Abstract
Estrogen receptor (ER) α variants have been identified in an array of nonendothelial cells. We previously demonstrated that estrogen rapidly induces nitric oxide release via a phosphatidylinositol 3-kinase/Akt/endothelial nitric-oxide synthase (eNOS) pathway in EA.hy926 cells (immortalized human endothelial cells), which express a 46-kDa ER. We now confirm that, due to alternative splicing, the 46-kDa endothelial cell protein (ER46) is an amino-terminal truncated product of full-length ERα (ER66). ER46 is expressed in the plasma membrane, cytosol, and nucleus of resting, estrogen-deprived cells. Flow cytometric and immunofluorescence microscopic analyses demonstrated that the ER46 C but not N terminus is Ab-accessible in the plasma membrane. Inhibition of palmitoylation with tunicamycin and [3H]palmitic acid labeling demonstrated an estrogen-induced, palmitoylation-dependent plasma membrane ER46 recruitment, with reorganization into caveolae. In reconstituted, estrogen-stimulated COS-7 (ER-null) cells, membrane ER46 more efficiently triggered membrane eNOS phosphorylation than ER66. Conversely, ER66 more efficiently mediated estrogen response element reporter-gene transactivation than ER46. These results demonstrate that ER46 is localized and further dynamically targeted to the plasma membrane in a palmitoylation-dependent manner. ER46 more efficiently modulates membrane-initiated estrogen actions, including eNOS activation, than full-length ER66. These findings may have important implications in vascular-specific targeting of estrogen receptor agonists.
Estrogen receptor (ER) is traditionally defined as a ligand-dependent transcription factor located in the cytosol or nucleus. The presence of a plasma membrane functional ER, which correlates with rapid estrogen signaling, has been debated for decades and remains unresolved (1, 2), due in part to the lack of unequivocal molecular identity of this receptor. Expression of truncated ER variants in mammalian tissues is not uncommon. A 46-kDa isoform has been biochemically identified in human tissues including MCF-7 cells (3), osteoblasts (4), and vascular endothelial cells (ECs; ref. 5). This protein (ER46) is devoid of the N terminus (A and B domains) but otherwise shares identity with ERα, one of the three cloned ERs (ERα, ERβ, and ERγ). It is capable of forming a heterodimer with a full-length ERα (ER66) in vitro and in vivo (3, 4). In ER-negative tissues, transfection of ER46 promotes nuclear transactivation of ER66-responsive genes, and in ER66-positive tissues, ER46 dimerizes with ER66 and serves as a competitive inhibitor of ER66 for DNA binding (3, 4). Although the genomic actions of ER46 have been suggested in the context of cell proliferation, the role of ER46 in rapid, nongenomic signaling has not been defined. We previously reported that in 17β-estradiol (E2)-deprivation conditions, both primary human umbilical vein ECs (HUVECs) and immortalized vascular ECs (EA.hy926) express a 46-kDa protein that immunoreacts with selective ERα Abs and seems capable of transducing E2-triggered rapid signaling. EA.hy926 cells phenotypically represent fully differentiated vascular ECs. However, unlike HUVECs that express both ER46 and ER66 (5), EA.hy926 cells exclusively express ER46 in our culture conditions and are unable to support E2-dependent estrogen response element (ERE) transactivation (5). By contrast, this cell line is highly responsive to membrane-impermeant E2 within minutes as demonstrated by NO release and cGMP production through an E2-stimulated, ligand-dependent phosphatidylinositol 3- kinase/Akt/endothelial NO synthase (eNOS) pathway (5, 6). We thus hypothesize that ER46 is preferentially membrane-associated and responsible for E2-triggered rapid signaling in the endothelium. In this report, we present the molecular and cellular details to demonstrate that ER46 is an ERα isoform and is capable of mediating eNOS activation specifically from its membrane site in EC.
Materials and Methods
Materials.
F10 (E/F domains), anti-GDI, and anti-c-myc Abs were purchased from Santa Cruz Biotechnology. H222 (E domain) Ab was obtained from Abbott. Anti-calnexin Ab was purchased from StressGen Biotechnologies (Victoria, Canada); anti-β-COP Ab was from Affinity Bioreagents (Neshanic Station, NJ); anti-E2 antiserum was from OEM Concepts (Toms River, NJ); 1D5 (A/B domains) Ab was from DAKO; antiphospho-eNOS (Ser-1177) Ab was from Cell Signaling (Beverly, MA); and anti-TBP, anti-eNOS, and anti-caveolin-1 (cav-1) Abs were from Transduction Laboratories (Lexington, KY). The hER19Pu (obtained from P. Chambon, Université Louis Pasteur, Strasbourg, France) insert ER cDNA encodes an ER that excludes the first two amino acids translated from the alternative translation start site, and what would be considered the complete NH2 terminus of ER46. For simplification, the recombinant ER expressed as a consequence of hER19Pu transfections will be subsequently referred to as “ER46.” All other chemicals and reagents were purchased from Sigma unless otherwise indicated.
Cellular and Biochemical Assays.
Cell maintenance, transient transfection with Lipofectamine Plus reagent (GIBCO/BRL), immunoblot, immunoprecipitation, fluorescence-activated cell sorter analysis, and eNOS activity assays all were performed identically to those described (5–7). Protein mass on immunoblot was determined by Rainbow molecular weight markers (Amersham Pharmacia).
Subcellular Fractionation.
Plasma membranes were isolated by a cationic colloidal silicon isolation technique (8), and caveolae isolation was performed according to the detergent-free method (9, 10).
Confocal Microscopy.
Cells plated onto gelatin-coated coverslips were fixed in 4% paraformaldehyde/PBS for 30 min, mildly permeabilized with a low concentration (5 μg/ml) of digitonin (11) for 5 min, blocked in 1% normal goat serum/0.1% Tween 20 in Tris-buffered saline for 1 h, and incubated with primary Abs and secondary Abs (GAM-Alexa-488 or GAR-Alexa-568, 1:50 dilution; Molecular Probes) for 2 h, respectively. Cytosolic proteins were partially removed by extensive washes (12), and cells were microphotographed in 1-μm sections by using a Bio-Rad MRC confocal system with a Zeiss Axiovert microscope, and processed in photoshop (Adobe Systems, Mountain View, CA).
RT-PCR.
Total cellular RNAs were extracted by TRIzol reagent (GIBCO/BRL), and RT-PCR was carried out as described (13). The 5′ primers for ERα are 5′-GGGCGCCGCCTACGAGTT (460–477 bp), 5′-GCCCTACTACCTGGAGAA (676–693 bp), 5′-CCAGGGTGGCAGAGAAAG (766–783 bp); and the 3′ primer for ERα is 5′-CTCTCAGACTGAGGCAGGGAAACC (2,060–2,075 bp).
Cell Surface Biotinylation and Cross-Linking.
Cell monolayers were biotinylated with 0.2 mg/ml EZ-link-sulfo-NHS-LC-biotin (Pierce) prepared in 10 mM Hepes (pH 8.0), 150 mM NaCl, 0.2 mM MgCl2, and 0.2 mM CaCl2 at 0°C for 30 min. The reaction was terminated by washes with 10 mM Tris⋅HCl (pH 7.5). For cross-linking, cells were incubated with 2 mM BS3 (noncleavable) in PBS on ice for 30 min, followed by extensive washes with PBS.
Radiolabeling of ER46 with [3H]Palmitate and [3H]Myristate.
EA.hy926 cells were E2-deprived for 48 h in phenol red- and fatty acid-free DMEM in the presence of 10% gelding horse serum, and quiesced for 12 h in the identical DMEM containing 0.5% BSA. Cells were washed in PBS, labeled for 4 h with 100 μCi/470 ng of palmitate/35-mm dish [9.10(n)-3H]palmitate (1.0 mCi/ml), or 100 μCi/420 ng of myristate/35 mm-dish [9.10(n)-3H]myristate (1.0 mCi/ml) in DMEM supplemented with 0.25% BSA and 18 μM cycloheximide, and treated with vehicle alone or 30 nM E2 for 10 min. After extensive washes in PBS, cells were harvested by a nonenzymatic method as described (5) and further subjected to subcellular fractionation, immunoprecipitation with F10 Ab, autoradiography of 3H-labeled ER for 8 weeks at −80°C, and immunodetection of total ER with F10 Ab.
E2-Affinity Precipitation.
Cells were chemically disrupted in hypotonic solution [10 mM Tris⋅HCl (pH 7.5), 1 mM EDTA, 1 mM DTT, 1 mM MgCl2, 1% Nonidet P-40, and protease inhibitor mixture; Roche Applied Science), precleared with 6-CMO-ω-hexylamide-agarose for 4 h and with anti-actin Abs plus protein A/G agarose beads for 4 h and subsequently incubated overnight with 20 mM E2-6-CMO-ω-hexylamide-agarose made by conjugating 10 ml of ω-aminohexyl-agarose with 414 μM N-hydroxysuccinimide, dicyclohexylcarbodiimide, and E2-6-CMO in tetrahydrofuran. The beads were then washed with high-salt buffer (2 M NaCl/2.7 mM KCl/1.5 mM KH2PO4/1 mM Na2HPO4, pH 7.2) for 30 min at 4°C and with PBS three times.
Results
Conditional ERα Transcripts in EA.hy926 Cells.
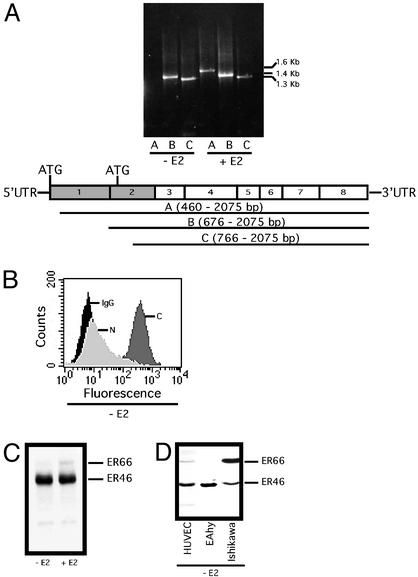
The human ERα gene has at least three translation initiation sequences and is also demonstrated to undergo alternative splicing and differential promoter activation in a tissue-specific manner (3, 14). The ERα-positive MCF-7 cells are reported to produce both a full-length (ER66) and short-form (ER46) ERα. This ER46 results from the ERα transcript alternatively spliced from exon 1E to exon 2 and is translated from the internal start codon (+752 bp, +758 bp). We therefore examined ERα transcripts in EA.hy926 with a focus on exons 1–3. Determined by RT-PCR, EAhy.926 cells after E2 deprivation (48 h) and serum starvation (12 h) expressed a short-form ERα transcript apparently lacking exons 1 and 2 (1–475 bp, Fig. 1A). The short-form transcript confines the translation initiation to the internal ATG codons with an anticipated 46-kDa product, if any. As shown in Fig. 1 B and C, the 46-kDa protein was immunoreactive with anti-C terminus-specific ERα Ab, but not anti-A/B domain Abs. These results suggest that ER46 is generated by alternative splicing of the ERα gene. Notably, the addition of 30 nM E2 to the culture for 24 h stimulated the expression of a long-form ERα mRNA with a restored N-terminal sequence (Fig. 1A). DNA sequencing verified ERα identity (data not shown). This change is consistent with the appearance of ER66 in immunoblots (Fig. 1C), suggesting that EA.hy926 cells possess a complete ERα gene and are inducible to express full-length ERα. In addition, ER46 was also found in primary vascular EC including human umbilical vein EC (Fig. 1D), human microvascular EC (data not shown), human coronary artery EC (data not shown), and ER-enriched human endometrial carcinoma Ishikawa cells by immunoblot (Fig. 1D). These data indicate that ER46 is strongly expressed in both ECs and non-ECs.
Figure 1.
Expression of ER46 as an ERα variant. Cells were E2-deprived for 48 h and serum-starved for 12 h, referred to as “pretreated,” and then incubated with vehicle (−E2) or 30 nM E2 (+E2) for 24 h. (A) RT-PCR of ERα transcripts in EA.hy926 cells. A–C are PCR products, corresponding to the use of the denoted upstream primers. (B) Fluorescence-activated cell sorter analysis of ER46 expression in EA.hy926 cells with the anti-C-terminal F10 (C), 1D5 (N), and irrelevant OKT3 (IgG) Abs. (C and D) ER immunoblot in EA.hy926 (C) and noted cells (D) using F10 Ab.
Subcellular Localization of ER46.
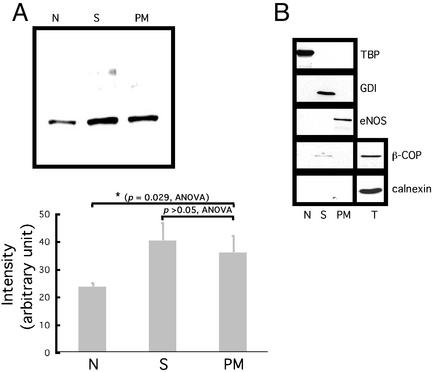
ER66 localizes to the cytosol, nucleus, and plasma membrane (minor fraction) in both MCF-7 cells and ER66 transfectants (15). To determine the ER46 compartmentalization in EA.hy926 cells, subcellular fractionation was performed to isolate nuclei, plasma membranes, and the cytosolic components (Fig. 2A). ER46 was identified by immunodetection from the cytosol, plasma membrane, and nucleus. To exclude cross-contamination of the plasma membrane fraction by cytosol, Golgi apparatus, and endoplasmic reticulum, the fraction purity was assessed by immunoblot with the marker proteins TATA box-binding protein (nucleus), GDP dissociation inhibitor (cytosol), eNOS (membrane), β-COP (Golgi), and calnexin (endoplasmic reticulum). The results confirmed that the presence of ER46 in the plasma membrane was not due to interfractional contamination (Fig. 2B).
Figure 2.
Subcellular localization of ER46. (A) Immunoblot of ER46 among different organelles with F10 Ab. Organelles were isolated from 106 pretreated cells. Proteins were precipitated by ethanol from each fraction before use. N, nucleus; S, cytosol; PM, plasma membrane. Densitometric values were determined from three independent experiments and expressed as means ± SD, with P values of compared means as noted. (B) Examination of fraction purity by immunoblotting various protein markers from the N, S, PM, and total lysate (T). TBP, TATA-box binding protein; GDI, GDP dissociation inhibitor.
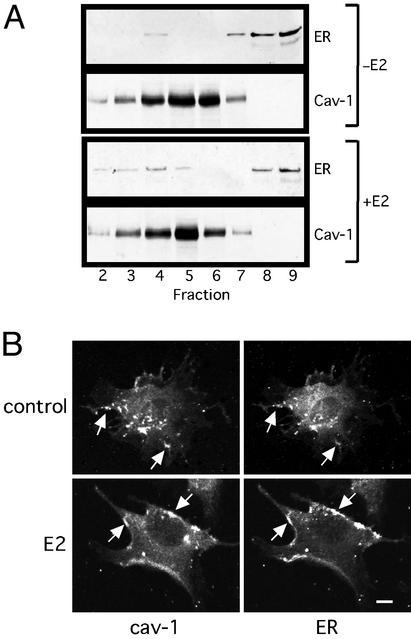
Caveolae are flask-shaped vesicular invaginations (50–100 nm) of the plasma membrane that consist of cav-1, signaling molecules, and enzymes including eNOS (16). We proposed that membrane ER46 was localized to caveolae because isolated caveolae from native vascular ECs and ER66 overexpressors contain ER66 and respond to E2 in vitro with eNOS activation (17). Thus, caveolae were biochemically isolated, followed by immunodetection of cav-1 and ER46 (Fig. 3A). The majority of ER46 sedimented to the high-density fractions that mostly contained cytosol as well as some mitochondria and Golgi. As expected, cav-1 was highly concentrated in lighter fractions (2–7). ER46 was detectable in fraction 4 as well although the signal was weak. The results suggest that only a minor fraction of ER46 resides in caveolae in resting cells. However, E2 induced the appearance of ER46 in the cav-1-enriched intermediate fractions 3 and 4. To evaluate further the possibility that ER46 resides in caveolae in situ, confocal microscopy was performed on digitonin-permeabilized, fixed cells. Fig. 3B demonstrates that at least a subset of cav-1 and ER46 colocalized in E2-treated cells. Merged images of cav-1 and ER46 in E2-treated cells confirmed substantial colocalization in the plasma membrane with a “patch-like” plasma membrane pattern (data not shown), consistent with the fractionation results.
Figure 3.
Localization of membrane ER46 in caveolae. (A) Immunoblot of ER46 and cav-1 from isolated caveolae. Pretreated EA.hy926 cells were incubated with vehicle alone (−E2) or 30 nM E2 (+E2) for 10 min before fractionation. Fractions 2–9 were subjected to immunoblotting with anti-Cav-1 and F10 Abs. (B) Confocal microscopy of cell-surface cav-1 and ER46 with anti-Cav-1 and F10 Abs. Arrow, overlapped signals. (Bar = 10 μm.)
Topology of Membrane ER46.
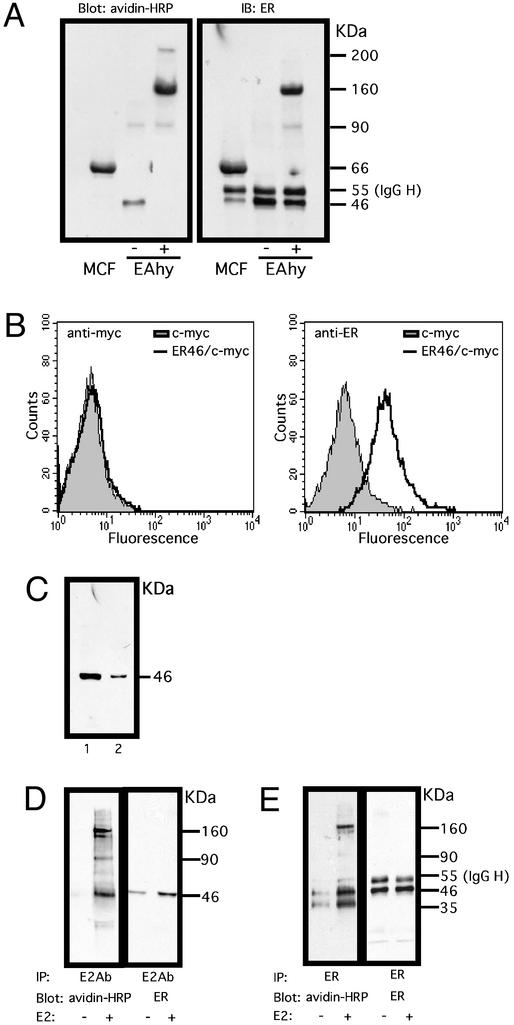
Integral membrane proteins (18) and some periplasmic proteins (19) can be chemically modified at primary amine groups of lysine residues by extracellular biotin, a conventional hapten that is widely used for cell surface labeling. To determine the presence of biotin-accessible membrane ER46 in situ, cell-impermeant sulfo-succinimidyl biotin ester was added to cultures at 0°C, at which temperature the energy-dependent exocytosis, endocytosis, internalization, and intracellular trafficking were markedly retarded. Biotin–ER was immunoprecipitated with anti-C-terminal ERα Ab, gel-electrophoresed, and blotted with avidin-horseradish peroxidase. Immunoprecipitated, biotinylated proteins (or complexes) had apparent molecular masses of 46 and 90 kDa (Fig. 4A, EAhy −). Only the 46-kDa protein was detectable by reblotting with anti-ER Abs, suggesting the presence of membrane ER46. Utilization of cell-impermeant cross-linkers (EAhy +) confirmed that ER46 biotinylation occurred at the cell surface, because this promoted disappearance of the 46-kDa band and revealed 90-, 160-, and 200-kDa proteins (complexes). It is likely that both 90- and 160-kDa proteins, which were immunoreactive with anti-ER Abs in the cross-linked samples, are nonmonomeric ERs. They may result from biotin-induced aggregation of ER with lipid moieties, the formation of SDS-resistant homo- or heterooligomers (20), or ER46-irrelevant proteins. The 200-kDa protein could be ER-irrelevant, or an ER-related higher molecular mass complex present in amount too small to be identified by chemoilluminescence. Membrane ER-positive MCF-7 cells that served as a positive control validated the specificity of membrane ER detection. Intracellular protein biotinylation was excluded by the lack of actin-biotin detection (data not shown).
Figure 4.
The topology of membrane ER46. (A) Biotinylation of ER46 in EA.hy926 and MCF-7 cells. Surface proteins of the pretreated cells were biotinylated by EZ-link-sulfo-NHS-LC-biotin. Proteins were immunoprecipitated from the total cell lysate with F10 Ab, revealed by ligand-blot with avidin-horseradish peroxidase (HRP), and reblotted with F10 Ab. −, without cross-linker BS3; +, with cross-linker BS3. (B) Fluorescence-activated cell sorter analysis of surface expression of c-myc-tagged ER46 in nonpermeabilized COS-7 cells. COS-7 cells were transfected with pCMV or ER46/pCMV (N-terminal myc epitope-tag) for 48 h. c-myc and F10 Abs were used to distinguish the N and C termini of the receptor, respectively. The N-terminal c-myc epitope tag was Ab-reactive when cells were permeabilized (data not shown). (C) Binding of ER46 to E2-affinity matrix. Pretreated EA.hy926 cell lysates were subjected to E2-affinity precipitation and the selected fractions, i.e., total cell lysate (1) and the matrix after several washes (2), were immunoblotted for ER proteins with F10 Ab. (D and E) Binding of E2 to surface ER46. The pretreated cells were treated with vehicle alone or 30 nM E2 for 10 min then biotinylated. Proteins were immunoprecipitated with E2 antiserum (D) or F10 Ab (E), revealed by ligand-blot with avidin-HRP, and reblotted with F10 Ab.
Fig. 4B demonstrates fluorescence-activated cell sorter profiles of COS-7 cells transfected with the pCMV-tag3C-ER46 (N-terminal myc epitope tag) construct, stained with either an anti-c-myc epitope or anti-C-terminal ER Ab. Because the cells were not permeabilized before staining, only that portion of the recombinant molecule which is Ab-accessible, i.e., extracellular, yielded a fluorescent signal. Only the C terminus of ER46 was Ab-reactive. Thus, the A/B domain-deleted ER46 appears to have a C-terminal ectodomain and an N terminus that is not Ab-accessible.
To simply document that ER46 was a receptor for E2, EA.hy926 lysates were incubated with an E2-affinity matrix, and E2-bound molecules were released from the matrix after high-salt (1.5 M KCl) washes. Fig. 4C is an immunoblot demonstrating a 46-kDa anti-ER (F10)-reactive protein in total cell lysates (lane 1) and, more importantly, from E2-affinity matrix-bound material (lane 2). The resistance to high-salt elution is consistent with an E2–ER46 in vitro interaction of high affinity.
To document membrane ER46 binding sites for E2, immunoprecipitation of E2-bound proteins from E2-treated, biotinylated cells, using an E2 antiserum, revealed 46-, 90-, and 160-kDa proteins that were also detected with anti-ER Abs (Fig. 4D). A similar pattern was observed when ER Abs were used for immunoprecipitation (Fig. 4E), although an additional, unknown 35-kDa protein was inconsistently detected as well. These studies demonstrate the ability of ER46 to bind E2 in situ, and the ligand-induced recruitment of ER46 or ER46-associated proteins, consistent with the results displayed in Fig. 3.
Posttranslational Modification of Membrane ER46.
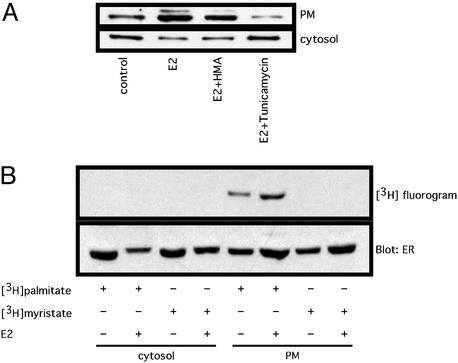
findmod and scanprosite algorithms predict that ER46 is potentially palmitoylated and myristoylated. To determine whether palmitoylation and/or myristoylation are required for ER46 membrane anchorage, appearance of membrane ER46 was monitored in the presence of fatty-acylation chemical inhibitors. By competing for the binding of palmitoyl-CoA to the protein fatty acyltransferase, tunicamycin is one of two known cell-permeant inhibitors for palmitoylation. In addition, tunicamycin is also able to block N-linked glycosylation by inactivating UDP-GlcNAc:dolichyl-phosphate GlcNAc-1-phosphate transferase. To distinguish the effects of tunicamycin on glycosylation and pamitoylation, we pretreated the cells with the protein synthesis inhibitor cycloheximide, which, because a majority of N-linked glycosylation is endoplasmic reticulum-localized and cotranslational, also disrupts both N-linked glycosylation and cotranslational palmitoylation without affecting posttranslational protein palmitoylation (21). Fig. 5A demonstrates that E2 treatment or EA.hy926 cells stimulated ER46 recruitment from the cytosol to the plasma membrane. This translocation was unaffected by the myristate antagonist (2-hydroxymyristic acids) but abrogated by tunicamycin beyond any effect seen with cycloheximide. In fact, tunicamycin also diminished basal (control) plasma membrane localization of ER46. Furthermore, metabolic labeling of EA.hy926 cells with 3H-labeled fatty acids demonstrated palmitate but not myristate incorporation into plasma membrane ER46, and that this palmitoylation is augmented in response to E2 (Fig. 5B). These findings strongly suggest that ER46 palmitoylation is a required posttranslational modification for its membrane localization.
Figure 5.
Palmitoylation of membrane ER46. (A) Effects of 2-hydroxymyristic acid (HMA) and tunicamycin on membrane localization of ER46. In the presence of 18 μM cycloheximide, the pretreated cells were incubated with or without (control) HMA (0.5 mM) or tunicamycin (30 μM) for 2 h. Vehicle (control) or 30 nM E2 was added for 10 min. The plasma membrane (PM) and cytosolic fractions were immunoblotted with F10 Ab. (B) Determination of palmitate or myristate incorporation into ER46. Pretreated cells were labeled in serum-free DMEM supplemented with 0.25% BSA, 100 μCi of [3H]palmitate, or 100 μCi of [3H]myristate for 4 h in the presence of 18 μM cycloheximide, followed by vehicle alone (−E2) or 30 nM E2 (+E2) treatment, subcellular fractionation, immunoprecipitation with F10 Ab, autoradiography of labeled ER for 8 weeks at −80°C, and immunodetection of total ER with F10 Ab.
Comparative Roles of ER46 and ER66 in Nongenomic vs. Genomic Functions.
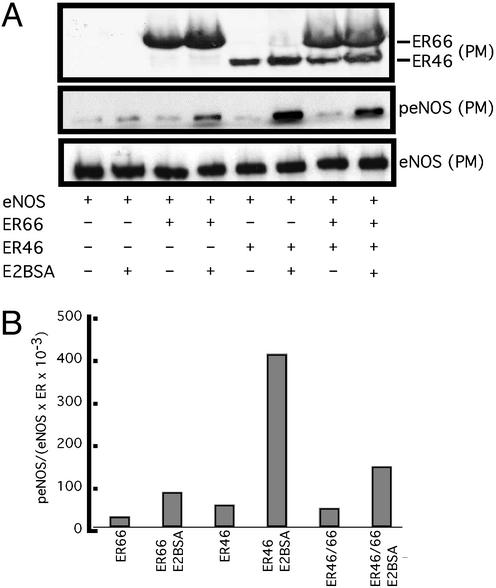
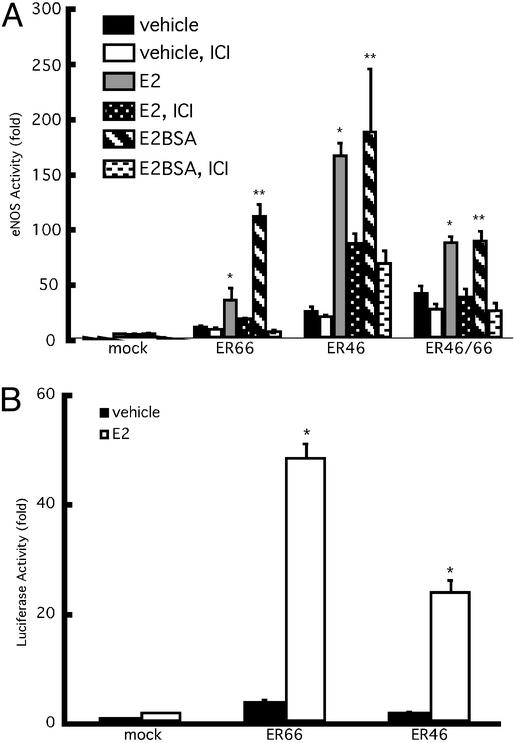
Our data (previous section) and those accumulating in the literature suggest that membrane ERs are more likely to transduce rapid activation signals, whereas full-length, traditional ERs may transactivate ERE-driven genes more efficiently. To test this hypothesis, COS-7 cells were transfected with eNOS and ER46 or ER66, stimulated with cell-impermeant E2BSA, and levels of activated (Ser-1177-phosphorylated), plasma membrane eNOS were determined. Fig. 6A demonstrates a greater level of E2BSA-stimulated eNOS phosphorylation in membranes isolated from ER46- than ER66-transfected cells. This greater efficiency could be the result of intrinsically more favorable coupling with signaling partners or preferential membrane localization (total levels of cellular ER46 and ER66 in COS-7 transfectants were equivalent; data not shown). Of note is that when coexpressing ER46 and ER66, membrane eNOS phosphorylation in response to E2BSA was quantitatively less (Fig. 6B), suggesting a repressive and/or competitive effect of ER66 on ER46 function. Similar results were obtained in eNOS activity assays of plasma membranes isolated from transfectants stimulated with vehicle, E2, or E2BSA (Fig. 7A). Fig. 7A demonstrates that the largely ER antagonist-inhibitable augmentation in eNOS activity was greater in ER46- than ER66-transfected cells, and that ER46/66 cotransfectants were less capable of responding to estrogen than singly ER46-expressing cells. E2BSA consistently stimulated an equal or greater level of eNOS activation than E2, supporting membrane receptor-mediated signaling events.
Figure 6.
Effect of ER46 on eNOS phosphorylation. COS-7 cells were transfected with eNOS and β-galactosidase in combination with ER66/pSG5, ER46/pSG5, or both for 24 h. The transfectants were treated with vehicle alone or 80 ng/ml E2BSA (equivalent to 30 nM E2) for 10 min. Immunoblots were performed with F10 Ab in the isolated plasma membrane (A), followed by densitometric determination (B). p-eNOS densitometric values are normalized to those of total membrane eNOS and of total membrane ERs.
Figure 7.
Effects of ER46 on eNOS activity and ERE reporter gene transactivation. Cells were pretreated and then incubated with vehicle alone, 30 nM E2, or 80 nM E2BSA for 24 h. Some received 10 μM ICI 182, 780 (ICI) for 30 min before the incubation with the previously listed agents. eNOS activity was assessed by measuring the conversion of l-[3H]arginine to l-[3H]citrulline (A) in the absence of exogenous calmodulin and calcium. Data are normalized to β-galactosidase (β-gal) activities and given as means ± SD (n = 4) of eNOS activity fold increase over mock-transfected vehicle control. (B) EA.hy926 cells were transfected with β-gal and ERE-Luc with and without ER66/pSG5 or ER46/pSG5 for 24 h, followed by the treatment with vehicle alone or 30 nM E2 for 24 h. Luciferase activity was then measured in cell lysates and normalized to β-gal. Values are expressed as means ± SD (n = 4). *, E2 vs. vehicle (ANOVA, within group), P < 0.05; **, E2BSA vs. vehicle (ANOVA, within group), P < 0.05.
We have shown that endogenous levels of ER46 are insufficient to generate E2-stimulated transactivation of an ERE-driven gene (5). However, Fig. 7B demonstrates that overexpression of ER46 does confer E2 responsiveness with regard to ERE-driven luciferase gene activation. More importantly, E2-stimulated luciferase activity in ER66 transfectants was significantly higher than in ER46 transfectants, despite the fact that total ER46 levels were higher than ER66 levels in the ER46 and ER66 transfectants, respectively, due to endogenous ER46 (data not shown). These data support that ER66 more efficiently effects estrogen-driven genomic functions, whereas ER46 is more capable of rapid signal transduction.
Discussion
Accumulating evidence indicates that ER46 is expressed in ovariectomized wild-type and ERα-neo knockout mice (22). ERα is demonstrated to encode ER46 by alternative splicing in non-ECs and by internal ribosome entry in vitro (3, 23). We now confirm that vascular ECs also express ER46, which is inherently associated with the plasma membrane (Figs. 1 and 2). This finding is in agreement with the k-nearest neighbors (psort ii) program that predicts 26.1%, 30.4%, 17.4%, and 26.1% of ER46 is localized to the nucleus, cytoplasm, mitochondria, and membrane fractions, respectively. By contrast, 73.9%, 8.7%, 0.1%, and 17.3% of ER66 carries the comparative predictions. Thus, the differential compartmentalization of ER46 and ER66 in the cell suggests that the functional site and primary role of each receptor may be different.
Membrane ER46 is constitutively palmitoylated, possessing an “N-in” and “C-out” topology (Figs. 4 and 5). ER46 and ER66 have no polybasic stretches of amino acids, apical membrane attachment domains, glycosylphosphatidylinositol-anchor sites, or PDZ domains. Thus, chemical modification, the interplay with other membrane lipids and proteins, or an atypical hydrophobic segment become candidate mechanisms for ER coupling to the membrane. In this study, we found that posttranslational palmitoylation is required for ER46 membrane localization and for E2-induced recruitment of ER46, most likely to caveolae. ER46 has 12 cysteines that could be acylated by palmitate. The chemical susceptibility of thioester bond formation between palmitate and cysteine residue(s) is known to permit a rapid and reversible acylation of proteins (24) that facilitate molecular shuttling (25) and membrane receptor clustering (26). Therefore, it is possible that by timely palmitoylation (27), ER46 is dynamically recruited from the cytoplasmic pool to plasmalemmal caveolae in which it could encounter other palmitoylated molecules including eNOS, the G protein α-subunit, Src kinases, Ha-Ras, and cav-1. The interaction between palmitate tails of ER46 and other caveolae modules may emerge as a sorting signal for the directional recruitment and rearrangement of the receptor, analogous to the engagement and retention of eNOS, Gi1α, P59hck, and Src kinase in caveolae. Thus, palmitoylation seems to be a common regulatory mechanism shared by a growing list of caveolae proteins (i.e., receptors, enzymes, signaling molecules) that could be locally controlled to form an eNOS-centered signaling complex. Whether ER46 is modified by nonenzymatic palmitoylation, autopalmitoylation, or an unidentified membrane palmitoyltransferase (27) remains to be determined.
The importance of endothelial ER46 is underscored by our finding that membrane ER46 is superior to ER66 for eNOS activation (Fig. 6 A and B). The difference results, at least in part, from the enhanced ability (4-fold greater than ER66) of membrane ER46 to phosphorylate eNOS on Ser-1177 (Fig. 6A). The possible difference in the orientation of the ligand-binding pocket [with differential sensitivities to the antagonist ICI 182,780 (Fig. 6B)] may explain this hierarchy in eNOS activation. In the aqueous phase, ligand binding requires an “open” conformation of ER (28), whereas as little as one amino acid (Y537) mutation can render an unfolded ER that even possesses some ligand-independent activity (29, 30) and is potentiated by cav-1 (31). The short-form ER46 thus may favor an open conformation that facilitates the kinetics of ligand–receptor interaction manifested by rapid ligand binding and release. It could potentially accelerate the “fire” of signal transduction that enables a considerable pool of eNOS to be recruited and activated by the given amount of ligands and receptors. This possibility could also explain some constitutive activity of ER46 as observed in this case (Fig. 6B). Furthermore, focal engagement of signaling regulators to ER may be different for ER46 and ER66. Indeed, the structure-sensitive binding of coactivators and corepressors to ER has been proven as one mechanism underlying the tissue-specific actions of selective ER modulators (32). Although E2 stimulates a phosphatidylinositol 3-kinase/Akt/eNOS pathway in either ER46 (6)- or ER66 (33)-expressed EC, whether ER66 and ER46 have selectively engaged distinct proximal elements in this pathway is unknown. Any such differences could bias the efficiency of eNOS activation, as a result of the net balance of all positive and negative regulators of the loop.
We have also observed that ER66 and ER46 are differentially efficient with regard to gene transactivation (Fig. 6C). This difference may be due to the absence of AF-1 of ER46, which, on ER66, generally augments the activity of ligand-bound ER as a transcriptional enhancer, or to less efficient nuclear targeting. By forming a heterodimer with ER66, ER46 is reported to act as a competitive inhibitor of ER66 for DNA binding in the genomic regulation of breast cancer cell proliferation (3, 4). In our study, coexpression of ER66 and ER46 in the plasma membrane confers less eNOS activity than that of ER46 alone, suggesting that membrane ER66 and ER46, if coexisting, are not independent. As with the ligand-activated transcription factor, the requirement of dimeric rather than monomeric ERs in the active membrane complex is possible. It is conceivable that an ER46:ER66 dimer may recruit and incorporate required intermediary signaling components, quantitatively and qualitatively different from that of an ER46 dimer. Alternatively, if both receptors separately transmit hormone signals, ER46 and ER66 may have to share a limiting number of key signaling modules. Furthermore, if the stability and fluidity of ER66 and ER46 per se in the membrane are different, this could control the sensitivity and the transducing efficiency of the receptor as well. Collectively, these receptor biological differences could determine the ultimate effectiveness of eNOS activation.
Our present investigations demonstrate that ER46 modulates eNOS activity from the cell surface. All human ECs that we have studied to date, from multiple vascular beds (macro- and microvascular; data not shown), coexpress ER46 and ER66. Our data support that the balance of these two receptors dictates the efficiency of estrogen-stimulated eNOS activation and NO release. Defining approaches to manipulate this balance may provide new avenues for cardiovascular protection.
Acknowledgments
We thank Drs. Richard Hochberg (Yale University) for providing E2-6-CMO-ω-hexylamide-agarose, P. Chambon for hER0Pu (ER66/pSG5) and hER19Pu (179-595AA ERα/PSG5) constructs, C. Klinge (University of Louisville, Louisville, KY) for PGL3-pro-3(EREc38)-luciferase constructs, W. Sessa (Yale University) for eNOS/pSG5 constructs, and Mrs. D. Brenckle for manuscript preparation. This work was supported by National Institutes of Health Grant R01HL61782 (to J.R.B.).
Abbreviations
- ER
estrogen receptor
- E2
17β-estradiol
- cav-1
caveolin-1
- eNOS
endothelial NO synthase
- ERE
estrogen response element
- EC
endothelial cell
References
- 1.Razandi M, Pedram A, Greene G L, Levin E R. Mol Endocrinol. 1999;13:307–319. doi: 10.1210/mend.13.2.0239. [DOI] [PubMed] [Google Scholar]
- 2.Pietras R J, Szego C M. Nature. 1977;265:69–72. doi: 10.1038/265069a0. [DOI] [PubMed] [Google Scholar]
- 3.Flouriot G, Brand H, Denger S, Metivier R, Kos M, Reid G, Sonntag-Buck V, Gannon F. EMBO J. 2000;19:4688–4700. doi: 10.1093/emboj/19.17.4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denger S, Reid G, Kos M, Flouriot G, Parsch D, Brand H, Korach K S, Sonntag-Buck V, Gannon F. Mol Endocrinol. 2001;15:2064–2077. doi: 10.1210/mend.15.12.0741. [DOI] [PubMed] [Google Scholar]
- 5.Russell K S, Haynes M P, Sinha D, Clerisme E, Bender J R. Proc Natl Acad Sci USA. 2000;97:5930–5935. doi: 10.1073/pnas.97.11.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haynes M P, Sinha D, Russell K S, Collinge M, Fulton D, Morales-Ruiz M, Sessa W C, Bender J R. Circ Res. 2000;87:677–682. doi: 10.1161/01.res.87.8.677. [DOI] [PubMed] [Google Scholar]
- 7.Russell K S, Haynes M P, Caulin-Glaser T, Rosneck J, Sessa W C, Bender J R. J Biol Chem. 2000;275:5026–5030. doi: 10.1074/jbc.275.7.5026. [DOI] [PubMed] [Google Scholar]
- 8.Jacobson B S, Schnitzer J E, McCaffery M, Palade G E. Eur J Cell Biol. 1992;58:296–306. [PubMed] [Google Scholar]
- 9.Song K S, Li S, Okamoto T, Quilliam L A, Sargiacomo M, Lisanti M P. J Biol Chem. 1996;271:9690–9697. doi: 10.1074/jbc.271.16.9690. [DOI] [PubMed] [Google Scholar]
- 10.Sowa G, Pypaert M, Sessa W C. Proc Natl Acad Sci USA. 2001;98:14072–14077. doi: 10.1073/pnas.241409998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottlieb R A, Granville D J. Methods. 2002;26:341–347. doi: 10.1016/S1046-2023(02)00040-3. [DOI] [PubMed] [Google Scholar]
- 12.Dammai V, Subramani S. Cell. 2001;105:187–196. doi: 10.1016/s0092-8674(01)00310-5. [DOI] [PubMed] [Google Scholar]
- 13.Collinge M, Pardi R, Bender J R. J Immunol. 1998;161:1589–1593. [PubMed] [Google Scholar]
- 14.Flouriot G, Griffin C, Kenealy M, Sonntag-Buck V, Gannon F. Mol Endocrinol. 1998;12:1939–1954. doi: 10.1210/mend.12.12.0209. [DOI] [PubMed] [Google Scholar]
- 15.Marquez D C, Pietras R J. Oncogene. 2001;20:5420–5430. doi: 10.1038/sj.onc.1204729. [DOI] [PubMed] [Google Scholar]
- 16.Shin J S, Abraham S N. Science. 2001;293:1447–1448. doi: 10.1126/science.1061079. [DOI] [PubMed] [Google Scholar]
- 17.Chambliss K L, Yuhanna I S, Mineo C, Liu P, German Z, Sherman T S, Mendelsohn M E, Anderson R G, Shaul P W. Circ Res. 2000;87:E44–E52. doi: 10.1161/01.res.87.11.e44. [DOI] [PubMed] [Google Scholar]
- 18.Gottardi C J, Dunbar L A, Caplan M J. Am J Physiol. 1995;268:F285–F295. doi: 10.1152/ajprenal.1995.268.2.F285. [DOI] [PubMed] [Google Scholar]
- 19.Hospital V, Chesneau V, Balogh A, Joulie C, Seidah N G, Cohen P, Prat A. Biochem J. 2000;349:587–597. doi: 10.1042/0264-6021:3490587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cullen B M, Halliday I M, Kay G, Nelson J, Walker B. Biochem J. 1992;283:461–465. doi: 10.1042/bj2830461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patterson S I, Skene J H. Methods Enzymol. 1995;250:284–300. doi: 10.1016/0076-6879(95)50079-0. [DOI] [PubMed] [Google Scholar]
- 22.Pendaries C, Darblade B, Rochaix P, Krust A, Chambon P, Korach K S, Bayard F, Arnal J F. Proc Natl Acad Sci USA. 2002;99:2205–2210. doi: 10.1073/pnas.042688499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barraille P, Chinestra P, Bayard F, Faye J C. Biochem Biophys Res Commun. 1999;257:84–88. doi: 10.1006/bbrc.1999.0334. [DOI] [PubMed] [Google Scholar]
- 24.Dunphy J T, Linder M E. Biochim Biophys Acta. 1998;1436:245–261. doi: 10.1016/s0005-2760(98)00130-1. [DOI] [PubMed] [Google Scholar]
- 25.Galbiati F, Volonte D, Meani D, Milligan G, Lublin D M, Lisanti M P, Parenti M. J Biol Chem. 1999;274:5843–5850. doi: 10.1074/jbc.274.9.5843. [DOI] [PubMed] [Google Scholar]
- 26.El-Husseini Ael D, Schnell E, Dakoji S, Sweeney N, Zhou Q, Prange O, Gauthier-Campbell C, Aguilera-Moreno A, Nicoll R A, Bredt D S. Cell. 2002;108:849–863. doi: 10.1016/s0092-8674(02)00683-9. [DOI] [PubMed] [Google Scholar]
- 27.Mumby S M. Curr Opin Cell Biol. 1997;9:148–154. doi: 10.1016/s0955-0674(97)80056-7. [DOI] [PubMed] [Google Scholar]
- 28.Carlson K E, Choi I, Gee A, Katzenellenbogen B S, Katzenellenbogen J A. Biochemistry. 1997;36:14897–14905. doi: 10.1021/bi971746l. [DOI] [PubMed] [Google Scholar]
- 29.Weis K E, Ekena K, Thomas J A, Lazennec G, Katzenellenbogen B S. Mol Endocrinol. 1996;10:1388–1398. doi: 10.1210/mend.10.11.8923465. [DOI] [PubMed] [Google Scholar]
- 30.Zhong L, Skafar D F. Biochemistry. 2002;41:4209–4217. doi: 10.1021/bi0121095. [DOI] [PubMed] [Google Scholar]
- 31.Schlegel A, Wang C, Pestell R G, Lisanti M P. Biochem J. 2001;359:203–210. doi: 10.1042/0264-6021:3590203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonnell D P, Norris J D. Science. 2002;296:1642–1644. doi: 10.1126/science.1071884. [DOI] [PubMed] [Google Scholar]
- 33.Hisamoto K, Ohmichi M, Kurachi H, Hayakawa J, Kanda Y, Nishio Y, Adachi K, Tasaka K, Miyoshi E, Fujiwara N, et al. J Biol Chem. 2001;276:3459–3467. doi: 10.1074/jbc.M005036200. [DOI] [PubMed] [Google Scholar]