Abstract
Gravitropism allows plant organs to direct their growth at a specific angle from the gravity vector, promoting upward growth for shoots and downward growth for roots. Little is known about the mechanisms underlying gravitropic signal transduction. We found that mutations in the ARG1 locus of Arabidopsis thaliana alter root and hypocotyl gravitropism without affecting phototropism, root growth responses to phytohormones or inhibitors of auxin transport, or starch accumulation. The positional cloning of ARG1 revealed a DnaJ-like protein containing a coiled-coil region homologous to coiled coils found in cytoskeleton-interacting proteins. These data suggest that ARG1 participates in a gravity-signaling process involving the cytoskeleton. A combination of Northern blot studies and analysis of ARG1-GUS fusion-reporter expression in transgenic plants demonstrated that ARG1 is expressed in all organs. Ubiquitous ARG1 expression in Arabidopsis and the identification of an ortholog in Caenorhabditis elegans suggest that ARG1 is involved in other essential processes.
Plants use the gravity vector as a directional guide to growth, thereby positioning roots and shoots below and above ground. The complexity of this process is evident when one recognizes that primary and lateral organs must have distinct responses to gravity to become spatially separated. Furthermore, roots and shoots grow in opposite directions within the gravity field in an environmentally regulated manner (1).
Gravitropism was first identified nearly 200 years ago by Knight (2) and was later characterized by Darwin (3). To date, few details of the underlying mechanisms have been resolved. Higher plants have been shown to perceive gravity primarily by the sedimentation of starch-containing amyloplasts, which are located in the columella cells of root tips as well as in the starch sheath cells of shoots (4–6). An unknown process converts the physical movement of amyloplasts into a physiological signal. It has been hypothesized that the second messenger Ca2+ as well as the cytoskeleton are involved in gravitropic signal transduction (7–9).
After stimulation, a concentration gradient of auxin and apoplastic Ca2+ is believed to develop across the root tip and to be transmitted to the distal and main elongation zones, where it would promote differential cellular elongation on opposite flanks, resulting in organ bending (10–12). The AUX1 and AGR1 genes appear to encode components of the auxin influx and efflux carriers involved in the signal-transmission phase of gravitropism (13–15).
To gain insight into the poorly understood transduction phase of gravitropism, we identified and characterized two mutant alleles at a new locus in Arabidopsis thaliana named ARG1 (Altered Response to Gravity). We show that arg1 mutants are specifically altered in root and hypocotyl gravitropism and lack the hormonal-response phenotypes found in mutants affected in polar auxin transport and organ bending. We also demonstrate that ARG1 encodes a DnaJ-like protein whose structure suggests an interaction with the cytoskeleton.
MATERIALS AND METHODS
Plant Stocks and Manipulation.
Wild-type A. thaliana seeds of the ecotype Wassilewskija (WS) were provided by Timothy Caspar (DuPont). Wild-type Landsberg erecta (Ler), Columbia (Col), and No-O seeds, as well as seeds from the Feldmann collection of T-DNA insertional mutants (16), were provided by the Arabidopsis Biological Resource Center (ABRC; Ohio State University, Columbus, Ohio). arg1–1 and arg1–2 were isolated from the DuPont and Feldmann collections of T-DNA insertional mutants, respectively, by using the reorientation and root-waving assays described in refs. 17 and 18.
All techniques and incubation conditions aimed at growing and manipulating A. thaliana seeds, seedlings, and plants were described in ref. 19.
Quantification of Root and Hypocotyl Gravitropism.
Seedlings were grown embedded in vertically oriented 0.8% agar-containing germination media [GM, half-strength Murashige–Skoog (MS) salts and 1.5% sucrose (20)] in square Petri dishes wrapped in aluminum foil (to confer darkness) and incubated within a Conviron (Asheville, NC) TC16 growth chamber (22°C, 75% relative humidity). Plates were unwrapped and pictures were taken and digitized. Digitized images were used to determine the angle from vertical of a surface tangential to the hypocotyl tip below the apical hook, as described (19). Hypocotyl response to gravistimulation was analyzed essentially as described above, except that plates were rotated 90° after 4 days of growth and incubated for 3 more days before being analyzed. Root gravitropism was analyzed as described (19).
Analysis of Hypocotyl Phototropism.
The kinetics of hypocotyl phototropism were determined by growing wild-type and mutant seedlings on vertically oriented 0.8% agar-containing GM plates wrapped in black paper and aluminium foil. After 4 days of growth (time 0), the plates were unwrapped, photographed, and rewrapped on all sides but one. The unwrapped side was exposed to a horizontal light source (≈1 μE⋅m−2⋅sec−1 from cool-white fluorescent tubes). At the specified times, the plates were unwrapped, photographed, and rewrapped to continue the experiment. The angles between planes tangential to the hypocotyl tips and vertical planes were measured and analyzed as described above.
Analysis of Starch Accumulation in Wild-Type and Mutant Columella and Hypocotyl Endodermis Cells.
Wild-type, arg1–1, and arg1–2 seedlings were grown for 7 days on the surface of a vertical 0.8% agar-containing GM medium in the light (19). They were then stained for 15 minutes with the iodine/potassium iodide (IKI) solution described in ref. 21 and analyzed under a Nikon Optiphot-2 microscope equipped with Nomarski optics (22). pgm mutant seedlings were used as negative-staining controls in these experiments (23).
Analysis of Root Growth in the Presence or Absence of Phytohormones or Auxin-Transport Inhibitors.
Wild-type and mutant seedlings were grown on vertically oriented 0.8% agar-containing GM plates in the Conviron for 4 days before being transferred onto vertically oriented 0.8% agar-containing GM plates supplemented with the phytohormones or auxin-transport inhibitors cited in the text at the indicated concentrations. Phytohormones and auxin-transport inhibitors were purchased from Sigma, prepared as 10 mM stock solutions in diluted NaOH/EtOH or water (15), and added to the pH-buffered medium at the concentrations defined in the text. Plates were incubated vertically under constant light (≈75 μE⋅m−2⋅sec−1; E, Einstein = 1 mol of photons) at 22°C for 5 days, with pictures taken every 12 hours. Root lengths were measured on digitized images and statistically analyzed (19).
Extraction and Analysis of Nucleic Acids from Plant Tissues.
Procedures used to extract, digest, and Southern hybridize DNA from plants were as described (24). Restriction-fragment sizes were estimated by comparing their electrophoretic migration with that of molecular weight markers (1-kb DNA molecular weight standards, GIBCO/BRL). All DNA cloning procedures also were described (25, 26).
RNA was extracted from plant tissues and analyzed as described (24). Transcript sizes also were estimated by comparing their electrophoretic migration with that of RNA molecular weight markers (0.24–9.5-kb RNA ladder, GIBCO/BRL).
Probes used in both Southern and Northern blot analyses were 32P-labeled by random priming (26).
ARG1 Mapping by Bulked Segregant Analysis.
Two mapping populations were generated by crossing arg1–2 plants (WS ecotype) with either Col or Ler plants. The corresponding F1 plants were self-fertilized. Segregating F2 progeny were obtained and also self-fertilized to generate F3 families. Seedlings (37 days old) from each of 29 homozygous wild-type and 40 homozygous arg1–2 mutant F3 families were grouped into two pools. DNA was extracted from each pool and PCR-amplified with cleaved amplified polymorphic sequence (CAPS) PCR-primer pairs identifying polymorphic loci scattered throughout the entire genome (27). PCR-amplified DNA was restriction-digested and gel-electrophoresed (25–27). Data interpretation was as described (28).
Generation of a Collection of Plants Carrying Recombined Polymorphic Chromosome 1S with Recombination Breakpoints in Proximity of ARG1.
Ecotype polymorphisms (pCITd117, cDNA5, and ETR1) were found to be linked to ARG1 by performing an restriction fragment length polymorphism (RFLP) analysis of genomic DNAs extracted from the segregating F3 families described above (29, 30). For pCITd117 and cDNA5, a DNA fragment which, when used as a probe, identified restriction fragment length polymorphisms between WS and Col ecotypes, was sequenced from the ends (31). PCR primers flanking the polymorphism were then made from the following sequences: for pCITd117, 5′-TTA GTT GTC ACC CAT ATC TCC ATC-3′ and 5′-ATC ACC CAA GGC CAA GAA C-3′; for cDNA5: 5′-GGG CCG AGC AAC CAA TAA TAG G-3′ and 5′-TTC CCC GTT TCG ATC TCC TCT TTA-3′). The primers were used to amplify genomic DNA from the two ecotypes. The resultant PCR products were restriction enzyme-digested (XbaI for pCITd117, DdeI for cDNA5) and run on an agarose gel to confirm the presence of the polymorphism. The ETR1 fragment length polymorphism, located 1.4 kb upstream from the ETR1 translational start site, was identified by sequencing the cloned ETR1 upstream sequences from the WS and Col ecotypes (31). PCR primers were designed flanking that polymorphism (5′-GTA TCT GCC CCC ACT CTT-3′ and 5′-AGC CTA TCT CGA ACT GAA TC-3′). Template DNA for PCR analysis was isolated from cotyledon tissue by using the method described in ref. 32.
Chromosome Walking.
A contig of bacterial artificial chromosome (BAC) genomic DNA clones derived from the Texas A & M University BAC library (obtained from the ABRC, Ohio State University) and overlapping the ARG1 locus was constructed by using standard procedures (33–35). DNA fragments were subcloned and analyzed as described (25, 26, 28). The DNA clones were purified from bacteria by using the QIAprep spin miniprep kit (Qiagen, Chatsworth, CA).
Complementation of arg1–2 by a Cloned Wild-Type Genomic DNA Fragment.
BAC DNA fragments were subcloned into the Agrobacterium pBIN19 binary vector and transformed into Agrobacterium strain GV3101. arg1–2 plants were transformed with these clones by using vacuum infiltration, and the primary transformants were identified by kanamycin selection (36). T2 seeds were harvested from the primary transformants and plated on 1.5% agar-containing germination medium to test for wavy root growth (19).
Cloning and Sequencing of Genomic Fragments and cDNAs from ARG1, arg1–1, and arg1–2.
Random-primed 32P-labeled genomic fragment A (Fig. 2C) was used as a probe to screen a cDNA library in a λ vector (37) by using standard procedures (26). Both ARG1 genomic and cDNA clones were sequenced by the Blattner laboratory (University of Wisconsin-Madison) using shotgun cloning and ABI sequenator-based sequencing strategies (31). Sequence assembly and initial analysis were performed by using the lasergene program (DNAstar, Madison, WI). Database searches for homologous sequences were carried out by using the Washington University-Basic Local Alignment Search Tool (wu-blast) and the National Center for Biotechnology Information (NCBI) blast programs (38, 39). Predictions of potential transmembrane domains in the protein were made by using the tmpred, toppred2, and das programs (40–42). Coiled-coil domains were predicted by using the coils and paircoils programs (43, 44).
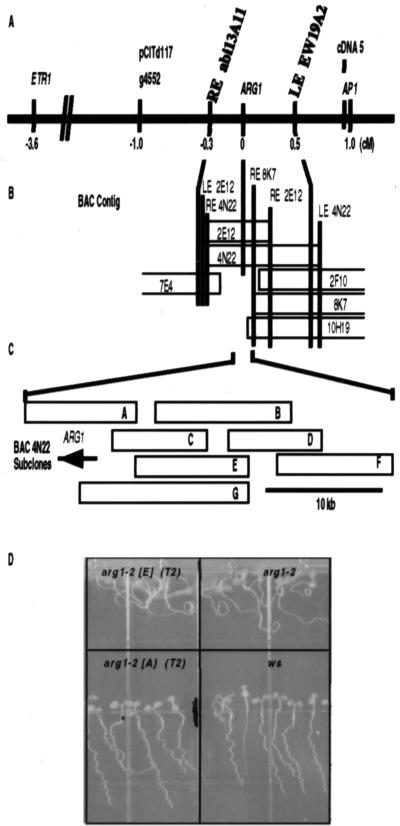
Figure 2.
Genetic and physical map of the genomic region spanning ARG1. (A) Genetic map of a region of chromosome 1 between the genes ETR1 and APETELA 1 (AP1). Genetic and molecular markers are listed above the solid line, whereas their distances (in cM) from ARG1 are listed below. The YAC contig described in ref. 47 stretches from the RFLP marker g4552 to AP1. ARG1 was mapped between the YAC-end markers RE abi13A11 and LE EW19A2 by RFLP analysis. (B) Depiction of the BAC contig (drawn to a cM scale) assembled to cover the chromosomal region between YAC-end markers RE abi13A11 and LE EW19A2. Vertical bars represent BAC- and YAC-end probes that hybridized to DNA isolated from the BAC clones (open rectangles). ARG1 was mapped on BACs 4N22 and 2E12 by RFLP analysis. (C) The relative positions of BAC 4N22 subclones (open rectangles labeled A–G). The ARG1 gene is represented by an arrow indicating the direction of its transcription. (D) Fragment A rescues the wavy-root growth phenotype of arg1–2. Wild-type WS seedlings (Lower Right), arg1–2 seedlings (Upper Right), T2 progeny of a fragment A-transformed arg1–2 plant (Lower Left), and T2 progeny of a fragment E-transformed arg1–2 plant (Upper Left) were subjected to the wavy-root growth assay described in Materials and Methods. The asterisk indicates a segregating T2 progeny that develops a mutant root-waving phenotype; others develop a wild-type root-wave phenotype that cosegregates with the transgene (not shown).
arg1–1 and arg1–2 genomic sequences were PCR-amplified by using an ARG1-specific primer pair (5′-CAT ATA AAC AAG ACC TTC TCA AGC CAA AAG TCG TA-3′ and 5′-CGC CGA AAT AAA ATA AAT CTG GAT TGA AGC AAG TC-3′) following conventional procedures (26). They were sequenced as described above.
Generation and Analysis of Transgenic Plants Carrying a GUS-ARG1 Expression-Reporter Construct.
A fragment of DNA carrying the β-Glucuronidase gene (GUS) ORF followed by a Nos terminator sequence (GUS-Nos fragment, derived from the pBI101.3 vector; CLONTECH) was fused in-frame within the StuI site of the fourth exon of ARG1, within the 9.6-kb fragment A (Fig. 2C) in the pBIN19 vector. Fragment A contains 3.6 kb of genomic DNA sequences upstream from the ARG1 translation start site as well as 3.0-kb downstream from the ARG1 translation terminator. This construct was transformed into wild-type Arabidopsis plants (ecotype WS) as described (36). GUS staining was performed by immersing seedlings, or plant parts, into 50 mM sodium phosphate buffer (pH 7.0) containing 0.5 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-Gluc, Research Organics) overnight at 37°C. Transgenic plants carrying an identical construct, with the GUS-Nos fragment in the opposite orientation, were used as negative controls.
RESULTS AND DISCUSSION
Phenotypic Characterization of arg1–1 and arg1–2 Seedlings.
arg1–1 and arg1–2 were identified while screening collections of T-DNA insertional mutants for seedlings with root and/or hypocotyl gravitropism defects (17, 19, 45). Analysis of vertically grown 4-day-old arg1–1 and arg1–2 seedlings revealed that their root tips and hypocotyls were oriented like those of wild-type with respect to the gravity vector, albeit with wider distributions of growth vector angles from the vertical (Fig. 1 A and B). After 90° rotation and further growth, arg1–1 and arg1–2 organs were found to reorient much slower to the gravity vector than wild-type organs (Fig. 1 A and B). The distribution of organ tip angles from the vertical remained larger in arg1 mutant seedlings even after they were allowed to grow for a long enough period of time to allow for complete gravitropic reorientation (7 days; data not shown). Furthermore, the poor responsiveness of arg1 hypocotyls was not caused by a global problem with organ bending (46) because arg1–1 and arg1–2 hypocotyls could reorient toward a light source with wild-type bending kinetics (Fig. 1C).
Figure 1.
Phenotypic characterization of arg1 seedlings (28). (A) Average hypocotyl tip angles from the horizontal of dark-grown 4-day-old wild-type (WS) and arg1–1 and arg1–2 mutant seedlings, 0 and 72 hours after rotating the plates by 90°. The two series of measurements were done on separate plates and involved 96 and 112 plants, respectively. SDs are shown by thin lines on the top of each bar. A legend to A–D is provided in F. (B) The kinetics of root bending on gravistimulation. Seedlings (4 days old) were reoriented 90° at time zero. Root-tip angles from the horizontal were measured over time, and average angles along with SD (vertical bars) were calculated (n = 57–78). (C) The kinetics of hypocotyl bending on exposure to a horizontal light source. Seedlings (4 days old) were exposed to a horizontal light source at time zero. Hypocotyl tip angles from the vertical were measured over time, and average angles along with SDs (vertical bars) were calculated (n = 23 and 30). (D) The effect of 2,4-D on average root growth of wild-type WS and of mutant arg1–1 and rgr1 (auxin-resistant control; ref. 17) seedlings. Seedlings (4 days old) were placed on media containing the indicated concentrations of 2,4-D (x axis) at time zero. Average root growth rates (mm per 12 hours) along with SDs (vertical bars) were calculated and plotted (n = 12–19). (E) arg1–1 and arg1–2 seedlings accumulate starch in their statocytes like wild type. Wild-type WS (Center) and arg1–1 (Left) and arg1–2 (Right) mutant seedlings (7 days old) were stained with IKI and analyzed under a light microscope equipped with Nomarski optics (×20).
Because many agravitropic mutants exhibit altered sensitivities to exogenously applied auxin and/or ethylene (plant hormones involved in the cellular growth processes accompanying organ bending), we grew arg1–1, arg1–2, and wild-type seedlings on media containing various concentrations of 2,4-dichlorophenoxyacetic acid (2,4-D, a synthetic auxin) or indoleacetic acid (IAA, a naturally occurring auxin) or placed them in chambers containing various concentrations of ethylene. arg1–1 seedlings responded like wild-type to different concentrations of 2,4-D in the medium, unlike the auxin-resistant rgr1 control seedlings, which demonstrated a higher rate of root growth than wild-type in the presence of 0.05, 0.1, or 0.5 μM 2,4-D (Fig. 1D). arg1–1 and arg1–2 also responded like wild-type to the plant hormones IAA (1 × 10−6 and 1 × 10−7 M), abscisic acid (1 × 10−5 and 1 × 10−6 M), gibberellins (GA4 and GA7; 1 × 10−5 and 1 × 10−6 M), or benzylaminopurine (1 × 10−5, 5 × 10−6, and 1 × 10−6 M) added to the medium, to ethylene added in the growth chamber (0.1, 1, 10, or 100 ppm), or to the auxin-transport inhibitors naphthylphthalamic acid (5 × 10−5, 1 × 10−5, and 10−6 M) or 2,3,5-triiodobenzoic acid (TIBA; 5 × 10−5, 1 × 10−5, and 1 × 10−6 M) added to the medium (data not shown).
To verify whether arg1 mutations affect starch accumulation in the root-cap columella or hypocotyl endodermal cells (statocytes), we stained 7-day-old wild-type and mutant seedlings with IKI, and examined them under a light microscope. Results shown in Fig. 1F indicated that both wild-type and mutant seedlings contained starch in their root-cap columella cells. Similar results were obtained when hypocotyl endodermal cells were analyzed (data not shown), strongly suggesting that ARG1 is not essential for starch accumulation in these cells.
Because arg1 mutations did not affect the hypocotyl phototropic bending abilities, the root growth response to phytohormones and auxin-transport inhibitors, or the accumulation of starch in the statocytes of Arabidopsis seedlings, we hypothesize that ARG1 is involved in the signal-transduction phase of gravitropism within the roots and hypocotyls of A. thaliana (46).
Molecular Cloning of ARG1.
Even though both arg1–1 and arg1–2 were isolated from separate collections of T-DNA insertional mutants (Materials and Methods), we found that neither mutation was T-DNA-tagged (data not shown). Therefore, we identified the ARG1 gene by using a positional cloning approach. arg1–2 plants (ecotype WS) were crossed to wild-type plants of either the Col or the Ler ecotype (mapping crosses), and the corresponding F1 plants were self-fertilized. The resultant F2 plants were also self-fertilized, and the F3 progeny from individual F2 plants were scored for gravitropism (19). By using bulk segregant analysis (28), we found that ARG1 was linked to the CAPS 1–3 and CAPS 1–4 PCR markers on the lower arm of chromosome 1 (27). RFLP analysis placed ARG1 3.6 centimorgans (cM) distal to ETR1 and 1 cM proximal to AP1 on a contig of genomic DNA clones in a yeast artificial chromosome (YAC) vector (29, 47) (Fig. 2A). By using YAC end-fragments as probes, we identified genomic DNA clones in a BAC vector, which we then assembled into a contig spanning ARG1 (Fig. 2B; refs. 33 and 34). We designed PCR-based polymorphic markers flanking ARG1, and used them to screen for recombinations between polymorphic chromosomes in the vicinity of ARG1, analyzing 2015 F2 mapping-cross plants (Materials and Methods). Further RFLP analysis, using subcloned BAC fragments as probes, placed ARG1 within a 37-kb segment of genomic DNA on BAC 4N22 (Fig. 2B). Seven overlapping fragments within this interval, named A–G (Fig. 2C), were cloned into the pBin19 Agrobacterium binary vector and transformed into arg1–2 plants (Fig. 2C; ref. 36). One of these clones, which contained a 9.6-kb genomic DNA insert (fragment A), complemented the agravitropic phenotype of arg1–2 plants: All T1 transformants exhibited a wild-type root-wave phenotype. That phenotype was transmitted as a single dominant trait in the T2 progeny (21 < n < 48) derived from self-crossing six of these T1 transformants (Fig. 2D; χ2 probability values of 0–0.4). On the other hand, all of the T2 seedlings (50 for each primary transformant) derived from selfing the primary transformants carrying one of the fragments B–G exhibited an altered root-wave phenotype (Fig. 2D; data not shown).
We sequenced fragment A and used the sequence to perform database searches, identifying perfect matches with three Arabidopsis expressed sequence tag clones (48) (http://genome www.stanford.edu/Arabidopsis/). All three mapped within a 1.1-kb DNA sequence in the middle of fragment A. We also used fragment A as a probe to screen a cDNA library (37) and identified six cDNA clones. All six were encoded by the same gene as the aforementioned expressed sequence tags. Sequence analysis of these clones and the corresponding genomic DNA identified a 1,233-bp ORF encoded by a gene 2.9 kb in size that contained 11 exons and 10 introns. We PCR-amplified this gene from the arg1 mutants and found the arg1–1 allele to contain a single C residue inserted near the end of the seventh exon, whereas the arg1–2 allele contained a single C residue deletion 53 bp downstream from the translational start site (Fig. 3A). Because both of these frameshift mutations are predicted to result in premature translational termination and no other candidate genes were identified on fragment A, we concluded that this gene was indeed ARG1.
Figure 3.
Structure of the ARG1 protein. (A) A scale-drawn representation of the putative domains (shaded rectangles) within the ARG1 protein (open rectangle). The locations of the arg1–1 and arg1–2 mutations are depicted by arrows. TM, transmembrane domain. (B–C) Amino acid sequence alignments of the ARG1 J domain (B) and coiled-coil region (C) with the corresponding domains from other proteins (49–53, 58–63). The name of the protein and the contributing organism is to the right of the sequences, whereas the beginning positions of the sequences within the protein are listed to the left. Amino acid residues within the black boxes are identical to the corresponding ARG1 residue, whereas shaded residues are conserved, as determined by the blosom 62 substitution matrix (64). Alignments were made by using the DNAstar program dna*. H. Ch., Heavy Chain.
ARG1 Encodes a DnaJ-Like Protein.
The ARG1 gene is predicted to encode a 410-aa polypeptide with a molecular mass of 45.5 kDa (Fig. 3A). Database searches revealed that the NH2 terminus shares strong sequence similarity with the highly conserved J domain of DnaJ-like molecular chaperone proteins found in prokaryotes and eukaryotes (Fig. 3 A and B; refs. 49–53). DnaJ-like proteins are involved in a variety of processes including protein folding, protein partitioning into organelles, signal transduction, and targeted protein degradation (54). The J domain has been shown to interact directly with Hsp70, thereby regulating its ATPase activity, which affects protein binding and folding. The Saccharomyces cerevisiae DnaJ-like protein YDJ1, together with Hsp70 and Hsp90, participates in a number of signal-transduction pathways involving steroid hormones as well as tyrosine kinases and serine-threonine kinases (55). Interestingly, these heteroligomeric complexes bind actin filaments in a Ca2+/calmodulin-regulated manner (56).
Computer modeling programs predicted that a stretch of 15–18 hydrophobic amino acids located adjacent to the J domain could form a transmembrane helix (Fig. 3A) and that a 70-aa region near the COOH terminus of ARG1 forms a coiled-coil structure (P > 95%; Fig. 3A). Coiled-coil domains are found in a wide variety of proteins, where they bring about specific protein oligomerization (57). Database searches revealed that this coiled-coil region shares significant sequence similarity to portions of coiled coils found in a number of cytoskeleton-interacting proteins including Rho-associated protein kinase, tropomyosin, myosin and kinesin heavy chain proteins, and inner centromere protein (INCENP) (Fig. 3C; refs. 58–63). These proteins are involved in various processes including signal transduction, actin-filament reorganization and stabilization, protein trafficking, and cytokinesis. Interestingly, the deletion of the coiled-coil domain in INCENP, a protein involved in the assembly of cytoskeletal structures during mitosis, abolished its ability to associate with microtubules while not affecting its ability to associate with the spindle (63).
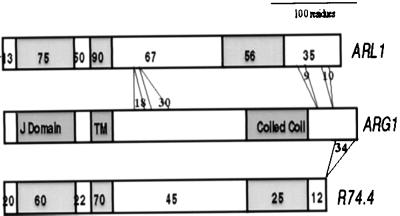
ARG1 also was found to share 50% and 32% sequence identities with hypothetical proteins from A. thaliana and C. elegans, respectively, identified by the two global genome sequencing initiatives (Fig. 4). These proteins are collinear, contain the same above-mentioned domains, and also share homologies with ARG1 across regions located outside of these domains (Fig. 4). A functional overlap between ARG1 and this Arabidopsis paralog (we named it ARL1, for ARG1-Like gene) could explain why arg1 mutants are not completely agravitropic.
Figure 4.
Representation of amino acid sequence alignments between ARG1 and its putative orthologs in A. thaliana [ARL1, GenBank accession no. AC002396 (F3I6.4)] and C. elegans [R74.4, GenBank accession no. Z36238 (R74.4)]. The ARG1 sequence is represented by an open box at the middle of the figure, whereas the A. thaliana and C. elegans orthologs are represented by open boxes at the top and bottom of the figure, respectively. The conserved J, potential transmembrane and coiled coil domains are represented by shaded boxes in all cases. The percentage of amino acids found identical within each domain between each ortholog and ARG1 is indicated by a number inserted in that domain of the ortholog. Sequence alignment gaps including more than five residues are represented by grey lines joining the two orthologs. The number of amino acids involved in each gap is represented by a number inserted between the corresponding grey lines.
ARG1 Is Expressed Ubiquitously in Plants.
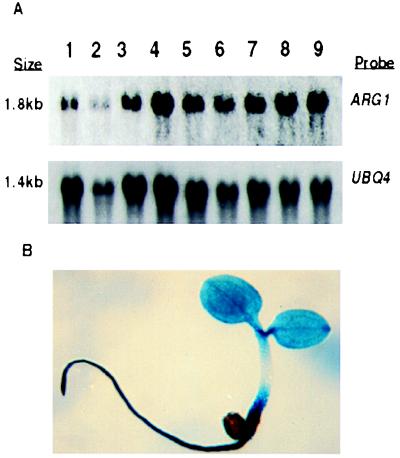
To determine the spatial expression pattern of ARG1, total RNA was extracted from several plant organs and subjected to a Northern blot analysis, using the ARG1 cDNA as a probe. Results shown in Fig. 5A (Upper) demonstrated that ARG1 mRNA is present in roots and hypocotyls plus cotyledons of 8-day-old light-grown seedlings, in hypocotyls and cotyledons of etiolated 5-day-old seedlings, and in roots, rosette leaves, cauline leaves, stems, flowers, and siliques of soil-grown adult plants. When the amounts of UBQ4 transcripts detected on the same blots were used to normalize for sample-loading differences (Fig. 5A, Lower), these tissues were found to contain similar amounts of ARG1 mRNA (Fig. 5A).
Figure 5.
The ARG1 gene is expressed ubiquitously in A. thaliana plants. (A) Northern blot analysis of total RNAs extracted from roots (lane 1) or hypocotyls and cotyledons (lane 2) of 8-day-old light-grown seedlings, from hypocotyls and cotyledons of etiolated 5-day-old seedlings (lane 3), or from roots (lane 4), rosette leaves (lane 5), cauline leaves (lane 6), inflorescence stems (lane 7), flowers (lane 8), or siliques (lane 9) of 24-day-old soil-grown plants. The Northern membrane was first hybridized with a 32P-labeled full-length ARG1 cDNA probe (Upper). After exposure, the blot was stripped and rehybridized with a UBQ4 probe (65) to control for loading differences (Lower). (B) Cytochemical detection of GUS activity in a 3-day-old transgenic A. thaliana seedling carrying the GUS gene fused in-frame within the fourth exon of ARG1.
Arabidopsis seedlings carrying a GUS reporter gene fused in-frame within the fourth exon of ARG1 were also analyzed, revealing GUS staining throughout the entire seedling including the root, hypocotyl, cotyledons, and leaves (Fig. 5B). GUS staining also was observed in pollen from these transgenic plants, as well as along their inflorescences (data not shown). Because gravitropism occurs in discreet regions of the root tip and along the hypocotyl and inflorescence stem, this extensive expression pattern suggests that ARG1 is involved in one or more additional processes.
CONCLUSIONS
Because arg1 mutants are affected specifically in gravitropism and lack any other obvious abnormalities, including those associated with processes that affect differential cellular growth, it is likely that ARG1 is involved in gravitropic signal transduction. Interestingly, the ARG1 protein contains a highly conserved J domain found in several other proteins that also act in signal-transduction pathways (54, 55). Furthermore, sequence similarities between the putative coiled-coil region of ARG1 and coiled coils found in cytoskeleton-interacting proteins suggest that ARG1 interacts with the cytoskeleton.
Indirect evidence suggests that the cytoskeleton is involved in gravity sensing. Sedimenting amyloplasts appear to be enmeshed in a dense network of short and dynamic microfilaments that may transmit the signal derived from statolith sedimentation to plasma-membrane or endoplasmic-reticulum receptors (9). Therefore, it is possible that ARG1 facilitates the transmission of gravity signals to these receptors (9). The putative transmembrane domain carried by ARG1 could allow for its interaction with the membrane at cytoskeleton attachment points. Alternatively, ARG1 could promote the transduction of gravity signals by participating in the formation of a signal-transducing complex in proximity of the cytoskeleton (12, 55, 56).
ARG1 is expressed in all tissues of the plant. Furthermore, an ARG1 ortholog is found in C. elegans. These surprising results suggest that ARG1 may play functions other than simply regulating gravitropism. The lack of pleiotropic phenotypes may reflect the fact that we may not have analyzed the right environmental response and/or developmental processes yet. Alternatively, other ARG1 functions may be masked by functional redundancy from ARG1 paralogs in Arabidopsis, such as ARL1.
Acknowledgments
We gratefully acknowledge Robert Rutherford for identifying the arg1–2 mutant and Tim Caspar, Pablo Scolnick, Patricia Russell, Carl Simmons, and Kathleen Carroll for assistance in mutant screening and initial characterization. We also thank Usha Vijayraghavan and John Bowman for the YAC clones and YAC end fragment clones, John Bowman for the cDNA5 clone, the Arabidopsis Biological Resource Center for supplying the BAC filters and BAC clones as well as the pCITd117 and g4552 clones, Frans Tax for the ETR1 clone, and Todd Richmond and John Satterlee for the cDNA library. This work was supported in part by grants from the National Institutes of Health (R01-GM 48053) and the National Aeronautic and Space Administration (NAG5-4596) and by a Packard fellowship in Science and Engineering to P.H.M.; J.S. was supported in part by a Predoctoral fellowship in Genetics (National Institutes of Health Training Grant 5-T32-6M07133) and from a National Science Foundation/Department of Energy/U.S. Department of Agriculture Training Grant fellowship (BIR 92-2033). This is paper no. 3518 of the Laboratory of Genetics.
ABBREVIATIONS
- CAPS
cleaved amplified polymorphic sequence
- RFLP
restriction fragment length polymorphism
- Col
Columbia
- Ler
Landsberg erecta
- WS
Wassilewskija, GUS, β-glucuronidase
- IAA
indoleacetic acid
- cM
centimorgan
- 2
4-D, 2,4-dichlorophenoxyacetic acid
- YAC
yeast artificial chromosome
- BAC
bacterial artificial chromosome
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF089810).
References
- 1.Masson P H. BioEssays. 1995;17:119–127. doi: 10.1002/bies.950170207. [DOI] [PubMed] [Google Scholar]
- 2.Knight T A. Phil Trans R Soc. 1806;99:108–120. [Google Scholar]
- 3.Darwin C. The Power of Movement in Plants. London: John Murray; 1880. [Google Scholar]
- 4.Sack F D. Int Rev Cytol. 1991;127:193–252. doi: 10.1016/s0074-7696(08)60695-6. [DOI] [PubMed] [Google Scholar]
- 5.Kuznetsov O A, Hasenstein K H. Planta. 1996;198:87–94. doi: 10.1007/BF00197590. [DOI] [PubMed] [Google Scholar]
- 6.Blancaflor E B, Fasano J M, Gilroy S. Plant Physiol. 1998;116:213–222. doi: 10.1104/pp.116.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J S, Mulkey T J, Evans M L. Plant Physiol. 1983;73:874–876. doi: 10.1104/pp.73.4.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sievers A, Busch M B. Planta. 1992;188:619–622. doi: 10.1007/BF00197057. [DOI] [PubMed] [Google Scholar]
- 9.Baluska F, Hasenstein K H. Planta. 1997;203:S69–S78. doi: 10.1007/pl00008117. [DOI] [PubMed] [Google Scholar]
- 10.Lee J S, Mulkey T J, Evans M L. Science. 1983;220:1375–1376. doi: 10.1126/science.220.4604.1375. [DOI] [PubMed] [Google Scholar]
- 11.Estelle M. Curr Biol. 1996;6:1589–1591. doi: 10.1016/s0960-9822(02)70780-x. [DOI] [PubMed] [Google Scholar]
- 12.Cox D N, Muday G K. Plant Cell. 1994;6:1941–1953. doi: 10.1105/tpc.6.12.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett M J, Marchant A, Green H G, May S T, Ward S P, Millner P A, Walker A R, Schulz B, Feldmann K A. Science. 1996;273:948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- 14.Luschnig C, Gaxiola R A, Grisafi P, Fink G R. Genes Dev. 1998;12:2175–2187. doi: 10.1101/gad.12.14.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen R, Hilson P, Sedbrook J, Rosen E, Caspar T, Masson P. Proc Natl Acad Sci USA. 1998;95:15112–15117. doi: 10.1073/pnas.95.25.15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldmann K A. T-DNA Insertion Mutagenesis in Arabidopsis: Seed Infection/Transformation. Teaneck, NJ: World Scientific; 1992. [Google Scholar]
- 17.Simmons C, Migliaccio F, Masson P, Caspar T, Söll D. Physiol Plant. 1995;93:790–798. [PubMed] [Google Scholar]
- 18.Rutherford R, Gallois P, Masson P H. Plant J. 1998;16:145–155. doi: 10.1046/j.1365-313x.1998.00279.x. [DOI] [PubMed] [Google Scholar]
- 19.Rutherford R, Masson P. Plant Physiol. 1996;111:987–998. doi: 10.1104/pp.111.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 21.Caspar T, Huber S C, Somerville C. Plant Physiol. 1985;79:11–19. doi: 10.1104/pp.79.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiss J Z, Wright J B, Caspar T. Physiol Plant. 1996;97:237–244. doi: 10.1034/j.1399-3054.1996.970205.x. [DOI] [PubMed] [Google Scholar]
- 23.Kiss J Z, Hertel R, Sack F D. Planta. 1989;177:198–206. [PubMed] [Google Scholar]
- 24.Sedbrook J C, Kronebusch P J, Borisy G G, Trewavas A J, Masson P H. Plant Physiol. 1996;111:243–257. doi: 10.1104/pp.111.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 26.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 27.Konieczny A, Ausubel F M. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 28.Michelmore R W, Paran I, Kesseli R V. Proc Natl Acad Sci USA. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang C, Bowman J L, DeJohn A W, Lander E S, Meyerowitz E M. Proc Natl Acad Sci USA. 1988;85:6856–6860. doi: 10.1073/pnas.85.18.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang C, Kwok S F, Bleecker A B, Meyerowitz E M. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- 31.Hensel M, Shea J E, Baumler A J, Gleeson C, Blattner F, Holden D W. J Bacteriol. 1997;179:1105–1111. doi: 10.1128/jb.179.4.1105-1111.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klimyuk V I, Carroll B J, Colwyn M T, Jones J D G. Plant J. 1993;3:493–494. doi: 10.1111/j.1365-313x.1993.tb00169.x. [DOI] [PubMed] [Google Scholar]
- 33.Choi S, Creelman R A, Mullet J E, Wing R A. Mol Biol Rep. 1995;13:124–129. [Google Scholar]
- 34.Woo S-S, Jiang J, Gill B S, Paterson A H, Wing R A. Nucleic Acids Res. 1994;22:4922–4931. doi: 10.1093/nar/22.23.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibson S I, Somerville C. Chromosome Walking in Arabidopsis thaliana Using Yeast Artificial Chromosomes. Teaneck, NJ: World Scientific; 1992. [Google Scholar]
- 36.Bechtold N, Ellis J, Pelletier G. C R Acad Sci. 1993;316:1194–1199. [Google Scholar]
- 37.Kieber J J, Rothenberg M, Roman G, Feldmann K A, Ecker J R. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- 38.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 39.Gish W, States D J. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 40.Hofmann K, Stoffel W. Biol Chem Hoppe-Seyler. 1993;347:166. doi: 10.1515/bchm3.1992.373.1.187. [DOI] [PubMed] [Google Scholar]
- 41.von Heijne G. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 42.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 43.Lupas A, Van Dyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 44.Berger B, Wilson D B, Wolf E, Tonchev T, Milla M, Kim P S. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okada K, Shimura Y. Science. 1990;250:274–276. doi: 10.1126/science.250.4978.274. [DOI] [PubMed] [Google Scholar]
- 46.Liscum E, Briggs W R. Plant Physiol. 1996;112:291–296. doi: 10.1104/pp.112.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vijayraghavan U, Siddiqi I, Meyerowitz E. Genome. 1995;38:817–823. doi: 10.1139/g95-105. [DOI] [PubMed] [Google Scholar]
- 48.Newman T, deBruijn F J, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M, et al. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bustard K, Gupta R S. J Mol Evol. 1997;45:193–205. doi: 10.1007/pl00006219. [DOI] [PubMed] [Google Scholar]
- 50.Behrens S, Narberhaus F, Bahl H. FEMS Microbiol Lett. 1993;114:53–60. doi: 10.1016/0378-1097(93)90141-n. [DOI] [PubMed] [Google Scholar]
- 51.Kaneko T, Tanaka A, Sato S, Kotani H, Sazuka T, Miyajima N, Sugiura M, Tabata S. DNA Res. 1995;2:153–166. doi: 10.1093/dnares/2.4.153. [DOI] [PubMed] [Google Scholar]
- 52.Blumberg H, Silver P A. Nature (London) 1991;349:627–630. doi: 10.1038/349627a0. [DOI] [PubMed] [Google Scholar]
- 53.Zinsmaier K E, Hofbauer A, Heimbeck G, Pflugfelder G O, Buchner S, Buchner E. J Neurogenet. 1990;7:15–29. doi: 10.3109/01677069009084150. [DOI] [PubMed] [Google Scholar]
- 54.Caplan A J, Cyr D M, Douglas M G. Mol Biol Cell. 1993;4:555–563. doi: 10.1091/mbc.4.6.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kimura Y, Yahara I, Lindquist S. Science. 1995;268:1362–1365. doi: 10.1126/science.7761857. [DOI] [PubMed] [Google Scholar]
- 56.Nishida E, Koyasu S, Sakai H, Yahara I. J Biol Chem. 1986;261:16033–16036. [PubMed] [Google Scholar]
- 57.Lupas A. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 58.Matsui T, Amano M, Yamamoto T, Chihara K, Nakafuku M, Ito M, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. EMBO J. 1996;15:2208–2216. [PMC free article] [PubMed] [Google Scholar]
- 59.Leung T, Manser E, Tan L, Lim L. J Biol Chem. 1995;270:29051–29054. doi: 10.1074/jbc.270.49.29051. [DOI] [PubMed] [Google Scholar]
- 60.Weston D S, Kemp W M. Exp Parasitol. 1993;76:358–370. doi: 10.1006/expr.1993.1044. [DOI] [PubMed] [Google Scholar]
- 61.Saez C G, Myers J C, Shows T B, Leinwand L A. Proc Natl Acad Sci USA. 1990;87:1164–1168. doi: 10.1073/pnas.87.3.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patel N, Thierry-Mieg D, Mancillas J R. Proc Natl Acad Sci USA. 1993;90:9181–9185. doi: 10.1073/pnas.90.19.9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mackay A M, Eckley D M, Chue C, Earnshaw W C. J Cell Biol. 1993;123:373–385. doi: 10.1083/jcb.123.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Henikoff S, Henikoff J G. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burke T J, Callis J, Vierstra R D. Mol Gen Genet. 1988;213:435–443. doi: 10.1007/BF00339613. [DOI] [PubMed] [Google Scholar]