger J, Matsumoto T, Vanwildemeersch M, Sasaki T, Timpl R, Claesson-Welsh L, Spillmann D, Lindahl U. EMBO J. 2002;21:6303–6311. doi: 10.1093/emboj/cdf638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim Y M, Hwang S, Pyun B J, Kim T Y, Lee S T, Gho Y S, Kwon Y G. J Biol Chem. 2002;277:27872–27879. doi: 10.1074/jbc.M202771200. [DOI] [PubMed] [Google Scholar]
- 27.Wickstrom S A, Alitalo K, Keski-Oja J. Cancer Res. 2002;62:5580–5589. [PubMed] [Google Scholar]
- 28.Urbich C, Reissner A, Chavakis E, Dernbach E, Haendeler J, Fleming I, Zeiher A M, Kaszkin M, Dimmeler S. FASEB J. 2002;16:706–708. doi: 10.1096/fj.01-0637fje. [DOI] [PubMed] [Google Scholar]
- 29.Jiang L, Jha V, Dhanabal M, Sukhatme V P, Alper S L. Am J Physiol Cell Physiol. 2001;280:C1140–C1150. doi: 10.1152/ajpcell.2001.280.5.C1140. [DOI] [PubMed] [Google Scholar]
- 30.Dixelius J, Larsson H, Sasaki T, Holmqvist K, Lu L, Engstrom A, Timpl R, Welsh M, Claesson-Welsh L. Blood. 2000;95:3403–3411. [PubMed] [Google Scholar]
- 31.Maeshima Y, Colorado P C, Kalluri R. J Biol Chem. 2000;275:23745–23750. doi: 10.1074/jbc.C000186200. [DOI] [PubMed] [Google Scholar]
- 32.Tsukaguchi H, Sudhakar A, Le T C, Nguyen T, Yao J, Schwimmer J A, Schachter A D, Poch E, Abreu P F, Appel G B, et al. J Clin Invest. 2002;110:1659–1666. doi: 10.1172/JCI16242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Folkman J, Haudenschild C. Nature. 1980;288:551–556. doi: 10.1038/288551a0. [DOI] [PubMed] [Google Scholar]
- 34.Ruoslahti E, Pierschbacher M D. Science. 1987;238:491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 35.Zachary I, Rozengurt E. Cell. 1992;71:891–894. doi: 10.1016/0092-8674(92)90385-p. [DOI] [PubMed] [Google Scholar]
- 36.Shiojima I, Walsh K. Circ Res. 2002;90:1243–1250. doi: 10.1161/01.res.0000022200.71892.9f. [DOI] [PubMed] [Google Scholar]
- 37.Hemmings B A. Science. 1997;275:628–630. doi: 10.1126/science.275.5300.628. [DOI] [PubMed] [Google Scholar]
- 38.Miron M, Verdu J, Lachance P E, Birnbaum M J, Lasko P F, Sonenberg N. Nat Cell Biol. 2001;3:596–601. doi: 10.1038/35078571. [DOI] [PubMed] [Google Scholar]
- 39.Downward J. Curr Opin Cell Biol. 1998;10:262–267. doi: 10.1016/s0955-0674(98)80149-x. [DOI] [PubMed] [Google Scholar]
- 40.Pause A, Belsham G J, Gingras A C, Donze O, Lin T A, Lawrence J C, Jr, Sonenberg N. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 41.Lee M J, Thangada S, Paik J H, Sapkota G P, Ancellin N, Chae S S, Wu M, Morales-Ruiz M, Sessa W C, Alessi D R, Hla T. Mol Cell. 2001;8:693–704. doi: 10.1016/s1097-2765(01)00324-0. [DOI] [PubMed] [Google Scholar]
- 42.Clark E A, Brugge J S. Science. 1995;268:233–239. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- 43.Furuta Y, Ilic D, Kanazawa S, Takeda N, Yamamoto T, Aizawa S. Oncogene. 1995;11:1989–1995. [PubMed] [Google Scholar]
- 44.George E L, Georges-Labouesse E N, Patel-King R S, Rayburn H, Hynes R O. Development (Cambridge, UK) 1993;119:1079–1091. doi: 10.1242/dev.119.4.1079. [DOI] [PubMed] [Google Scholar]
- 45.Yang J T, Rayburn H, Hynes R O. Development (Cambridge, UK) 1993;119:1093–1105. doi: 10.1242/dev.119.4.1093. [DOI] [PubMed] [Google Scholar]
- 46.Cary L A, Han D C, Polte T R, Hanks S K, Guan J L. J Cell Biol. 1998;140:211–221. doi: 10.1083/jcb.140.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roberts M S, Woods A J, Shaw P E, Norman J C. J Biol Chem. 2003;278:1975–1985. doi: 10.1074/jbc.M208607200. [DOI] [PubMed] [Google Scholar]
- 48.Becker P M, Verin A D, Booth M A, Liu F, Birukova A, Garcia J G. Am J Physiol Lung Cell Mol Physiol. 2001;281:L1500–L1511. doi: 10.1152/ajplung.2001.281.6.L1500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.