Abstract
A subset (γ2) of late herpes simplex virus 1 genes depends on viral DNA synthesis for its expression. For optimal expression, a small number of these genes, exemplified by US11, also requires two viral proteins, the α protein infected cell protein (ICP) 22 and the protein kinase UL13. Earlier we showed that UL13 and ICP22 mediate the stabilization of cdc2 and the replacement of its cellular partner, cyclin B, with the viral DNA polymerase processivity factor UL42. Here we report that cdc2 and its new partner, UL42, bind a phosphorylated form of topoisomerase IIα. The posttranslational modification of topoisomerase IIα and its interaction with cdc2–UL42 proteins depend on ICP22 in infected cells. Although topoisomerase II is required for viral DNA synthesis, ICP22 is not, indicating a second function for topoisomerase IIα. The intricate manner in which the virus recruits topoisomerase IIα for post-DNA synthesis expression of viral genes suggests that topoisomerase IIα also is required for untangling concatemeric DNA progeny for optimal transcription of late genes.
Herpes simplex virus 1 (HSV-1) encodes at least 84 unique ORFs. Its genes are expressed in a coordinately regulated, sequentially ordered manner (1, 2). α genes are expressed first, and their products enable the expression of β (early) and γ (late) genes. The latter genes are classified further as γ1 or γ2 genes. Whereas γ2 gene expression requires viral DNA synthesis, the expression of γ1 is enhanced by but is not totally dependent on viral DNA synthesis. Studies have shown that primary human cell strains and some animal cell lines accumulate grossly reduced amounts of a subset of γ2 proteins exemplified by the products of US11, UL38, or UL41 after infection with mutants lacking the genes encoding infected cell protein (ICP) 22 or the UL13 protein kinase (3, 4). An apparent involvement of cellular factors in the expression of this subset of viral genes emerged from studies of cell cycle proteins. In these studies it was noted that the cyclin-dependent kinase cdc2 (cdk1) was posttranslationally modified, stabilized, and activated 4–12 h after infection (5). Concurrently, cyclin B, a partner of cdc2, was degraded. Mapping studies with viral mutants revealed that both the posttranslational modification of cdc2 and the degradation of cyclin B depended on the presence of ICP22 and UL13 protein kinase. Further studies reinforced the apparent connection between the phenotype of mutant viruses from which either α22 or UL13 mutants were deleted and the activation of cdc2 in infected cells. Thus cells transfected with a dominant negative (dn) mutant of cdc2 and infected with wild-type virus expressed representative α, β, and γ1 proteins but failed to express the γ2 US11 protein (6). These studies linked the stabilization of cdc2 with the expression of the US11 gene and indicated that ICP22 and UL13 mediate the accumulation of the subset of γ2 proteins represented by US11 by inducing the posttranslational modification of cdc2, but they left open the target of the activated cdc2.
A clue as to the possible role of cdc2 in the course of HSV-1 infection emerged from studies showing that cdc2 actively phosphorylated its substrate even though its natural partner was degraded. Studies based on the hypothesis that cdc2 had to acquire a new, viral partner to compensate for the loss of cyclin B revealed that cdc2 interacted physically and functionally with the viral DNA polymerase processivity factor encoded by the UL42 ORF (7). Taken together, these studies indicated that activated cdc2 played a role in late viral gene expression but left unanswered the question of the role of the cdc2–UL42 complex in this process.
In the search for a potential target of the cdc2–UL42 complex, we took cognizance that in uninfected cells topoisomerase IIα is modified in a cell cycle-dependent manner. Thus cdc2 interacts with topoisomerase II, and, moreover, proliferating cell nuclear antigen, the cellular homolog of UL42, mediates cyclin-dependent kinase substrate phosphorylation (8–12). Topoisomerase II is of particular interest because it is one of the key enzymes required for viral DNA synthesis that is not encoded by herpes viruses, yet members of the α, β, and γ herpes viruses (i.e., HSV-1, cytomegalovirus, and Epstein–Barr virus) all have been reported to require this enzyme for viral DNA synthesis (13–16). Here we report that the cdc2–UL42 complex is associated with a phosphorylated form of topoisomerase IIα and that in infected cells this interaction required ICP22.
Materials and Methods
Cells and Viruses.
HEp-2 cells were obtained from American Type Culture Collection and maintained in DMEM with 10% newborn calf serum. Primary human foreskin fibroblasts (pHFF) transformed with telomerase were provided by T. Shenk (Princeton University, Princeton) (17). Rabbit skin cells (RSC) were provided initially by J. McClaren (University of New Mexico, Albuquerque). HSV-1(F) is the prototype HSV-1 wild-type strain used in this laboratory (18). The HSV-1 mutant R325, lacking the carboxyl-terminal domain of ICP22, has been described (19).
Cell Infection and Immunoblotting.
The procedures for infection of cells, cell harvesting, electrophoresis of cell lysates in denaturing polyacrylamide gels, and reaction of the electrophoretically separated proteins with appropriate antibodies were described elsewhere (6). At times after infection indicated in Results, the cells were harvested and solubilized in high-salt lysis buffer (20 mM Tris, pH 8.0/1 mM EDTA/0.5% Nonidet P-40/400 mM NaCl/0.1 mM sodium orthovanadate/10 mM NaF/2 mM DTT/100 μg each of PMSF and tolylsulfonyl phenylalanyl chloromethyl ketone per ml/2 μg each of aprotonin and leupeptin per ml). Equivalent amounts of protein per sample were separated by electrophoresis in denaturing polyacrylamide gels. The electrophoretically separated proteins were transferred to a nitrocellulose sheet and reacted with antibodies to topoisomerase IIα (Calbiochem), influenza hemagglutinin (HA) tag (Santa Cruz Biotechnology), or UL42 protein (20).
Two-Dimensional Gel Electrophoresis.
Cells were harvested for two-dimensional electrophoresis 12 h after infection as described in detail elsewhere (21). Briefly, cells were lysed in solution containing 8 M urea, 4% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), and 40 mM Tris base. First-dimension isoelectric focusing was done in an IPGPhor electrophoresis unit with immobilized pH 3–10 linear gradient strips (Amersham Biosciences). The immobilized strips were electrophoretically separated in denaturing polyacrylamide gels and reacted with antibody to topoisomerase IIα.
Topoisomerase II Activity Assays.
Cells were harvested 12 h after infection and rinsed in TEMP buffer (10 mM Tris, pH 7.4/1 mM EDTA/4 mM MgCl2/0.5 mM PMSF), resuspended in 3 ml of TEMP buffer, incubated on ice for 10 min, and then homogenized in a Dounce glass homogenizer. The nuclei were pelleted by centrifugation, rinsed with TEMP buffer, resuspended in TEP buffer (10 mM Tris, pH 7.4/1 mM EDTA/0.5 mM PMSF) and an equal volume of 1 M NaCl, and stored on ice for 1 h. The samples were then spun at 16,000 × g for 15 min in a microfuge at 4°C. Topoisomerase II activity contained in the supernatant fluid fraction was measured with the aid of catenated kinetoplast DNA (kDNA, TopoGEN, Columbus, OH). Specifically, 200 ng of catenated kDNA was incubated with nuclear lysate at 30°C. The reaction was terminated by the addition of gel loading buffer, and the reaction mixture was resolved on 1% agarose gels. Decatenated products were quantified by using an Eagle Eye Video System (Stratagene).
Transient Transfection.
The construction of a UL42-expressing plasmid in the multiple coding site of pcDNA3.1(+) has been described elsewhere (7). A plasmid encoding HA-tagged dn cdc2 (cdc2-dn-HA) kinase was provided by S. van den Heuvel (Massachusetts General Hospital Cancer Center, Charlestown, MA) (22). HEp-2 cells were transfected with 2 μg of total plasmid DNA as described in Results by using LipofectAMINE Plus (GIBCO/BRL) as described elsewhere (6). Cells were harvested 36 h after transfection and assayed for protein production and topoisomerase II activity.
GST Pull-Down.
GST fusion proteins encoding full-length (aa 1–488), the amino half (N′, aa 1–244), or the carboxyl half (C′, aa 226–488) of UL42 were produced and used as described (7). Cell lysates used as a source of pull-down material were derived from asynchronous or nocodazole-arrested HEp-2 cells. The lysate of nocodazole-treated cells also was digested with calf intestinal alkaline phosphatase in the presence or absence of phosphatase inhibitors (sodium orthovanadate and NaF) for 30 min at 34°C and reacted with GST–UL42. GST pull-downs were immunoblotted with antibody to topoisomerase IIα.
Immunoprecipitation.
Immunoprecipitation with antibody to UL42 was done as described (7). The electrophoretically separated proteins in the precipitate were reacted with antibodies to topoisomerase IIα and UL42.
Results
Topoisomerase II Is Active and Modified in HSV-1-Infected Cells.
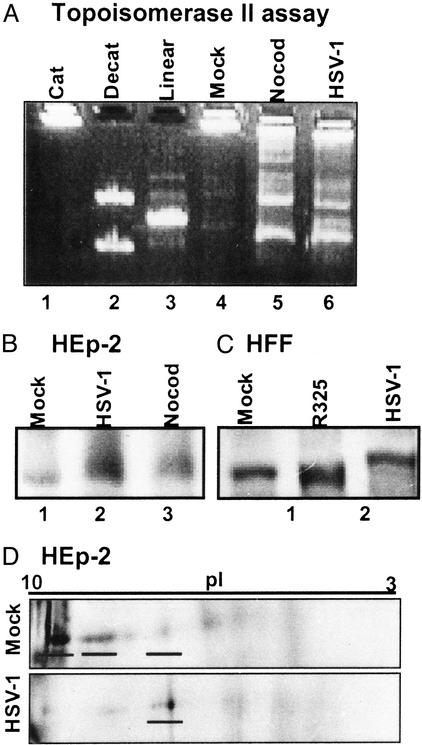
Because topoisomerase IIα is modified in a cell cycle-dependent manner and HSV-1 activates cdc2 kinase 8–12 h after infection, the first series of experiments determined the fate of topoisomerase II in HSV-1-infected cells. HSV-1(F)-infected cells were compared with either mock-infected cells (cells handled in an analogous manner to HSV-1-infected cells but without exposure to the virus) or nocodazole-treated cells (cells arrested in G2/M with active cdc2). Topoisomerase II activity was measured as the ability to decatenate concatameric kDNA. Catenated kDNA was reacted with nuclear fractions of mock-infected (with or without nocodazole treatment) or HSV-1-infected HEp-2 cell lysates and resolved by agarose gel electrophoresis (Fig. 1A ). Assay controls are shown in lanes 1–3. Catenated kDNA (Cat, lane 1) remained in the loading well, whereas the decatenated kDNA (Decat, lane 2) yielded two products of topoisomerase II activity, an upper nicked, open circular kDNA and a lower band representing relaxed kDNA. Lane 3 shows the migration of linearized kDNA. Nuclear lysates (2 μg each) were assayed for their ability to decatenate kDNA (Fig. 1A). Mock-infected lysates (Mock, lane 4) had minimal topoisomerase II activity, whereas lysates of nocodazole-treated (Nocod, lane 5) or HSV-1-infected (HSV-1, lane 6) cells contained abundant decatenated kDNA products, indicating elevated topoisomerase II activity compared with that of mock-infected cell lysates. HSV-1(F)-infected cell lysate also had nuclease activity because it also contained linear kDNA. Treatment of viral infected cells with topoisomerase II inhibitors (etoposide and ICRF-193) inhibited the accumulation of decatenated kDNA products but not of the linearized form of kDNA (S.J.A. and B.R., unpublished data).
Figure 1.
Topoisomerase II is active and modified in HSV-1-infected cells. (A) Topoisomerase II activity assay. Assay controls are shown in the first three lanes: catenated kDNA (Cat), decatenated kDNA (Decat), and linearized kDNA (Linear). Nuclear fractions (2 μg) of mock-infected, nocodazole-arrested, and HSV-1-infected HEp-2 cell lysates were assayed with catenated kDNA. (B and C) Topoisomerase IIα immunoblot of lysates from cycling HEp-2 cells (B) and contact-inhibited pHFF (C). Cells were mock-infected or infected with wild-type HSV-1 or the HSV-1 mutant R325. Also, HEp-2 cells were treated with nocodazole. (D) Two-dimensional gel electrophoresis of lysates of mock- or HSV-1-infected HEp-2 cells immunoblotted for topoisomerase IIα.
Given that HSV-1-infected cells and nocodazole-treated cells both contained abundant topoisomerase II activity, we determined whether HSV-1-infected cells modified topoisomerase IIα in a manner analogous to that of nocodazole-treated cells. Lysates of cycling HEp-2 cells or contact-inhibited pHFF harvested 12 h after infection with HSV-1 were immunoblotted with anti-topoisomerase IIα antibody. As shown in Fig. 1 B and C, the lysates of HSV-1-infected HEp-2 cells or pHFF contained a slower migrating form of topoisomerase IIα than did those of mock-infected cells. Because the mock-infected HEp-2 cells were asynchronous, HEp-2 cells also were treated with nocodazole to block cells in G2/M. The lysates of nocodazole-treated HEp-2 cells had a slow migrating form of topoisomerase IIα compared with asynchronous, mock-infected HEp-2 cells (Fig. 1B, lanes 1 and 3). The electrophoretic mobility of topoisomerase IIα from nocodazole-treated HEp-2 cells was similar to that of the enzyme present in infected cell lysates (Fig. 1B, lanes 2 and 3).
To further delineate the modifications of topoisomerase IIα in infected cells, the lysates of mock-infected and of infected HEp-2 cells were separated by electrophoresis in two-dimensional gels. In mock-infected HEp-2 cell lysate, the majority of topoisomerase IIα isoelectrically focused near a pI = 10 consistent with its predicted basic pI (Fig. 1D). In contrast, 12 h after HSV-1(F) infection, the pI of the major isoform of topoisomerase IIα was more acidic. The major isoform observed in HSV-1(F)-infected cells also was observed in lysates of mock-infected cells, albeit at drastically lower levels.
We conclude that topoisomerase II is active, and topoisomerase IIα is posttranslationally modified in both resting and dividing cells infected with HSV-1(F). Because the pHFF were contact-inhibited at the time of infection, the results indicate that the modification of topoisomerase IIα is HSV-1-specific and not dependent on the phase of cells at the time of infection.
UL42 Interacts with Topoisomerase IIα in a cdc2-Dependent Manner.
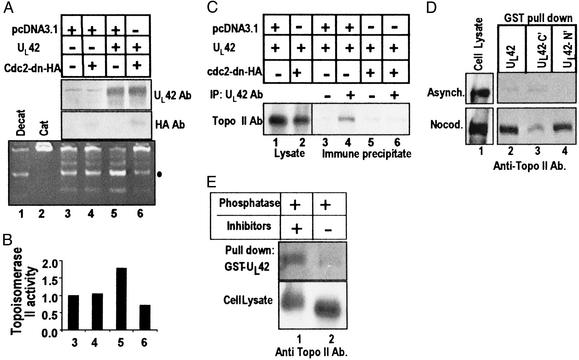
The results shown in Fig. 1 indicated that in infected or nocodazole-treated cells, topoisomerase IIα was posttranslationally modified. Previously, we showed that the viral DNA polymerase accessory factor, UL42, associated with cdc2 and induced histone H1 kinase activity (7). The question then arose whether UL42 could mediate topoisomerase II activity secondary to its association with cdc2. To address this issue, the following series of experiments was done. HEp-2 cells were transfected with plasmids encoding UL42 and/or cdc2-dn-HA. The total amount of plasmid DNA to which the cells were exposed was fixed at 2 μg with empty vector pcDNA3.1. Cells were harvested and lysed 36 h after transfection. The lysates were tested for topoisomerase II activity with catenated kDNA and for protein expression by immunoblotting of electrophoretically separated lysates. Transfection of UL42 resulted in enhanced topoisomerase II activity compared with that of HEp-2 cells transfected with the vector or cdc2-dn-HA DNA alone (Fig. 2A, lanes 3–5). Quantification of the decatenated nicked, open circular kDNA band (●) showed a 1.8-fold increase in the UL42 transfected cells compared with vector or cdc2-dn-HA transfected cells (Fig. 2B). However, cotransfection of UL42 with cdc2-dn-inhibited, UL42-mediated topoisomerase II activation (Fig. 2 A and B, lanes 5 and 6). Coexpression of cdc2-dn had no effect on the amount of UL42 present in the lysate (Fig. 2A, lanes 5 and 6, upper immunoblot).
Figure 2.
Interaction among UL42, topoisomerase IIα, and cdc2. (A) HEp-2 cells were transfected with plasmids encoding UL42, cdc2-dn-HA, and empty vector pcDNA3.1. Nuclear fractions were assayed for topoisomerase II activity after 36 h. Immunoblots of the transfected lysates were done for UL42 and HA to verify protein expression. The decatenated nicked, open circular kDNA (●) was quantified. (B) The activity of lysates of cells transfected with the vector only was assigned a value of 1. (C) Cells transfected with UL42 with and without the cdc2-dn construct were immunoprecipitated with UL42 antibody and immunoblotted for topoisomerase IIα. (D) GST pull-downs with lysates from asynchronous or nocodazole-arrested HEp-2 cells. GST fusion proteins included full-length UL42 (aa 1–488), UL42 amino-terminal half (N′, aa 1–244), and UL42 carboxyl-terminal half (C′, aa 226–488). Pull-downs were immunoblotted for topoisomerase IIα. Aliquots of the whole cell lysates are shown on the left. (E) Nocodazole-arrested HEp-2 cell lysates were treated with alkaline phosphatase in the presence or absence of phosphatase inhibitors and then reacted with GST fusion protein encoding the full-length UL42 protein. Samples were immunoblotted for topoisomerase IIα. Aliquots of the whole cell lysates are shown in the lower row.
Earlier it was reported that topoisomerase IIα and UL42 localize to viral DNA replication centers (23). We next investigated whether UL42 and topoisomerase IIα physically interacted. Immunoprecipitations were done from cells that had been transfected with plasmids encoding UL42 and/or cdc2-dn-HA. Cell lysates were immunoprecipitated with antibody to UL42 and immunoblotted for topoisomerase IIα. As shown in Fig. 2C, the anti-UL42 antibody pulled down topoisomerase IIα from lysates of cells transfected with UL42 but not from cells transfected with UL42 and cdc2-dn-HA (lanes 4 and 6). Because dn cdc2 inhibited the association between UL42 and topoisomerase IIα, we next determined whether this interaction was mediated by phosphorylation by using GST pull-down assays.
GST fusion proteins encoding full-length (aa 1–488), the amino half (N′, aa 1–244), or the carboxyl half (C′, aa 226–488) of UL42 were reacted with HEp-2 cell lysate from asynchronous or nocodazole-treated cells and immunoblotted with anti-topoisomerase IIα antibody (Fig. 2D). Although GST–UL42 proteins did not pull down topoisomerase IIα from asynchronous cells, GST fusion proteins encoding the full-length or amino half of UL42 pulled down topoisomerase IIα from nocodazole-arrested cell lysate. The carboxyl-terminal half of UL42 was less effective than the amino-terminal half in pulling down topoisomerase IIα from nocodazole-arrested lysates.
Because GST–UL42 bound topoisomerase IIα from nocodazole-treated lysates, compared with asynchronous lysates, lysates from nocodazole-arrested HEp-2 cells were treated with alkaline phosphatase in the presence or absence of phosphatase inhibitors and reacted with GST encoding the full-length UL42 (Fig. 2E). Phosphatase treatment of the cell lysate ablated the ability of topoisomerase IIα to interact with full-length GST–UL42. In the presence of phosphatase inhibitors, full-length GST–UL42 could pull down topoisomerase IIα. The results also showed that phosphatase treatment of the lysate in the absence of inhibitors resulted in a faster migrating topoisomerase IIα.
We conclude that UL42 DNA interacts with phosphorylated topoisomerase IIα, in a cdc2-dependent manner.
The Association of UL42 and Topoisomerase IIα in HSV-1-Infected Cells Depends on ICP22.
Earlier we reported that the activation of cdc2 depended on the viral regulatory protein ICP22 (5). Because topoisomerase IIα and UL42 interact in a cdc2-dependent manner, we determined whether topoisomerase IIα and UL42 association depended on ICP22 in the context of HSV-1 infection. These studies were done in pHFF and RSC. In confluent, resting primary human cells or RSC infected with a HSV-1 mutant (R325) lacking the coding sequences of the carboxyl-terminal domain of ICP22, viral DNA synthesis ensues, but the levels of accumulated US11 are grossly reduced (3). This mutant also failed to activate cdc2 (S.J.A. and B.R., unpublished observations). To test the hypothesis that ICP22 initiates the cycle of events resulting in the recruitment of topoisomerase IIα to the UL42–cdc2 complex, two experiments were done comparing R325 and wild-type HSV-1. First we determined whether the posttranslational modification of topoisomerase IIα depended on ICP22. As shown in Fig. 1C, topoisomerase IIα migrated slowly in HSV-1-infected pHFF compared with mock-infected cells by SDS gel electrophoresis (lanes 1 and 3). In pHFF infected with R325, topoisomerase IIα migrated faster than in wild-type HSV-1-infected cells (Fig. 1C, lanes 2 and 3). Although the migration of topoisomerase IIα was similar in mock- or R325-infected pHFF, there seemed to be quantitatively more topoisomerase IIα in R325-infected cell lysates compared with mock- or HSV-1-infected pHFF lysates.
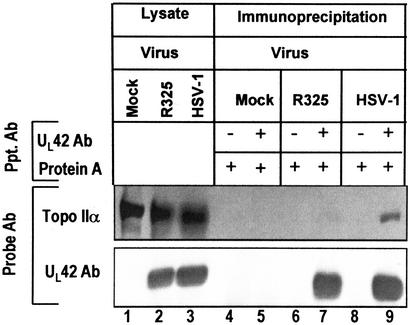
In the second experiment, we ascertained whether ICP22 mediated the interaction of topoisomerase IIα with UL42. Lysates of RSC infected with the wild-type or R325 virus were reacted with antibody to UL42. The precipitates were immunoblotted for UL42 and topoisomerase IIα (Fig. 3). Aliquots of the whole cell lysates are shown in lanes 1–3. As expected, antibody to UL42 immunoprecipitated this protein from lysates of either R325- or wild-type virus-infected cell lysates (Fig. 3, UL42 Ab, lanes 7 and 9). However, topoisomerase IIα was coprecipitated more efficiently from lysates of wild-type virus-infected lysates than from those of R325-infected lysates (Fig. 3, Topo IIα, lanes 7 and 9).
Figure 3.
UL42 interacts with topoisomerase IIα in an ICP22-dependent manner in HSV-1-infected cells. RSC were mock-infected or infected with wild-type or R325 mutant HSV-1. R325 lacks the coding region for the carboxyl-terminal region of ICP22. Cell lysates were immunoprecipitated with antibody to UL42 and immunoblotted for UL42 and topoisomerase IIα 12 h after infection. Whole cell lysates are shown in lanes 1–3.
We conclude that in HSV-1-infected cells, ICP22 mediates the posttranslational modification of topoisomerase IIα and enables the interaction of the UL42 DNA processivity factor with topoisomerase IIα.
Discussion
The strategy of HSV-1 conquest of infected cells involves the recruitment of cellular proteins that are degraded, blocked, or subverted to serve the needs of the virus. Examples include the modulation of the proteasomal pathway mediated by a viral E3 ubiquitin ligase function encoded by ICP0, inhibition of the presentation of antigenic peptides by the interaction of ICP47 with the TAP1/TAP2 transporters of peptides to the endoplasmic reticulum, and redirection of protein phosphatase Iα by the γ134.5 protein to dephosphorylate the α subunit of the eukaryotic translation initiation factor eIF-2 (24–27). Among the most striking examples of subversion of cellular proteins is their involvement in regulation of expression of viral genes. Thus, at the initiation of infection, the α gene trans-inducing factor (αTIF or VP16) recruits the cellular proteins HCF-1 and Oct-1 to enhance the expression of α genes significantly above basal levels (28, 29). Here we report the virus's sequential recruitments to use topoisomerase IIα for optimal expression of a subset of late viral genes.
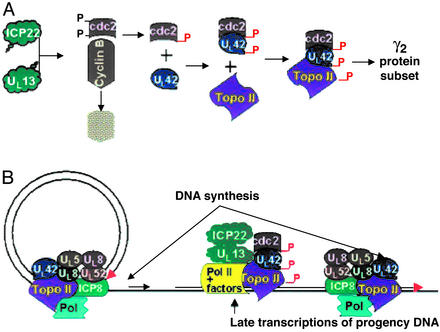
The sequence of events required for ample expression of the subset of γ2 genes exemplified by US11 is shown in Fig. 4. Earlier studies have shown that the carboxyl terminus of ICP22 and UL13 is required for the activation of cdc2. In an earlier report we have shown that cyclin B, a partner of cdc2, is degraded and that cdc2 acquires a new partner, the DNA polymerase processivity factor encoded by UL42 (5, 7). The UL42 protein physically interacts with cdc2 and is phosphorylated in an ICP22- and a cdc2-dependent manner. In this report we showed that the cdc2–UL42 complex recruits topoisomerase IIα. For the expression of a subset of γ2 genes requiring functional ICP22 and UL13, cdc2 mediates the interaction of topoisomerase IIα and UL42 (3–7). In essence, to express the subset of γ2 genes exemplified by US11, the virus must recruit topoisomerase IIα to UL42. To form the complex, the virus uses two viral proteins to stabilize and activate cdc2. It is noteworthy that both ICP22 and UL13 have been reported to modify RNA polymerase II and both are present in the replicative centers in which the transcription takes place (30). However, the modification of RNA polymerase II seems to be involved in global transcription (31).
Figure 4.
A model of the role of topoisomerase II in viral replication. (A) Schematic representation of the steps involved in the synthesis of the subset of γ2 proteins exemplified by the US11 protein. The α regulatory protein ICP22 and the UL13 protein kinase mediate the stabilization of cdc2 concurrently with the degradation of cyclin B. Cdc2 binds the viral protein UL42, and this complex recruits topoisomerase IIα. (B) The role of topoisomerase II in the viral life cycle. Shown is a schematic representation of viral DNA synthesis represented as a rolling circle and transcription of late genes off progeny DNA. Viral DNA synthesis requires topoisomerase II activity inasmuch as enzyme inhibitors effectively block this step in viral replication. The schematic diagram suggests a second role for topoisomerase II in the transcription of late genes from the progeny DNA. The topoisomerase IIα–UL42 complex associates with UL42 in a cdc2-dependent manner to enable efficient transcription of late viral genes from vast tangles of progeny viral DNA.
The studies presented here raise two questions. First, why does the virus encode numerous enzymes for viral DNA synthesis and yet fail to encode an enzyme as important as topoisomerase II (2, 32)? The second and no less intriguing question concerns the functions of the cdc2–UL42–topoisomerase IIα complex. The question arises from the observation that in RSC infected with R325, the mutant lacking the carboxyl-terminal domain of ICP22, viral DNA synthesis is not blocked (3). Thus cdc2 activates and recruits topoisomerase IIα for a function other than viral DNA synthesis, although in another context the enzyme may play an important role in the synthesis of viral DNA.
A hypothesis that could explain the role of the cdc2–UL42–topoisomerase IIα complex stems from the observation that viral DNA is made by the rolling circle model. The product consists of head-to-tail concatemers seen late in infection as huge tangles (33). Conceivably, the resulting tangles interfere with late transcription that is remedied by the presence of topoisomerase IIα. As illustrated in Fig. 4, topoisomerase IIα could have two functions: in viral DNA synthesis and in modification of progeny DNA to facilitate late transcription. The data shown in this report suggest that UL42 is involved in both DNA synthesis and in transcription complexes late in infection. Involvement in transcription complexes may explain the observation that UL42, known primarily as a DNA polymerase processivity factor, is more abundant than the viral DNA polymerase, hitherto its only known partner (2).
Acknowledgments
We thank R. Hagglund and L. Durand for invaluable discussion, S. van den Heuvel for plasmid encoding cdc2-dn, D. Tenney for antibody to UL42, and T. Shenk for the telomerase-transformed foreskin fibroblasts. These studies were aided by National Cancer Institute Grants CA87661, CA83939, CA71933, CA78766, and CA88860 and by the U.S. Public Health Service.
Abbreviations
- HSV-1
herpes simplex virus 1
- ICP
infected cell protein
- dn
dominant negative
- pHFF
primary human foreskin fibroblasts
- RSC
rabbit skin cells
- HA
hemagglutinin
- kDNA
kinetoplast DNA
- cdc2-dn-HA
HA-tagged dn cdc2
References
- 1.Honess R W, Roizman B. Proc Natl Acad Sci USA. 1975;72:1276–1280. doi: 10.1073/pnas.72.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roizman B, Sears A E. In: Fields Virology. Fields B N, Knipe D M, Howley P, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. New York: Lippincott-Raven; 1996. pp. 2231–2296. [Google Scholar]
- 3.Sears A E, Halliburton I W, Meignier B, Silver S, Roizman B. J Virol. 1985;55:338–346. doi: 10.1128/jvi.55.2.338-346.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purves F C, Ogle W O, Roizman B. Proc Natl Acad Sci USA. 1993;90:6701–6705. doi: 10.1073/pnas.90.14.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Advani S J, Brandimarti R, Weichselbaum R R, Roizman B. J Virol. 2000;74:8–15. [PMC free article] [PubMed] [Google Scholar]
- 6.Advani S J, Weichselbaum R R, Roizman B. Proc Natl Acad Sci USA. 2000;97:10996–11001. doi: 10.1073/pnas.200375297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Advani S J, Weichselbaum R R, Roizman B. J Virol. 2001;75:10326–10333. doi: 10.1128/JVI.75.21.10326-10333.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heck M M, Hittelman W N, Earnshaw W C. J Biol Chem. 1989;264:15161–15164. [PubMed] [Google Scholar]
- 9.Escargueil A E, Plisov S Y, Skladanowski A, Borgne A, Meijer L, Gorbsky G J, Larsen A K. FASEB J. 2001;15:2288–2290. doi: 10.1096/fj.00-0726fje. [DOI] [PubMed] [Google Scholar]
- 10.Taagepera S, Rao P N, Drake F H, Gorbsky G J. Proc Natl Acad Sci USA. 1993;90:8407–8411. doi: 10.1073/pnas.90.18.8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuccola H J, Filman D J, Coen D M, Hogle J M. Mol Cell. 2000;5:267–278. doi: 10.1016/s1097-2765(00)80422-0. [DOI] [PubMed] [Google Scholar]
- 12.Koundrioukoff S, Jonsson Z O, Hagan S, de Jong R N, van der Vliet P C, Hottiger M O, Hubscher U. J Biol Chem. 2000;275:22882–22887. doi: 10.1074/jbc.M001850200. [DOI] [PubMed] [Google Scholar]
- 13.Hammarsten O, Yao X, Elias P. J Virol. 1996;70:4523–4529. doi: 10.1128/jvi.70.7.4523-4529.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis A G, Cleator G M, Klapper P E, Templeton P P, Longson M. Res Virol. 1989;140:443–451. doi: 10.1016/s0923-2516(89)80122-0. [DOI] [PubMed] [Google Scholar]
- 15.Benson J D, Huang E S. J Virol. 1988;62:4797–4800. doi: 10.1128/jvi.62.12.4797-4800.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawanishi M. J Gen Virol. 1993;74:2263–2268. doi: 10.1099/0022-1317-74-10-2263. [DOI] [PubMed] [Google Scholar]
- 17.Bresnahan W A, Hultman G E, Shenk T. J Virol. 2000;74:10816–10818. doi: 10.1128/jvi.74.22.10816-10818.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ejercito P, Kieff E D, Roizman B. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 19.Post L E, Roizman B. Cell. 1981;25:227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- 20.Sheaffer A K, Hurlburt W W, Stevens J T, Bifano M, Hamatake R K, Colonno R J, Tenney D J. Virus Res. 1995;38:305–314. doi: 10.1016/0168-1702(95)00047-t. [DOI] [PubMed] [Google Scholar]
- 21.Advani S J, Hagglund R, Weichselbaum R R, Roizman B. J Virol. 2001;75:7904–7912. doi: 10.1128/JVI.75.17.7904-7912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van den Heuvel S, Harlow E. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 23.Ebert S N, Subramanian D, Shtrom S S, Chung I K, Parris D S, Muller M T. J Virol. 1994;68:1010–1020. doi: 10.1128/jvi.68.2.1010-1020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Sant C, Hagglund R, Lopez P, Roizman B. Proc Natl Acad Sci USA. 2001;98:8815–8820. doi: 10.1073/pnas.161283098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boutell C, Sadis S, Everett R D. J Virol. 2002;76:841–850. doi: 10.1128/JVI.76.2.841-850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. Nature. 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 27.He B, Gross M, Roizman B. Proc Natl Acad Sci USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stern S, Tanaka M, Herr W. Nature. 1989;241:624–630. doi: 10.1038/341624a0. [DOI] [PubMed] [Google Scholar]
- 29.Kristie T M, Sharp P A. Genes Dev. 1990;4:2383–2396. doi: 10.1101/gad.4.12b.2383. [DOI] [PubMed] [Google Scholar]
- 30.Long M C, Leong V, Schaffer P A, Spencer C A, Rice S A. J Virol. 1999;73:5593–5604. doi: 10.1128/jvi.73.7.5593-5604.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenkins H L, Spencer C A. J Virol. 2001;75:9872–9884. doi: 10.1128/JVI.75.20.9872-9884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehman I R, Boehmer P E. J Biol Chem. 1999;274:28059–28062. doi: 10.1074/jbc.274.40.28059. [DOI] [PubMed] [Google Scholar]
- 33.Jacob R J, Roizman B. J Virol. 1977;23:394–411. doi: 10.1128/jvi.23.2.394-411.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]