Abstract
Caffeine is an efficient inhibitor of cellular DNA repair, likely through its effects on ATM (ataxia telangiectasia mutated) and ATR (ATM and Rad3-related) kinases. Here, we show that caffeine treatment causes a dose-dependent reduction in the total amount of HIV-1 and avian sarcoma virus retroviral vector DNA that is joined to host DNA in the population of infected cells and also in the number of transduced cells. These changes were observed at caffeine concentrations that had little or no effect on overall cell growth, synthesis, and nuclear import of the viral DNA, or the activities of the viral integrase in vitro. Substantial reductions in the amount of host-viral-joined DNA in the infected population, and in the number of transductants, were also observed in the presence of a dominant-negative form of the ATR protein, ATRkd. After infection, a significant fraction of these cells undergoes cell death. In contrast, retroviral transduction is not impeded in ATM-deficient cells, and addition of caffeine leads to the same reduction that was observed in ATM-proficient cells. These results suggest that activity of the ATR kinase, but not the ATM kinase, is required for successful completion of the viral DNA integration process and/or survival of transduced cells. Components of the cellular DNA damage repair response may represent potential targets for antiretroviral drug development.
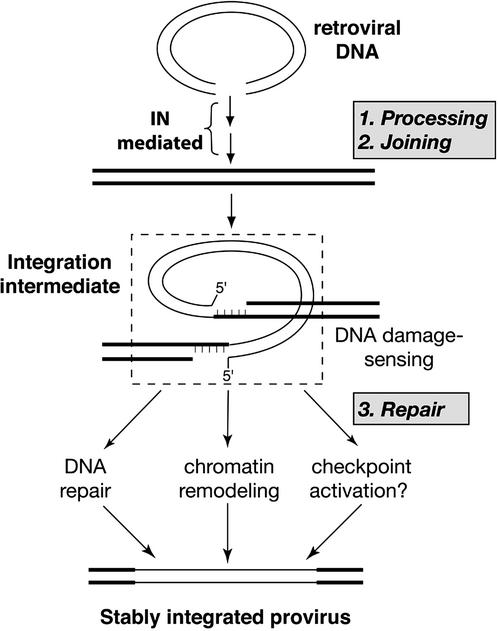
The retroviral DNA integration process comprises three distinct steps, the first two of which have been reconstituted in vitro with the purified retroviral enzyme integrase (1). In the first step, processing, integrase removes two nucleotides from the 3′ ends of the viral DNA. In the second step, joining, these newly created ends are joined to staggered phosphates in the host DNA in a concerted cleavage and ligation reaction. This reaction creates an integration intermediate with gaps in the flanking host DNA sequence (Fig. 1). In the last step of the integration process, repair, these gaps are filled and the 5′ ends of the viral DNA are joined to the host DNA, creating a stably integrated provirus. Repair can also be reconstituted in vitro by using combinations of various polymerases, ligases, and an endonuclease (2, 3), but the identity and mechanism of action of proteins responsible for repair of the integration intermediate in vivo are not yet known. Establishment of a stably integrated provirus is an essential step in retroviral replication and therefore the integrase protein is an attractive target for antiviral therapy (4–7). However, as integrase is virus-encoded, it is subject to a high mutation rate that can lead to drug resistance. This rapid evolution of resistance should not occur with drugs that can target cellular functions necessary to complete the integration process, without affecting cell viability.
Figure 1.
Steps in the retroviral DNA integration process.
We have recently observed that retroviral infection induces programmed death in scid lymphocytes that are deficient in the DNA repair protein, DNA-PK (DNA-dependent protein kinase) (8). Furthermore, this response to infection requires an active integrase (8). In addition to retrovirus-induced scid cell death, we have observed that the number of cells stably transduced by retroviral vectors (a measure of successful completion of all three steps in the integration process and cell survival; Fig. 1) is reduced in cells that are deficient in DNA-PK and other components of the nonhomologous end-joining pathway (8). These findings led us to propose that the integrase-mediated joining of retroviral and host DNA is sensed as damage by the host cell, and that DNA repair proteins, such as DNA-PK, may be required to facilitate stable integration. Components of the DNA damage response pathway(s) that participate in the final step of the integration process might therefore present novel targets for antiretroviral therapy.
DNA-PK belongs to a family of large, phosphatidylinositol 3-kinase-related protein kinases, that also includes ATM (ataxia telangiectasia mutated) and ATR (ATM and Rad3-related). The ATM and ATR kinases seem to have broader roles than DNA-PK in response to DNA damage, which include the regulation of cell cycle checkpoints. Detection of aberrant DNA and chromosome structures leads to the coordinate induction of checkpoint pathways and DNA repair systems by these proteins (9). Activation of a DNA damage checkpoint results in cell cycle arrest, allowing time for DNA repair or, in its absence, cell death.
Cells derived from ataxia telangiectasia patients and ATM−/− knockout mice are hypersensitive to ionizing radiation and display chromosomal instability and defects in cell cycle checkpoints. The regulation of checkpoints by ATM has been studied extensively, and a number of ATM substrates have been identified (10, 11). ATM also appears to play a direct role in DNA repair at sites of DNA damage (11). This ATM function may be mediated by modification of repair proteins, such as the phosphorylation of BRCA1, which is induced by ionizing radiation (12).
Studies with cells made null for ATR (13) and cells that express dominant-negative ATR proteins have also implicated ATR in cell cycle checkpoint control (14, 15). Furthermore ATR, like ATM, responds to DNA damage caused by ionizing radiation, and some data suggest that phosphorylation is sequential, with ATM kinase being activated first (11). ATR and ATM seem therefore to be operating in similar or overlapping pathways (10). However, the kinase activities of these proteins also have distinct functions; for example, BRCA1 is phosphorylated by ATR, but not ATM, in response to damage induced by UV light and stalled DNA replication forks (16).
In addition to the phosphorylation of checkpoint and DNA repair proteins, ATM and ATR share in vitro and in vivo sensitivity to the radiosensitizing agent caffeine (17–20). The IC50 values for kinase inhibition of these two proteins are similar and fall within 1–2 mM in vitro (20). We have observed that although ATM function is required for the residual retroviral transduction that occurs in cells that are deficient in nonhomologous end-joining proteins such as DNA-PK, retroviral transduction is normal in ATM-deficient cells (21). Of relevance to the experiments reported here, DNA-PK is not sensitive to caffeine (20). In the present study, we examined the effect of ATR on the retroviral integration process by using the inhibitor caffeine, as well as cells that express a dominant-negative, kinase-dead, ATRkd protein. The results suggest that successful completion of the integration process (Fig. 1) depends on ATR activity and establish a clear distinction between ATM and ATR function in response to this form of host DNA “damage.”
Materials and Methods
Cells and Viruses.
GM847/ATRkd is a simian virus 40-transformed human fibroblast cell line stably transfected with a mutant ATR gene under the control of a tet-inducible promoter (14). Upon addition of doxycycline (1–5 μg/ml) the cells overproduce a protein containing a D2475A substitution that inactivates the kinase activity. HeLa, 293T, and GM847/ATRkd cells were maintained in DMEM supplemented with 10% FBS and Pen/Strep. The vesicular stomatitis virus-G pseudotyped HIV-1 vector carrying the lacZ reporter was prepared as described (22). Preparation of the avian sarcoma virus (ASV) vectors has been described (8). Titers of infectious units of HIV and ASV vectors were determined by transduction assays. The titer of the ASV (IN−) vector was calculated by measuring reverse transcriptase activity in viral particles and comparing the results with activity in ASV (IN+) particles of known titer. AT221JE-T cells and derivative lines were maintained as described (21).
Chemicals.
Caffeine was obtained from Sigma and dissolved in water at 100 mM concentration. Doxycycline was obtained from CLONTECH and dissolved in water at 10 mg/ml. Etoposide (ETP) was obtained from Sigma and dissolved in DMSO (stock concentration 30 mg/ml). Stocks were stored at −20°C.
Colony Assays and Cell Growth in the Presence of Caffeine and Doxycycline.
To determine the effect of caffeine on cell growth, HeLa cells were plated at a density of 105 cells per 60-mm dish in the presence of caffeine. The drug was kept on cells for 24 h and then removed. At indicated intervals, cells were harvested and viable cells were counted. To determine the effect of caffeine on colony formation, 103 HeLa cells were plated on each 60-mm dish in the presence of caffeine; the drug-containing medium was removed after 24 h and colonies were counted 9 days later. To determine whether G418 treatment potentiates caffeine toxicity, G418-resistant HeLa cells were plated as above, in the presence of caffeine and 1 mg/ml G4178. Medium was replaced after 24 h with medium containing only G418, and colonies were counted 8 days later. To determine whether doxycycline treatment affects the growth of GM847/ATRkd cells, 105 cells were plated per 60-mm dish in the presence of the drug. After 48 h doxycycline-containing medium was replaced, and viable cells were counted at the indicated intervals.
Transduction Assays.
HeLa cells were plated at 105 per 60-mm dish (Fig. 2A) or 105 per well of a 24-well plate (Fig. 2C) and infected the next day with either the HIV-1- or ASV-based vectors for 2 h in the presence of 10 μg/ml DEAE dextran. The virus-containing medium was then replaced with fresh medium. Two to seven days after infection with the HIV-1 vector (as indicated) cells were stained with a β-galactosidase assay (Stratagene). Cells were selected for G418 resistance 1 day after infection with the ASV vector by addition of 1 mg/ml G418. Caffeine was added to cells together with the infecting virus and maintained in the medium until the next day (24 h), at which time caffeine-containing medium was replaced with fresh medium. After infection with ASV, G418 was also added.
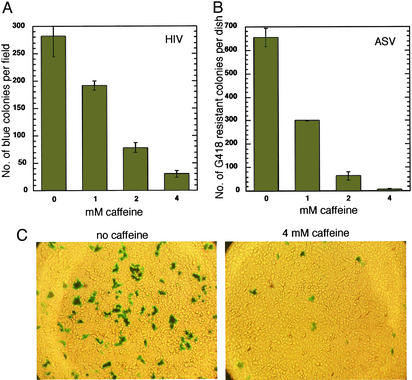
Figure 2.
Effect of caffeine on transduction by HIV-1- and ASV-based vectors. (A) HeLa cells were infected with the HIV-1(lacZ) vector and exposed to caffeine for 24 h, as described in Materials and Methods. Five days postinfection, cells were stained with a β-galactosidase assay and blue colonies were counted. (B) HeLa cells were infected with the ASV neor vector and exposed to caffeine for 24 h. After removal of caffeine, G418 was added to a final concentration of 1 mg/ml. G418-resistant colonies were counted 6 days postinfection. (C) Micrograph of HIV-1-transduced cells in the presence or absence of 4 mM caffeine. Cells were stained 3 days postinfection (see Materials and Methods). (Magnification: ×200.)
ATM-deficient AT22IJE-T cells and AT22IJE-T cells containing a vector-encoding ATM or an empty vector (12) were plated at 5 × 104 per well of a 24-well plate, two wells for each point, and infected the next day with the HIV-1 vector for 2 h in the presence of 10 μg/ml DEAE dextran. Caffeine was added to cells together with the HIV-1 vector and maintained in the medium until the next day (24 h). Cells were stained 3 days postinfection.
GM847/ATRkd cells were plated at 105 per 60-mm dish and infected the next day with either the HIV-1 or ASV vector for 2 h in the presence of 10 μg/ml DEAE dextran. Doxycycline was added at the time of plating and kept on cells for 24 h after addition of the virus. To determine the number of HIV-1 (lacZ) transductants, ATR-deficient cells were stained 48 h after infection.
Apoptosis Assay with ATRkd-Expressing Cells.
GM847/ATRkd cells were treated with 5 μg/ml doxycycline, and the next day aliquots were either mock-infected or infected with the ASV IN+ or IN− vectors [multiplicity of infection (moi) 10] in the presence of 5 μg/ml DEAE dextran. A fourth aliquot of these cells was treated with 50 μM ETP. Doxycycline was maintained in the medium after infection and ETP treatment. As a control, aliquots of GM847/ATRkd cells to which no doxycycline was added were treated in the same way. After 24 h, cells were harvested and stained with a terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) assay (In Situ Death Detection kit, Fluorescein, Roche Diagnostics). Stained cells were then quantitated by flow cytometry.
Western Blot Analyses.
For Western blot analysis of neor expression, the polyclonal population of G418-resistant HeLa cells (≈1,000 clones) was exposed to caffeine for 24 h. Cells were then harvested and lysed, and lysates were subjected to electrophoresis and analyzed by Western blot assays with a neomycin phosphotransferase II antibody (Upstate Biotechnology, Lake Placid, NY).
For detection of ATRkd, GM847/ATRkd cells were exposed to doxycycline (1 or 5 μg/ml) for 24 h. Cells were then harvested and Western blot analysis was performed with an anti-ATR antibody (Ab-2, Oncogene Science).
Real-Time PCR.
Extrachromosomal DNA from infected and uninfected cells was prepared by HIRT extraction. Real-time PCR amplification, data acquisition, and analysis were performed with the Cephid Smart Cycler, Sunnyvale, CA. Viral sequence primers directed against ASV pol were selected by using Primer Express (Applied Biosystems) and had the following sequences: forward primer, 5′-TCA GCG ATA GTC GTA ACT CAG CAT-3′; reverse primer, 5′-AGC CGT GGC CCA ATG AT-3′; probe, 5′-(FAM)CC GTG TTA CAT CGG TTG CTG CAC AA (BHQ)-3′. Results were normalized with those from primers against mtDNA (23). Each reaction contained 1× reaction buffer (20 mM Tris⋅HCl, pH 8.4/50 mM KCl), 0.25 mM each dNTP, 2.5 mM MgCl2, 400 nM primers, 200 nM probe, and 2.5 units of Platinum Taq polymerase (Invitrogen). Relative quantitation was calculated with the comparative cycle threshold method by using the untreated samples as the reference (Applied Biosystems Prism 7700 Sequence Detection System; Perkin–Elmer, P/N 4303859 Rev. A stock no. 77802-001). This method was validated with a 5-fold dilution curve of the untreated sample (slope was −0.0234). Similar results were obtained with calculations from a standard curve generated from a viral plasmid. Relative quantitation was averaged and standard deviations were determined between independent experiments.
PCR Detection of Circle Junctions in Vivo.
HeLa cells (106) were plated per dish and infected the next day with 1 ml of the undiluted ASV vector (titer ≈1 × 106 G418-resistant colony-forming units per ml), for 2 h, in the presence of 10 μg/ml DEAE dextran. Caffeine-containing medium was replaced after 24 h. Cells were harvested the next day and extrachromosomal DNA was extracted by using the HIRT method. DNA was dissolved in 50 μl per sample and 1–10 μl was used for PCR. Circle junctions were amplified by using primers comprising ASV LTR sequences. The sequence of the upstream primer was 5′-ACC AAT GTG GTG AAT GGT CAA-3′, and the sequence of the downstream was 5′-CTA CGA GCA CCT GCA TGA AGC-3′. PCR was run for 45 cycles, 94°C 30 s, 55°C 30 s, and 72°C 30 s. PCR products were analyzed on 1.5% agarose gels.
Alu-PCR.
We first established that infection at moi 0.01 produces a readout in the linear range for detection of host-viral DNA junctions. Cells were harvested 24 h after infection and chromosomal DNA was extracted. DNA concentrations of all samples were normalized by UV absorbance. PCRs (50 μl) contained 50 mM KCl, 20 mM Tris⋅HCl buffer (pH 8.4), 5 mM MgCl2, 200 μM dNTP, and 0.5 units of Taq polymerase (GIBCO/BRL). One hundred nanograms of chromosomal DNA was used in the first round of PCR with Alu primer 5′-GCC TCC CAA AGT GCT GGG ATT ACA G-3′ and ASV virus primer 5′-GGC TTC GGT TGT ACG CGG TTA GGA GT-3′. Samples were denatured at 92°C for 3 min, and then subjected to 20 PCR cycles of 92°C for 40 s, 65°C for 40 s, and 72°C for 1 min 30 s. Products of the first round were diluted 1/1,000 and used in the 25-cycle second round (nested) with viral LTR primers: 5′-AGG TGC ACA CCA ATG TGG TG-3′ and 5′-AAA AGC ACC GTG CAT GC-3′. Second-round PCR was cycled as follows: 92°C for 3 min; 25 cycles of 92°C for 40 s, 58°C for 40 s, 72°C for 40 s. All PCRs were performed in a Genius Techne Laboratories (Princeton) instrument. Products (10 μl) of the PCRs were loaded on the 2% agarose gel and subjected to Southern hybridization. The Southern probe was amplified from a plasmid with a cloned ASV genome by using LTR primers 5′-CAA ATG GCG TTT ATT GTA TCG-3′ and 5′-GAT TGG TGG AAG TAA GGT GG-3′ and labeled with [α-32P]dATP (ICN). Radioactive bands were detected overnight by exposure with Kodak BioMax MR film.
Results
The Number of HeLa Cells That Are Stably Transduced by HIV-1- and ASV-Based Vectors Is Reduced by Treatment with Caffeine.
To determine the effect of caffeine on the production of stable transductants by HIV-1, HeLa cells were infected with an HIV-1-based vector that expresses a lacZ reporter gene (22), in the presence of a range of concentration of the drug. Caffeine was added at the time of infection and removed after 24 h. The numbers of transductants were determined 5–6 days later by counting β-galactosidase-positive colonies. The results showed a significant reduction in the number of colonies stably transduced by the vector, with an IC50 for caffeine estimated at 1.5 mM (Fig. 2 A and C).
To determine whether caffeine would have a similar effect on transduction by another retrovirus, this experiment was repeated with an ASV-based vector (8) that carries a neor reporter, and colonies arising from stable transductants were selected after treatment with G418. As with HIV-1, we observed a reduction in the number of cells stably transduced by the ASV vector, with IC50 ≈0.8 mM (Fig. 2B).
Caffeine Does Not Inhibit HeLa Cell Growth, Colony Formation, or Expression of the Transduced Reporter Gene Under Conditions in Which the Number of Retroviral Transductants Is Reduced.
Other investigators have reported that caffeine has no detectable effect on cell growth at 0.5–10 mM concentration (24). We observed similar results at 2 or 4 mM concentration (Fig. 5A, which is published as supporting information on the PNAS web site, www.pnas.org). To test the effect of caffeine on colony formation, we plated uninfected HeLa cells at a low density and exposed them to the drug for 24 h. The results showed that such treatment with caffeine had no significant effect on either colony number or colony size (not shown) at up to 4 mM concentration (Fig. 5B, triangles). Finally, we examined the combined effects of G418 and caffeine treatment on colony formation by HeLa cells. An ASV-neor-transduced, G418-resistant population of HeLa cells was treated with the concentration of G418 used to select resistant colonies, together with the indicated concentrations of caffeine. We again observed no effect on colony formation by HeLa cells under these conditions (Fig. 5B, circles). Thus, the reduction in the number of cells that are stably transduced by the ASV vector cannot be attributed to the effect of caffeine on colony formation by the HeLa cells.
As we evaluated the effect of caffeine by expression of retrovirus-encoded reporters (Fig. 2), we considered the possibility that the observed reduction might be caused by suppression of reporter gene expression after stable retroviral DNA integration. Therefore, we investigated the effect of caffeine on production of the neomycin phosphotransferase protein in HeLa cells that had been stably transduced by the ASV vector. The results showed no reduction in the amounts of reporter protein after treatment of these cells for 24 h with up to 4 mM caffeine (Fig. 5C).
Effect of Caffeine on Amounts of Viral DNA and Its Nuclear Import, and Host-Viral DNA Junctions.
To determine whether caffeine affects the synthesis of viral DNA by reverse transcriptase, or other steps preceding reverse transcription, we infected HeLa cells with the ASV vector in the presence of caffeine and extracted extrachromosomal DNA at 24 h postinfection. The amount of viral DNA was determined by quantitative PCR. We found that addition of caffeine at up to 4 mM final concentration had only a modest effect on the amount of viral DNA detected (Fig. 3A), which could not account for the observed reduction in number of transduced cells. Consistent with this result, similar concentrations of caffeine had no detectable effect on the activity of reverse transcriptase in permeabilized ASV particles (data not shown).
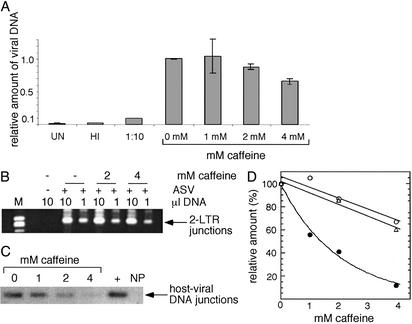
Figure 3.
Effect of caffeine on the synthesis of viral DNA, its nuclear import, and the yield of host-viral junction DNA. HeLa cells were infected with the ASV vector and treated with caffeine for 24 h, at which time cells were harvested. (A) Quantitation of viral DNA. Real-time PCR was used to determine the amount of viral DNA relative to mtDNA as described in Materials and Methods. Cells were infected at moi 0.1 and the average of two independent experiments analyzed in duplicate are shown. UN, uninfected cells, HI, cells infected with the heat-inactivated virus; 1:10, cells infected with 1:10 dilution of the virus. (B) Nuclear import of viral DNA determined by formation of LTR–LTR circle junctions. Detection of circle junctions in cells infected at moi 1.0 by PCR is described in Materials and Methods. The expected band is indicated by an arrow. Band intensity at 1 μl of DNA was analyzed by a PhosphorImager and is plotted in D as percentage intensity of circle junctions in the absence of caffeine. (C) Detection of host-viral junction DNA. Covalent joining of viral and cellular DNA after infection at moi 0.01 was evaluated by Alu-PCR as described in Materials and Methods. NP, no Alu primer in the first round of PCR (infection in the absence of caffeine); +, positive control (HeLa cells stably transduced by the ASV vector). Results were analyzed by using a PhosphorImager and are plotted in D as percentage intensity in the absence of caffeine. (D) Comparison of the affects of caffeine on the relative amounts of total, nuclear, and host-viral junction DNA after infection with the ASV vector based on quantitation of data in A–C. Open circles and top curve, total viral DNA; open triangles and middle curve, nuclear viral DNA (circle junctions); filled circles and bottom curve, host-viral DNA junctions. The latter percentages represent an average of two experiments, one of which is shown in C.
Caffeine could also conceivably affect the nuclear import of preintegration complexes, a step that precedes the integrase-mediated joining reaction. To test this possibility, we determined the effect of the drug on formation of viral DNA circle junctions, which serve as a marker for nuclear import of preintegration complexes (25). As shown in Fig. 3B, formation of circle junctions by the ASV vector was not affected significantly by 2 mM caffeine, and only a modest decrease, similar to that seen with total viral DNA, was observed at 4 mM caffeine.
To determine whether caffeine affects the formation or stability of viral-host DNA junctions in vivo, HeLa cells were infected with the ASV vector and treated with caffeine, and the DNA was analyzed 24 h later with Alu-PCR (see Materials and Methods). This method detects the covalent joining of viral and host DNA, irrespective of whether such junctions are repaired or unrepaired. The results showed a dose-dependent reduction in the relative amount of such joined DNA in the infected cultures, with an ≈10-fold (88%) decrease in the presence of 4 mM caffeine (Fig. 3 C and D), consistent with the observed reduction in the number of transductants shown in Fig. 2. A comparison of the relative amounts of total, nuclear, and host-viral junction DNA (Fig. 3D) illustrates the disproportionate reduction in junctions with increasing caffeine concentration. This difference could be explained either by selective inhibition of the integrase-catalyzed processing or joining reaction (Fig. 1) or instability of the integration intermediate in the presence of this DNA damage-sensitizing drug. Based on our earlier observations with nonhomologous end-joining-deficient cells (8, 21), it is also possible that unrepaired integration intermediates induce cell death, with a concomitant loss of these cells from the population. These possibilities are considered below.
Caffeine Does Not Inhibit the Activities of ASV and HIV-1 Integrases in Vitro.
It has been reported that retroviral integrase can be inhibited by treatment with caffeine-related compounds, such as dicaffeoylquinic acids (5, 26). These compounds also inhibit HIV-1 infection in cell culture (5). To determine whether caffeine inhibition of stable retroviral transduction is a consequence of its effect on retroviral integrases, we assayed the activities of HIV-1 and ASV integrases in vitro (27) in the presence of this drug. We observed that the processing and joining activities of ASV integrase and the processing activity of HIV-1 integrase were unaffected by caffeine at up to 10 mM concentration (not shown). It seems unlikely therefore that the reductions observed in the results of Figs. 2 and 3 can be explained by caffeine inhibition of the retroviral integrase.
Stable Transduction of ATM-Deficient and ATM-Proficient Cells Is Inhibited by Caffeine.
The major cellular targets of caffeine, with respect to the DNA damage response, are the ATM and ATR kinases (17–20). Because retroviral transduction is normal in ATM-deficient cells (21), we speculated that the caffeine effects observed were unlikely to be caused by a requirement for this protein. To test this hypothesis, we examined the effect of caffeine on HIV-1 transduction of ATM-deficient and ATM-proficient cells (Table 1). For these experiments we used an ataxia telangiectasia cell line in which ATM function was restored by stable transfection with an ATM-expressing plasmid; ataxia telangiectasia cells stably transfected with an empty vector served as a control. As we had observed previously, deficiency of ATM had no detectable effect on the efficiency of transduction by HIV-1 in the absence of caffeine. Furthermore, as retroviral transduction was inhibited similarly by caffeine, independent of the presence or absence of ATM, the drug's effect must be mediated through another protein kinase.
Table 1.
Effect of caffeine on transduction of ataxia telangiectasia cells that express an empty vector or a vector encoding ATM
| Cell line | No. of lacZ-transduced (blue) colonies at caffeine concentration, mM
|
||||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 4 | ||
| AT22IJE-T | (ATM−) | 284 | 171 (60) | 88 (31) | 40 (14) |
| AT22IJE-T/ATM | (ATM+) | 347 | 262 (76) | 144 (41) | 64 (18) |
| AT22IJE-T/empty | (ATM−) | 313 | 234 (75) | 132 (42) | 46 (14) |
Numbers in parentheses are percentages of colonies observed in the absence of caffeine. Cells were infected with the HIV-1 (lacZ) vector and Lac+ (blue) colonies detected as described in Materials and Methods; colony counts from two plates were averaged for each datum point.
Caffeine also inhibits a related kinase, mTOR, in vitro, at concentrations similar to those that inhibit ATM and ATR (20). To determine whether mTOR is involved in retroviral transduction, we infected HeLa cells and treated them with the mTOR-specific inhibitor, rapamycin. As we observed no effect of this inhibitor on retroviral transduction (data not shown) we conclude that mTOR is not required for this process. These data are consistent with the hypothesis that it is the inhibition of ATR, and not ATM or mTOR, that leads to a reduction in the number or stability of integrated viral genomes after treatment with caffeine.
A Reduced Number of Stable Transductants Is Observed When Cells Overexpress a Dominant-Negative ATR Mutant.
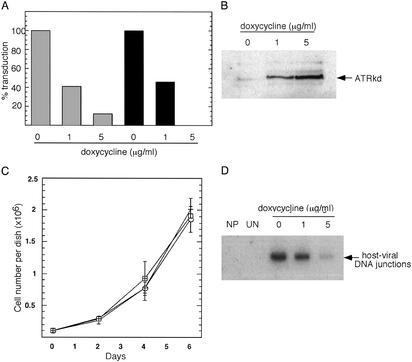
To examine the role of ATR more directly, we infected cells that are defective in ATR function. Knockout of ATR is embryonic lethal in mice, and cultured cells die rapidly after the ATR gene is excised (13, 28, 29). The absolute requirement for ATR for cell viability presents a technical barrier to analysis of the effect of its absence on the retroviral integration process. On the other hand, cells that overexpress the dominant-negative ATRkd protein (doxycycline-inducible expression) are viable and show deficiencies in DNA repair that are distinct from those of cells that lack ATM (10, 11, 14, 30). As illustrated in Fig. 4A, after infection of such cells we observed a dramatic, doxycycline-dependent reduction in the percentage of stable HIV-1 transductants that was the same with two concentrations of virus that differed 10-fold; similar results were observed with the ASV vector data (data not included). These effects cannot be attributed to doxycycline alone, as treatment of parental cells with this drug had no effect on the yield of stable transductants (data not shown). Furthermore, the reduction in the number of transduced cells was correlated with the amount of the dominant-negative ATRkd protein present (Fig. 4B). As shown in Fig. 4C, expression of ATRkd had no significant effect on the cell growth rate. Thus, reduction in the number of transductants produced after infection with this vector could not be caused by a general inhibition of cell growth. Finally, we observed that, as with caffeine treatment, the relative amount of host-viral junction DNA, detected by Alu-PCR, was reduced upon overexpression of ATRkd (Fig. 4D), even though the amount of viral DNA was similar to that in the uninduced control (not shown). One possible explanation for this result is that the integration process cannot be completed in the absence of normal ATR function and that all or some cells that contain unrepaired integration intermediates fail to survive the infection and are lost from the population. This interpretation would be consistent with other deficiencies in DNA repair reported for ATRkd-expressing cells (14–16, 30, 31).
Figure 4.
Inhibition of ATR function leads to a reduction in the number of retroviral transductants and the yield of host-viral junction DNA. (A) Effect of expression of a dominant-negative ATR gene (ATRkd) on transduction by the HIV-1 vector. GM847/ATRkd cells were treated with doxycycline and infected as described in Materials and Methods. Two days after infection, cells were stained with a β-galactosidase assay and blue cells were counted. Gray columns, cells infected with 10−2 dilution of the virus (moi <0.01); black columns, cells infected with 10−3 dilution of the virus. (B) ATRkd protein detected after treatment of GM847/ATRkd cells with 1 or 5 μg/ml doxycycline for 24 h. Cells were harvested and Western blot analysis was performed with an ATR antibody. (C) Growth of cells expressing ATRkd. GM847/ATRkd cells were plated at a density of 105 cells per 60-mm dish in the presence of doxycycline, which was left on the cells for 48 h and then removed. At indicated intervals, cells were harvested and viable cells were counted. Open circles, cells grown in the absence of doxycycline; crosses, cells treated with 1 μg/ml doxycycline; squares, cells treated with 5 μg/ml doxycycline. (D) Detection of host-viral junction DNA in cells that overexpress ATRkd. GM847/ATRKd cells were treated with doxycycline and infected with the ASV vector. Alu-PCR was performed 24 h postinfection as described in Materials and Methods. NP, no Alu primer in the first round of PCR (cells infected in the absence of doxycycline); UN, no virus (uninfected cells).
Infection of Cells That Express Dominant-Negative ATRkd Leads to Apoptosis.
We next examined the viability of cells that overexpress ATRkd after viral infection. In these experiments, GM857/ATRkd cells were treated for 24 h with doxycycline to induce ATRkd synthesis, and samples were then mock-infected or infected with the ASV vector (IN+) or an integrase-defective derivative (IN−). The IN− virus is competent for all early steps of viral replication, but defective for the integrase-mediated steps in the integration process (Fig. 1). A fourth sample was treated with ETP, a DNA topoisomerase II poison thought to generate DNA double-strand breaks throughout the cell cycle and shown to reduce the viability of ATRkd overexpressing cells (30). Samples of uninduced cells were treated similarly as a control. The cultures were harvested 24 h later and the percentage of apoptotic cells was determined by terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling and flow cytometry. The results (Table 2 and Fig. 6, which is published as supporting information on the PNAS web site) show significant increases in the percentage of apoptotic cells after infection with the IN+ ASV and ETP treatment, when the cells were induced to overexpress ATRkd. In contrast, less or no increase in the apoptotic fraction was detected in the induced cells that were mock-infected or infected with the IN− virus. These results support the hypothesis that formation of the retroviral integration intermediate triggers a cellular DNA damage response. They also suggest that the yields of host-viral junction DNA and stable transductants are reduced in ATRkd overexpressing cells because, lacking normal ATR function, a significant fraction of such cells do not survive the damage induced by formation of the integration intermediate.
Table 2.
Induction of apoptosis by retroviral infection of cells induced to overexpress ATRkd
| Mock-infected | ETP- treated | ASV(IN−)- infected | ASV(IN+)- infected | |
|---|---|---|---|---|
| Uninduced (− doxycycline) | 10.0 | 10.0 | 9.6 | 2.3 |
| Induced (+ doxycycline) | 17.6 | 25.5 | 7.1 | 36.1 |
| Difference | 7.6 | 15.5 | <0 | 33.8 |
Doxycycline was added or not to GM847/ATRkd cells, and the following day the cells were infected at moi 10 with either ASV vector (IN+) or with the integrase-deficient (IN−) vector, or treated with 50 μM etoposide, in the continued presence or absence of doxycycline. Twenty-four hours later, cells were stained using the TUNEL assay and analyzed as described in Materials and Methods. The percentage of cells that fell within the apoptotic window (see Fig. 6) is indicated.
Discussion
In this study, we show that treatment with the radiosensitizing agent caffeine results in a reduction in the number of cells that are stably transduced by HIV-1- and ASV-based retroviral vectors. The doses used have little or no effect on early steps that precede integrase-mediated joining of viral to host DNA, i.e., the synthesis of viral DNA and its nuclear import. In addition, caffeine has no effect on LTR-driven expression of a selectable reporter gene in vivo and does not inhibit integrase activities in vitro. However, caffeine treatment does cause a reduction in the amount of host-viral junction DNA, as determined by Alu-PCR. Because loss or inhibition of the caffeine-sensitive phosphatidylinositol 3-kinase-related kinases ATM and mTOR have no effect on the efficiency of retroviral transduction, we conclude that the relevant target of the drug is the related, caffeine-sensitive kinase ATR. This interpretation is supported by results from infection of cells that express a dominant-negative, kinase-dead ATRkd. This approach was necessary because cells are not viable if ATR is absent. An important caveat to the use of dominant-negative proteins is that effects observed may be caused by perturbation of other nontargeted proteins. However, because the results from caffeine treatment and ATRkd expression are consistent, it seems to us most likely that the effects we observe are attributable to loss of ATR function.
ATR has recently emerged as a major component for cellular DNA repair (10, 11). Its kinase activity is required in cellular response to ionizing and UV radiation, and collapsed replication forks (10, 11, 16). The findings that the integration process can be affected by caffeine and appears to require ATR function suggest that DNA repair proteins may be credible targets for therapeutic intervention in retroviral diseases such as AIDS (8). One concern with such an approach is that the inhibition of cellular proteins should not have a detrimental effect on the function of uninfected cells. As the observed reduction in the number of stable integrants occurs at drug concentrations below those that affect growth of uninfected cultured cells, these results support the concept that inhibitors of components of the DNA damage response may also serve as inhibitors of retroviral infection.
What aspect of the retroviral infection might require ATR function? The product of the first two integrase-catalyzed steps is an intermediate comprising the viral DNA flanked by gaps in host DNA sequence (Fig. 1). These DNA gaps, and perhaps discontinuities in the viral DNA, need to be repaired to create a stably integrated provirus. Furthermore, unrepaired gaps might become double-strand breaks when a cellular replication fork encounters such a lesion. Based on comparisons of experimental data with computer simulations, we have proposed that such replication fork damage, or a related event, triggers the apoptotic response in nonhomologous end-joining-deficient scid lymphocytes (32). In this context, the damage caused by an unrepaired integration intermediate may be similar to other replication fork catastrophes in which ATR has been proposed to play a pivotal role (16). Signaling through ATR-mediated phosphorylation of various targets might lead to a transient cell cycle arrest, as well as recruitment or activation of the proteins necessary for repair. Failure in either or both processes could render the integration intermediate unstable and could also lead to cell death. We have shown (Table 2 and Fig. 6) that infection of cells that overexpress the ATRkd protein induces an apoptotic response, similar to that induced upon addition of the genotoxic, DNA-topoisomerase II poison, ETP (30). No such response is observed if the infecting virus encodes a catalytically inactive integrase protein. In this model, therefore, the reduction in host-viral DNA junctions and number of transductants that we observe may reflect the loss of cells that cannot repair the damage caused by formation of the integration intermediate. Additional experiments will be required to determine the exact role of ATR in the integration process. The retroviral system may be especially useful for analysis of DNA repair-related functions as it provides a unique opportunity to distinguish the activities of the two DNA-damage responsive protein kinases, ATM and ATR.
Supplementary Material
Acknowledgments
We thank Dr. P. Adams for GM847/ATRkd cells and Drs. Adams and K. Scotto for reviewing the manuscript. This work was supported by National Institutes of Health Grants AI40385, CA71515, and CA06927 and an appropriation from the Commonwealth of Pennsylvania. The following Fox Chase Cancer Center Facilities were used in the preparation of this article: the Biochemistry and Biotechnology Facility and the Research Secretarial Services Facility.
Abbreviations
- ASV
avian sarcoma virus
- DNA-PK
DNA-dependent protein kinase
- ATM
ataxia telangiectasia mutated
- ATR
ATM and Rad3-related
- moi
multiplicity of infection
- ETP
etoposide
References
- 1.Skalka A M. In: Advances in Virus Research. Maramorosch K, Murphy F A, Shatkin A J, editors. Vol. 52. New York: Academic; 1999. pp. 271–459. [Google Scholar]
- 2.Brin E, Yi J, Skalka A M, Leis J. J Biol Chem. 2000;275:39287–39295. doi: 10.1074/jbc.M006929200. [DOI] [PubMed] [Google Scholar]
- 3.Yoder K E, Bushman F D. J Virol. 2000;74:11191–11200. doi: 10.1128/jvi.74.23.11191-11200.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazuda D J, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler J A, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller M D. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 5.Pommier Y, Neamati N. Adv Virus Res. 1999;52:427–458. doi: 10.1016/s0065-3527(08)60310-3. [DOI] [PubMed] [Google Scholar]
- 6.Pommier Y, Marchand C, Neamati N. Antiviral Res. 2000;47:139–148. doi: 10.1016/s0166-3542(00)00112-1. [DOI] [PubMed] [Google Scholar]
- 7.Katz R A, Skalka A M. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 8.Daniel R, Katz R A, Skalka A M. Science. 1999;284:644–647. doi: 10.1126/science.284.5414.644. [DOI] [PubMed] [Google Scholar]
- 9.Zhou B B, Elledge S J. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 10.Shiloh Y. Curr Opin Genet Dev. 2001;11:71–77. doi: 10.1016/s0959-437x(00)00159-3. [DOI] [PubMed] [Google Scholar]
- 11.Durocher D, Jackson S P. Curr Opin Cell Biol. 2001;13:225–231. doi: 10.1016/s0955-0674(00)00201-5. [DOI] [PubMed] [Google Scholar]
- 12.Cortez D, Wang Y, Qin J, Elledge S J. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 13.Cortez D, Guntuku S, Qin J, Elledge S J. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 14.Cliby W A, Roberts C J, Cimprich K A, Stringer C M, Lamb J R, Schreiber S L, Friend S H. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright J A, Keegan K S, Herendeen D R, Bentley N J, Carr A M, Hoekstra M F, Concannon P. Proc Natl Acad Sci USA. 1998;95:7445–7450. doi: 10.1073/pnas.95.13.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tibbetts R S, Cortez D, Brumbaugh K M, Scully R, Livingston D, Elledge S J, Abraham R T. Genes Dev. 2000;14:2989–3002. doi: 10.1101/gad.851000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou B B, Chaturvedi P, Spring K, Scott S P, Johanson R A, Mishra R, Mattern M R, Winkler J D, Khanna K K. J Biol Chem. 2000;275:10342–10348. doi: 10.1074/jbc.275.14.10342. [DOI] [PubMed] [Google Scholar]
- 18.Blasina A, Price B D, Turenne G A, McGowan C H. Curr Biol. 1999;9:1135–1138. doi: 10.1016/s0960-9822(99)80486-2. [DOI] [PubMed] [Google Scholar]
- 19.Hall-Jackson C A, Cross D A, Morrice N, Smythe C. Oncogene. 1999;18:6707–6713. doi: 10.1038/sj.onc.1203077. [DOI] [PubMed] [Google Scholar]
- 20.Sarkaria J N, Busby E C, Tibbetts R S, Roos P, Taya Y, Karnitz L M, Abraham R T. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- 21.Daniel R, Katz R A, Merkel G, Hittle J C, Yen T J, Skalka A M. Mol Cell Biol. 2001;21:1164–1172. doi: 10.1128/MCB.21.4.1164-1172.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 23.Butler S L, Hansen M S, Bushman F D. Nat Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 24.Asaad N A, Zeng Z C, Guan J, Thacker J, Iliakis G. Oncogene. 2000;19:5788–5800. doi: 10.1038/sj.onc.1203953. [DOI] [PubMed] [Google Scholar]
- 25.Coffin J M, Hughes S H, Varmus H E. Retroviruses. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [PubMed] [Google Scholar]
- 26.Zhu K, Cordeiro M L, Atienza J, Robinson W E, Jr, Chow S A. J Virol. 1999;73:3309–3316. doi: 10.1128/jvi.73.4.3309-3316.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katz R A, DiCandeloro P, Kukolj G, Skalka A M. J Biol Chem. 2001;276:34213–34220. doi: 10.1074/jbc.M104632200. [DOI] [PubMed] [Google Scholar]
- 28.Brown E J, Baltimore D. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 29.de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smith B, Carr A M, Lehmann A R, Hoeijmakers J H. Curr Biol. 2000;10:479–482. doi: 10.1016/s0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- 30.Cliby W A, Lewis K A, Lilly K K, Kaufmann S H. J Biol Chem. 2002;277:1599–1606. doi: 10.1074/jbc.M106287200. [DOI] [PubMed] [Google Scholar]
- 31.Tibbetts R S, Brumbaugh K M, Williams J M, Sarkaria J N, Gliby W A, Shieh S-Y, Taya Y, Prives C, Abraham R T. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daniel R, Litwin S, Katz R A, Skalka A M. J Virol. 2001;75:3121–3128. doi: 10.1128/JVI.75.7.3121-3128.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.