Abstract
A class of secreted poxvirus tumor necrosis factor (TNF)-binding proteins has been isolated from Tanapox-infected cell supernatants. The inhibitor bound to a TNF-affinity column and was identified as the product of the 2L gene. Sequence analysis of 2L family members from other yatapoxviruses and swinepox virus yielded no sequence homology to any known cellular gene. The expressed Tanapox virus 2L protein bound to human TNF with high affinity (Kd = 43 pM) and exhibits an unusually slow off-rate. However, 2L is unable to bind to a wide range of human TNF family members. The 2L protein can inhibit human TNF from binding to TNF receptors I and II as well as block TNF-induced cytolysis. Thus, Tanapox virus 2L represents an inhibitor of human TNF and offers a unique strategy with which to modulate TNF activity.
The tumor necrosis factor (TNF) superfamily of ligands plays critical roles in both adaptive and innate immunity. These ligands form a trimeric structure and bind to members of the TNF receptor (TNFR) superfamily that mediate the signaling events that regulate diverse responses, particularly inflammation and apoptosis (1). During a viral infection, TNF, the prototypic ligand for the family, is induced as an early response of the immune system to an invading pathogen (2). TNF is expressed by activated macrophages as a membrane-associated ligand, which subsequently is cleaved and secreted as a trimer. The proinflammatory effects of TNF are mediated through the cell surface TNFRI and, to a lesser extent, TNFRII (3). Binding of the TNF trimer causes conformational reorganization of the receptors and subsequent recruitment of signaling proteins such as TNFR-associated factor 2 (TRAF2) and Fas-associated death domain (FADD) (3). The TNF/TNFR signal transduction pathway is such an important aspect of the antiviral response that a number of viruses have evolved targeted strategies to specifically disrupt it.
Poxviruses employ a wide range of strategies to modulate, avoid, or diminish the various antiviral immune responses, including the secretion of cytokine- and chemokine-binding proteins and the production of cytokine and growth factor homologs (4–7). The best characterized of these strategies are the secreted homologs of the TNFRs that most likely were derived by the acquisition of an ancestral host TNFR cDNA and subsequent deletion of the transmembrane domain and cytoplasmic tail (8). The first characterized viral homolog of TNFR was the secreted T2 protein from Shope fibroma virus, which binds both TNF and lymphotoxin-α (LT-α) just as its cellular counterpart (9, 10). Deletion of the related T2 gene from myxoma virus greatly attenuated the virus in vivo (11).
Other poxvirus-secreted TNFRs were described for monkeypox virus, variola virus, vaccinia virus, ectromelia virus, and cowpox virus (4, 8, 12). Cowpox virus is atypical among poxviruses because it encodes multiple, unique, soluble TNFRs designated as cytokine response modifier (Crm)B (13), CrmC (14), CrmD (15), and CrmE (16). Each of these cowpox virus TNFRs has some sequence similarity with cellular TNFRs but differs with respect to its ligand-binding specificities.
The Yatapoxvirus genus of poxviruses are composed of Yaba-like disease virus (YLDV), Tanapox virus (TPV), and Yaba monkey tumor virus (YMTV). The Yatapoxviruses have a restricted host range, infecting only primates including humans. They produce a relatively mild, self-limiting infection in humans and monkeys (17, 18). Sequencing of the genomes of two members of the Yatapox genus, YLDV and YMTV, did not reveal any obvious TNFR homologs (ref. 19; C.R.B., H. Amano, Y. Ueda, T. Miyamura, T. Suzuki, X. Li, J.W.B., and G.M., unpublished results). Despite this, one member of the Yatapox genus, TPV, has been shown to express a TNF-binding activity that is detected in the supernatants of TPV infected cells (20). TPV is >98% identical to YLDV at the nucleotide level and is considered to be a strain of YLDV (19). We reasoned that if YLDV/TPV encode a secreted TNF inhibitor, it must be a member of a unique protein family because the genomes do not include any genes with similarity to the known TNFRs. Here, we describe the identification and characterization of this high-affinity TNF inhibitor secreted from TPV-infected cells, which forms the prototypic member of a previously uncharacterized class of pathogen-derived inhibitors for human TNF.
Materials and Methods
Reagents.
Recombinant human TNF, murine TNF, human IL-2, human IL-5, human lymphotoxin-α, and human IFN-γ were obtained from BioSource International (Camarillo, CA). Human TNFR1-Fc and human TNFR2-Fc were obtained from Apotech (Lausanne, Switzerland).
Viruses.
TPV was obtained from Joe Esposito (Centers for Disease Control and Prevention, Atlanta), and YMTV (VR587) was obtained from the American Type Culture Collection. TPV was propagated on OMK cells at 37°C, and YMTV was grown on CV1 cells at 34°C.
Preparation of Human TNF Column.
A human TNF affinity column was prepared by using Aminolink Plus coupling gel (Pierce) following manufacturer protocol.
Purification and Sequencing of the TPV TNF-Binding Protein.
OMK cells were infected with TPV at a multiplicity of infection of 50, and the cells were incubated at 37°C for 6 h. Cells were washed three times with serum-free medium and then incubated further for 18 h at 37°C in serum-free medium. The supernatants were collected and clarified by spinning for 30 min at 500 × g (IEC PR-6000; Damon Biotechnology, Needham, MA) followed by a 60-min centrifugation at 85,000 × g. The clarified supernatant was dialyzed overnight against deionized water at 4°C. The TPV supernatants then were applied to the human TNF affinity column at a rate of 1 ml/min. The column was washed with PBS, pH 7.4, and the TPV-2L protein was eluted with 0.2 M acetic acid. The acetic acid was neutralized with the addition of 1 M Tris base, and the material was dialyzed against PBS. SDS-gel loading buffer (50 mM Tris⋅Cl, pH 6.8/100 mM DTT/2% SDS/0.1% bromophenol blue/10% glycerol) (21) was added to the eluant from the human TNF column and the sample was boiled for 3 min. The proteins then were separated on a 10–20% Tris-glycine precast gel (NOVEX, San Diego). The gel was equilibrated with two 15-min washes with 25 mM Tris/40 mM norleucine, pH 9. Poly(vinylidene difluoride) (PVDF, 0.45 μM; Millipore) was washed in 100% methanol for 10 sec before rinsing in deionized water. Proteins were transferred to the PVDF membrane by electroblotting for 55 min at 1.5 mA constant current per centimeter squared of PVDF membrane. After transfer was complete, the blot was stained for 15 min with Coomassie blue followed by destaining in 10% acetic acid and 50% methanol. The membrane was air-dried, a 38- to 45-kDa protein band was excised, and N-terminal sequence analysis was performed by using automated Edman degradation.
Construction of Recombinant Baculoviruses Expressing TPV 2L.
TPV gene 2L was amplified by PCR by using TAQ polymerase and TPV DNA and primers that amplified the entire ORF. The 2L gene subsequently was cloned into the TA cloning vector pCR2.1 (Invitrogen) to produce pCR2.1–2L. In addition, a Myc-His-tagged version of TPV 2L was cloned by PCR by using TAQ polymerase, TPV DNA, the forward primer 5′-CCCAAGCTTCATGGATAAGTTACTATTATTTAGCAC-3′ (with the HindIII site underlined), and the reverse primer 5′-CCGCTCGAGGGTTTCCGTCTTCTTCATCCTCTTC-3′ (with the XhoI site underlined), which change the endogenous stop codon for 2L into a Gly residue. The resulting PCR product was cut with the restriction enzymes HindIII and XhoI and cloned into pcDNA3.1/myc/His ver C (Invitrogen) to produce pcDNA-2Lmyc/His. The genes 2L from pCR2.1–2L and 2Lmyc/His from pcDNA-2Lmyc/His were cloned into pFastBac-1 (Invitrogen), and recombinant baculoviruses referred to as AcTPV-2L and AcTPV-2Lmyc/His were produced by using the Bac-to-Bac system following manufacturer protocol (Invitrogen).
Generation of TPV 2L Antibodies.
TPV-2Lmyc/His was produced by infecting SF-21 cells with AcTPV-2Lmyc/His in SF-900 II serum-free medium (Invitrogen) and incubating at 27°C for 72 h. The medium was collected and concentrated 10-fold on a Nanosep 3K omega microconcentrator (Pall). The concentrated medium was separated on a 12% polyacrylamide gel, and the proteins were visualized with Coomassie blue. The band representing TPV-2Lmyc/His was excised washed with deionized water and then placed in PBS. New Zealand White rabbits were immunized four times with an emulsion of the polyacrylamide gel fragment containing the TPV-2Lmyc/His protein and Freund's adjuvant. Antiserum was collected from the rabbits and shown to recognize 2L protein by Western blotting.
Purification of TPV 2L.
TPV-2L was produced by infecting SF-21 cells with AcTPV-2L in SF-900 II serum-free medium (Invitrogen) and incubating at 27°C for 72 h. The medium was collected and dialyzed against PBS, pH 7.4, and concentrated by using a Nanosep 3K omega microconcentrator (Pall). The supernatant was passed over a human TNF affinity column at a rate of 1 ml/min. The column was washed with PBS, pH 7.4, and the TPV-2L protein was eluted with 0.2 M acetic acid. The acetic acid was neutralized with the addition of 1 M Tris base, and the material was dialyzed against PBS.
Western Blot of 2L Family Members.
Supernatants from YMTV, TPV, or AcTPV-2L infected cells grown in serum-free medium were concentrated 10-fold by using a Nanosep 3K omega microconcentrator (Pall). The concentrated proteins were mixed with an equal volume of SDS-gel loading buffer (50 mM Tris⋅Cl, pH 6.8/100 mM DTT/2% SDS/0.1% bromophenol blue/10% glycerol) (21) and boiled for 5 min followed by separation of the proteins on a 12% polyacrylamide gel. The proteins were transferred from the gel to Hybond C+ nitrocellulose membrane (Amersham Pharmacia) by using a semidry transfer apparatus and transfer buffer (20% methanol/25 mM Tris/250 mM glycine/0.1% SDS). The membrane was blocked in TBST (140 mM NaCl/3 mM KCl/24 mM Tris base, pH 7.4/0.2% Tween-20) + 5% powdered skim milk at 4°C overnight. A 1:10,000 dilution of polyclonal rabbit anti-TPV-2L antiserum in TBST + 5% powdered skim milk was applied to the membrane and incubated at room temperature for 1 h. The membrane was washed three times for 5 min each with TBST followed by a 1-h incubation at room temperature with goat anti-rabbit HRP (Jackson ImmunoResearch) diluted in TBST + 5% powdered skim milk. The membrane was washed three times for 5 min each, and the signal was detected by applying Chemiluminescence Reagent Plus (Perkin–Elmer) and exposing the membrane to x-ray film (Eastman Kodak).
Biomolecular Interaction Analysis Using Surface Plasmon Resonance (SPR).
Approximately 500 response units (300 pg/mm2) of purified baculovirus-produced TPV-2L was immobilized by using standard amine-coupling chemistry (22) onto a CM-5 BIAcore chip with a BIAcoreX biosensor (BIAcore, Uppsala). During the association phase, TNF, IL-2, IL-5, IFN-γ, or LT-α was diluted in running buffer HBS-EP [10 mM Hepes, pH 7.4/150 mM NaCl/3 mM EDTA/0.005% polysorbate 20 (vol/vol)] and was applied individually to the sensor chip in a 100-μl volume at a flow rate of 50 μl/min. During the dissociation phase, HBS-EP buffer was applied to the TPV-2L sensor chip at a flow rate of 50 μl/min. After completion of an experiment, the TPV-2L sensor chip was regenerated with 10 mM acetate, pH 4.0. To correct for natural drift that occurs in a BIAcoreX biosensor, a 100-μl injection of HBS-EP was made and the sensorgram produced from this injection was subtracted from the sensorgram for the test proteins. The data were analyzed globally with the BIAEVALUATION 3.0 software (BIAcore) by using a 1:1 mass transport model.
Production of Rabbit TNF.
Three 150-mm2 dishes of BGMK cells were infected with either vaccinia virus Western Reserve strain or a recombinant vaccinia virus expressing rabbit TNF (23) in serum-free medium and incubated at 37°C for 3 days. The supernatants from the cells were isolated and concentrated 10-fold on a Nanosep 3K omega microconcentrator (Pall).
Inhibition of TNF Binding to TNFR1 and TNFR2 by TPV-2L.
ELISA plates were coated with mouse anti-human IgG antibody (5 μg/ml in 50 mM sodium carbonate, pH 9.6) for 16 h at 37°C followed by blocking with PBS containing 4% powdered skim milk and 0.5% Tween 20 for 1 h at 37°C. The plates were incubated with hTNFR1-Fc or hTNFR2-Fc at 0.5 μg/ml for 1 h at 37°C in 100 μl of incubation buffer followed by the addition of Flag-hTNF or Flag-muTNF at 300 ng/ml (5 nM trimer) premixed with varying amounts of TVP-2L myc/His (5 μg/ml, i.e., 125 nM and 2-fold dilutions). After a 1-h incubation at 37°C, biotinylated anti-Flag M2 antibody (0.5 μg/ml; Sigma) was added and incubated at 37°C for 1 h. The presence of TNF was detected with the incubation of horseradish peroxidase-coupled streptavidin (1:4,000) (Jackson ImmunoResearch) for 1 h at 37°C, and the plates were washed, incubated with o-phenylenediamine (Sigma), and read at 490 nm.
Cytolytic Assays.
Mouse L929 cells in six-well plates were incubated in 1 ml of medium containing 5 μg/ml actinomycin D and either 3.5 nM purified TPV-2L or 3.5 nM purified myxoma T7 protein (a gift of Viron Therapeutics, London, Ontario), or no protein was added. The cells then were treated with increasing concentrations of purified human TNF, mouse TNF, or supernatants from wild-type or recombinant vaccinia virus expressing rabbit TNF-infected cells. The cells were incubated for 16 h at 37°C followed by staining with 0.5% crystal violet in methanol/water (1:4) for 5 min. The plates were rinsed in deionized water and allowed to air-dry before images were taken on an inverted microscope.
Results
Identification and Characterization of the TNF-Binding Activity of TPV.
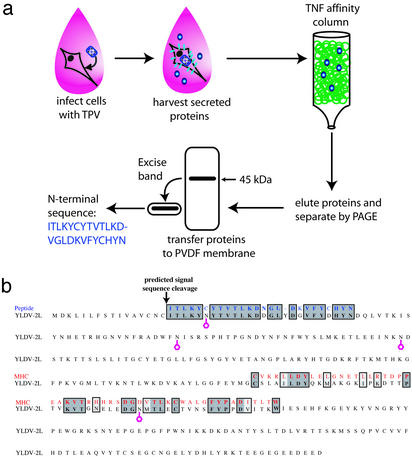
A previous report had suggested that TNF-binding protein was detectable in the supernatants of TPV-infected cells (20). To further characterize this activity, supernatants from TPV-infected cells were applied to a human TNF affinity column and proteins bound to the column were eluted and subjected to N-terminal sequencing (Fig. 1a). A comparison of the deduced N-terminal peptide sequence from the major eluted 45-kDa protein with amino acid sequences in the Swissprot database identified the putative TNF-binding protein as closely related to the product of the YLDV gene 2L (Fig. 1b). An alignment of the N-terminal peptide sequence with the YLDV gene 2L revealed that a closely related version of the N-terminal peptide sequence began immediately after the predicted signal sequence cleavage site (Fig. 1b), suggesting that the mature secreted protein has a cleavable signal sequence at the predicted location.
Figure 1.
TNF-binding activity is encoded by poxvirus orthologs of the YLDV 2L gene. (a) A schematic of the purification and sequencing strategy used to isolate the TNF-binding activity from TPV-infected cell supernatant. (b) Alignment of the YLDV 2L gene with the N-terminal sequence obtained from the TPV TNF-binding protein (blue) and amino acids 185–238 of murine MHC class I histocompatibility antigen H-2 Q5-k α-chain (red). The predicted signal sequence cleavage site is shown as well as four potential N-linked glycosylation sites.
The YLDV gene 2L is 338 aa long and predicted to encode a protein of 38.8 kDa with four potential N-linked glycosylation sites (ref. 19; Fig. 1b). YLDV 2L has no significant sequence homology to any cellular gene, including any known TNFRs, TNFR-related proteins such as TAC-I and BAFF-R, or any other TNF-binding proteins, indicating that this protein is a member of a previously uncharacterized protein family. A low level of sequence similarity to regions of the MHC class I heavy chain α2 and α3 domains was identified in a 54-aa stretch of the 2L gene (ref. 19; Fig. 1b), but the significance of this domain similarity is unclear.
To determine whether TPV encodes a close relative of the 2L protein, we first used PCR primers based on the flanking sequence of 2L from YLDV. YLDV and TPV are >98% identical at the nucleotide level (19); therefore, we tested whether PCR primers based on the YLDV sequence would amplify the corresponding TPV sequence. We were able to successfully PCR-amplify and sequence the TPV-2L gene ortholog and compare it with the other poxvirus 2L-like genes (Fig. 2; refs. 19, 24, and 25 and C.R.B., H.A., Y.U., T.M., T.S., X.L., J.W.B., and G.M., unpublished results). The four known 2L-family orthologs from YLDV, YMTV, TPV, and swinepox (SPV) share ≈31% amino acid identity, including six conserved cysteine residues that likely form three pairs of disulfide bonds (Fig. 2). However, there is considerable divergence between the SPV003/148 gene and the three yatapox 2L genes, making the identification of predicted domains that might be critical for TNF binding difficult by comparative sequence analysis alone.
Figure 2.
Alignment of the identified 2L orthologs from other poxviruses. An alignment of the vTNF-BP family members from TPV (TPV-2L), YLDV-2L, YMTV (YMTV-2L), and the two copies from SPV (SPV003 and SPV148). The blackened boxes denote conserved cysteine residues.
TPV and YMTV Each Express and Secrete 2L Protein.
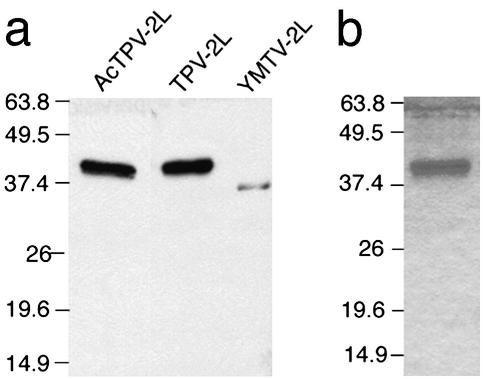
To confirm that TPV and YMTV express the predicted 2L protein, we produced an antiserum against a myc/His-tagged version of TPV 2L expressed from a baculovirus vector and used this antiserum in Western blots to test for the presence of the 2L protein in the supernatants of TPV- and YMTV-infected cells. In TPV-infected cell supernatants, a 45-kDa protein was detected that had a size similar to the TPV-2L protein produced by the recombinant baculovirus, confirming that the 2L protein indeed was expressed and secreted (Fig. 3a). In addition, a 37-kDa protein was detected in the supernatants of YMTV-infected cells, indicating that YMTV expresses a lower molecular mass variant of 2L and that the antiserum raised against TPV 2L crossreacts with YMTV 2L (Fig. 3a). The differences in molecular mass between the TPV and YMTV 2L genes likely results from differential glycosylation or processing of the two proteins. In this regard, YMTV 2L ORF includes three potential N-linked oligosaccharide sites whereas TPV 2L has four potential N-linked oligosaccharide sites.
Figure 3.
2L is produced and secreted during TPV and YMTV infections. (a) Supernatants from AcTPV-2L-infected SF-21 cells, TPV-infected OMK cells, or YMTV-infected CV1 cells were concentrated 10-fold, and the proteins were separated on a polyacrylamide gel. Proteins then were transferred to a nitrocellulose membrane that subsequently was probed with rabbit polyclonal antibodies raised against the TPV-2Lmyc/His protein. (b) Supernatants from AcTPV-2L-infected SF-21 cells were passed over a human TNF column. Proteins specifically binding to the column were eluted in 0.2 M acetic acid and subsequently separated on a polyacrylamide gel and visualized by Coomassie blue staining.
TPV-2L Exhibits High-Affinity Binding for Human TNF.
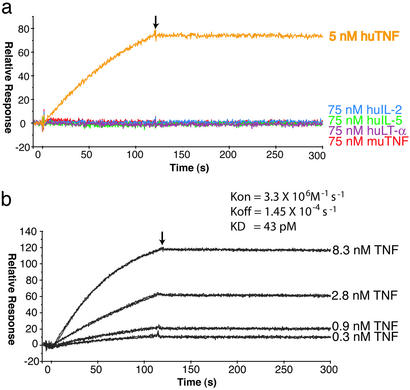
Previous studies had indicated that a 38-kDa TPV protein (termed gp38) had the ability to bind TNF, IL-2, IL-5, and IFN-γ (20, 26). Because TPV-2L binds to a TNF column, as does TPV gp38 (20), it is reasonable to presume that the gp38 protein previously described might be TPV-2L. We therefore tested TPV-2L to determine whether it possessed the ability to complex with human TNF, IL-2, IL-5, and IFN-γ by using SPR to measure the rates of association and dissociation between the ligands and TPV-2L. Purified baculovirus produced TPV-2L (Fig. 3b) was immobilized by amine-coupling on a BIAcore CM5 sensor chip, and human TNF, IL-2, IL-5, and IFN-γ were tested for their ability to bind to the 2L-chip. Five nanomoles of human TNF exhibited a fast association rate followed by a very slow dissociation rate with TPV-2L (Fig. 4a). In contrast, 75 nM human IL-2, human IL-5 (Fig. 4a), or human IFN-γ (data not shown) showed no detectable binding by SPR to TPV-2L. Therefore, TPV-2L possesses the TNF-binding activity reported previously for gp38 but lacks the other reported binding activities attributed to this species.
Figure 4.
TPV-2L binds to human TNF with high affinity but not murine TNF or human IL-2, IL-5, or LT-α. TPV-2L was immobilized on one flow cell of a CM-5 BIAcore sensor chip whereas the other flow cell acted as a blank control surface. Over the TPV-2L sensor chip was passed 100 μl of human TNF, IL-2, IL-5, LT-α, or murine TNF (a) or duplicate injections of 0.3, 0.9, 2.8, or 8.3 nM human TNF at a flow rate of 50 μl/min (b). The amount of protein bound to the surface was recorded as response units as a function of time. After completion of the injection phase (arrow), the dissociation phase was monitored during the injection of buffer alone (HBS-EP). The sensorgrams shown were normalized by subtracting the control surface sensorgram and a blank injection sensorgram. The binding kinetics were determined by using BIAEVALUATION 3.0 and a 1:1 mass transport model to determine the rate of association (Kon), the rate of dissociation (Koff), and the rate constant (Kd).
A number of poxvirus-encoded soluble TNFR homologs have the ability to complex with both TNF and LT-α (8). To determine the binding specificity for TPV-2L, we tested the ability of human LT-α and murine TNF to bind to TPV-2L by SPR. Even though human TNF binds to TPV-2L with high affinity, we were unable to detect any interaction between TPV-2L and 75 nM murine TNF (Fig. 4a). Similarly, 75 nM human LT-α was unable to bind to TPV-2L. In addition, TPV-2L was unable to bind to the human TNF family members FasL, APRIL, CD40L, TRAIL, EDA1, EDA2, BAFF, TL1A, Tweak, CD30L, 41BBL, Light, GITRL, RNAKL, OX40L, or mCD27L (see Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org), suggesting that TPV-2L has a very narrow ligand specificity that is restricted to human, and likely other primate, TNFs. The tropism of TPV-2L binding to human TNF is consistent with the tropism of TPV and YLDV, which normally infect only primates, including humans.
To determine the kinetics of interaction between TPV-2L and human TNF, SPR was performed by using varying concentrations of human TNF applied to the TPV-2L sensor chip. TPV-2L binds with high affinity to TNF with a Kd of 43 pM (Fig. 4b). TPV-2L binds with a relatively fast association rate of 3.38 × 106 M−1⋅s−1 and a very slow dissociation rate of 1.45 × 10−4 s−1. These data indicate that TPV-2L is a high-affinity binding protein for human TNF but not for murine TNF, human LT-α, human IL-2, human IL-5, or human IFN-γ. Furthermore, the 2L/human TNF complex is particularly stable, with an estimated half-life of ≈6 h on the BIAcore chip.
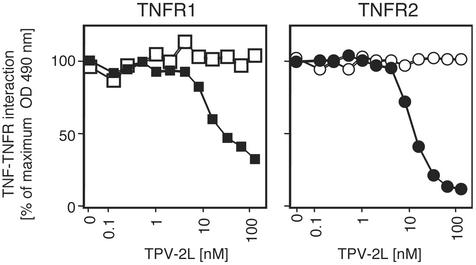
TPV-2L Blocks TNF Binding to TNFRI and TNFRII.
To determine whether TPV-2L could block the ability of human or murine TNF to bind to the TNFRs TNFRI and TNFRII, a myc/His-tagged version of TPV-2L was incubated with either Flag-tagged human or murine TNF. The TPV-2L/TNF mixture was applied to ELISA plates coated with either TNFRI or TNFRII, and the amount of TNF able to bind was quantified. Fig. 5 demonstrates that TPV-2L is able to bind and inhibit human but not murine TNF from binding to the TNFRs. TPV-2L inhibited the binding of hTNF to its receptors with an IC50 of 10–20 nM (of monomer). Because TNF was used at a concentration of 5 nM (of trimer), this indicates that the estimated ratio of TNF and TPV-2L in the complex may be close to 1:1 stoichiometry.
Figure 5.
TPV-2L inhibits TNF binding to both TNFRI and TNFRII. The binding of a fixed concentration of Flag-tagged human TNF (hTNF) (solid symbols) to human TNFRI (hTNFR1) (■ and □) or human TNFRII (hTNFR2) (● and ○) was inhibited by increasing amounts of TPV-2L. In contrast, binding of Flag-tagged murine TNF (muTNF) (open symbols) was unaffected by TPV-2L.
TPV-2L Inhibits the Activity of Human but Not Murine or Rabbit TNF.
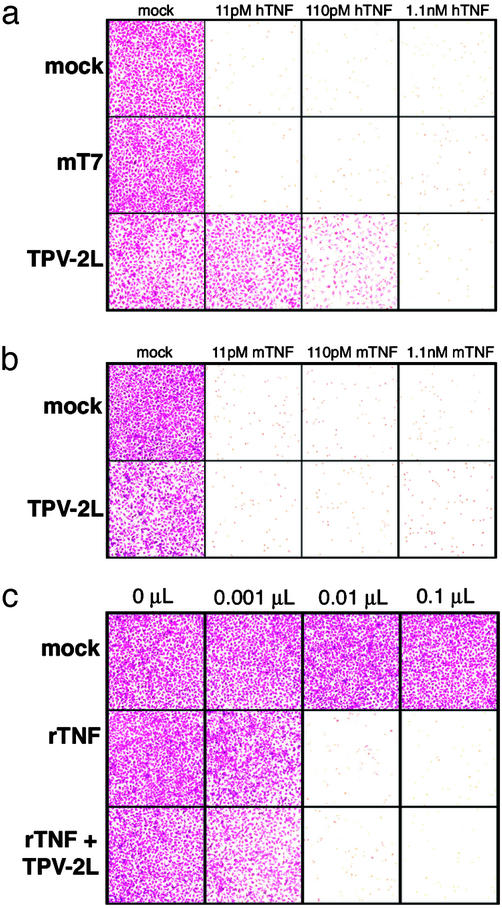
To determine whether TPV-2L could inhibit the biological activity of TNF, we carried out a cytotoxicity assay by using murine L929 cells. The addition of human or murine TNF induces murine L929 cells to undergo apoptosis (Fig. 6 a and b, mock). Similarly, the addition of 3.5 nM purified T7 protein, an IFN-γ-binding protein from myxoma virus (27, 28), as a control failed to inhibit TNF-induced apoptosis (Fig. 6a). In contrast, the addition of 3.5 nM TPV-2L was sufficient to dramatically inhibit cytolysis caused by 11 pM human TNF (Fig. 6a). It required a 30-fold molar excess of TPV-2L (3.5 nM) to TNF (110 pM) to achieve inhibition of TNF-induced apoptosis (Fig. 6a).
Figure 6.
TPV-2L inhibits human but not murine or rabbit TNF. L929 cells were treated with actinomycin D along with 11 pM, 110 pM, or 1.1 nM of either human (a) or murine (b) TNF to induce apoptosis. The cells then were treated immediately with PBS (mock), 3.5 nM purified myxoma T7 protein (mT7), or 3.5 nM purified TPV-2L. (c) L929 cells were treated with 0, 0.001, 0.01, or 0.1 μl of supernatants from wild-type vaccinia virus Western Reserve strain (mock) or vaccinia virus expressing rabbit TNF in the presence (rTNF+TPV-2L) or absence (rTNF) of 3.5 nM TPV-2L. All cells then were incubated for 16 h, and cell viability was determined by staining with crystal violet.
The species specificity for TNF inhibition by 2L protein was examined next. Murine TNF at a concentration of 1.1 pM was unable to induce apoptosis of L929 cells (data not shown) whereas 11 pM was sufficient, and 2L protein did not affect either threshold. These data suggest that 3.5 nM TPV-2L cannot inhibit 11 pM murine TNF. To test whether TPV-2L could inhibit rabbit TNF, supernatants from cells infected with a recombinant vaccinia virus, which secretes rabbit TNF into the medium (23), was used to induce apoptosis of L929 cells. Fig. 6c shows that titrated recombinant rabbit TNF induced apoptosis of L929 cells, whereas control supernatants from a wild-type vaccinia virus were unable to induce apoptosis (mock). The addition of TPV-2L had no effect on the induction of apoptosis triggered by any concentration of recombinant vaccinia virus expressing rabbit TNF. These data are consistent with the observation that TPV-2L possesses a high affinity and specificity for human TNF.
Discussion
We have identified and characterized a high-affinity inhibitor for human TNF encoded by the TPV gene 2L, with related family members present in YLDV, YMTV, and SPV. Unlike other poxvirus TNF-binding proteins, the protein encoded by TPV-2L shows no homology to cellular TNFRs and, hence, is referred to as vTNF-BP to denote its unique status. In fact, the only sequence similarity present in TPV-2L is to cellular MHC class I molecules, and this is restricted to a portion of the α2 and α3 domains. The α1 and α2 domains of MHC class I are responsible for the peptide-binding pocket, and the α3 domain is responsible for binding to β2-microglobulin (29). TPV-2L lacks a complete α2 or α3 domain, so it is unclear what significance, if any, this apparent similarity might represent. It is interesting to speculate that TPV-2L may have originated from the acquisition of a cellular MHC class I molecule followed by extensive sequence divergence, but it should be noted that the similarity between MHC class I and TPV-2L is relatively low and is restricted to a 54-aa stretch exhibiting ≈33% identity.
The 2L orthologs map near the termini for each of TPV, YLD (19), YMTV (C.R.B., H. Amano, Y. Ueda, T. Miyamura, T. Suzuki, X. Li, J.W.B., and G.M., unpublished results), and SPV (25). In SPV, the TPV-2L ortholog maps in the terminal inverted repeat, so that each genome of SPV contains two identical copies of the 2L ortholog, designated SPV003 and SPV148. In contrast, only the last three to six codons at the C terminus of the 2L gene at the left genomic end are found in the terminal inverted repeats of TPV, YLD, and YMTV, indicating that only one functional copy of vTNF-BP is present in each of these yatapoxvirus genomes.
With the identification of a novel class of vTNF-BPs in the yatapox and suipox genera, the importance of the TNF pathway as a significant antipoxviral strategy is underscored. To date, inhibitory proteins targeted against TNF have been identified in all poxvirus genera, with the exception of the capripoxviruses (30, 31) and molluscipoxviruses (32) (although molluscipoxvirus could interfere with TNF signaling via the vFLIP molecules). Whether capripoxviruses and molluscipoxviruses truly lack anti-TNF strategies or whether, as the yatapoxviruses and suipoxviruses, they utilize anti-TNF strategies that have yet to be characterized remains to be demonstrated.
TPV-2L binds to TNF with an ≈1:1 stoichiometry to human TNF. However, these figures rely strongly on the way concentrations were determined. We also do not know whether TPV-2L is a monomer, a dimer, or an oligomer although preliminary evidence suggests a monomeric form. Currently, x-ray crystallographic studies are underway to determine the precise stoichiometry of the complex as well as the contact points between TPV-2L and human TNF.
TPV-2L binds with a very high affinity, with a dissociation rate constant of 43 pM. What is particularly striking about the binding of TPV-2L to human TNF is the extremely slow rate of dissociation. The SPR sensorgrams of TPV-2L binding to human TNF shows negligible dissociation between TNF and TPV-2L. Thus, TPV-2L exhibits the strongest interaction reported to date between human TNF and any other characterized proteins. For example, murine TNFRs and cowpox CrmC bind to murine TNF with an affinity of ≈200 pM (14). The TNF antagonists currently in human clinical trials, infliximab (Remicade) and etanercept (Enbrel), bind human TNF with affinities of 450 and 1,150 pM, respectively (33).
Although TPV-2L binds to human TNF with high affinity, it failed to interact with human IL-2, IL-5, IFN-γ, LT-α, and murine TNF at least as measured by SPR. These latter ligands were tested at a 250-fold higher concentration than that used for human TNF. It remains possible that the interaction between TPV-2L and other cytokines is not detectable by SPR, but it is worth noting that murine TNF could not be inhibited by TPV-2L in an in vivo cytolysis assay, confirming the SPR-binding analysis.
Previous reports had identified a multicytokine-binding protein present in the supernatants of TPV-infected cells that neutralized TNF, IL-2, IL-5, and IFN-γ (20, 26). These activities were ascribed to the protein designated gp38, based on its molecular mass of 38 kDa. We show here that TPV-2L is a 45-kDa protein that binds and inhibits TNF but lacks any detectable IL-2, IL-5, or IFN-γ binding. There are a number of possible reasons to explain the differing binding activities between the predicted TPV gp38 species and what we have observed for TPV-2L. It is possible that within the supernatants of TPV infected cells, there are several proteins of approximately the same size that contribute to the multicytokine-binding activity, of which 2L is one. Alternatively, IL-2, IL-5, or IFN-γ simply may not bind to TPV-2L under the conditions used for SPR. However, crosslinking studies between IL-2 and TPV-2L failed to show any detectable interaction between these two proteins (D. Ricciuto, J.W.B., and G.M., data not shown). We conclude that TNF binding and inhibition are properties of TPV-2L but that the other cytokine-inhibitory properties originally reported for gp38 reside in other still-unidentified TPV proteins.
In conclusion, we describe a high-affinity inhibitor for human TNF expressed by the 2L gene of TPV. The 2L sequence is unrelated to any known TNFR or inhibitor and, hence, represents a unique inhibitory mechanism for human TNF. Solving the structure of the vTNF-BP/TNF complex should prove illuminating given its unusually long half-life, and vTNF-BP should provide a useful model with which to devise anti-TNF strategies for therapeutic purposes.
Supplementary Material
Acknowledgments
We thank Drs. James Johnston and Fuan Wang for critically reading the manuscript. This work was supported by the Swiss National Foundation (to P.S.), National Cancer Institute of Canada, and Viron Therapeutics (to G.M.). G.M. holds a Canada Research Chair in molecular virology. Part of this work is included in M.P.-M's Ph.D. thesis.
Abbreviations
- TNF
tumor necrosis factor
- YLDV
Yaba-like disease virus
- TPV
Tanapox virus
- YMTV
Yaba monkey tumor virus
- SPV
swinepox
- vTNF-BP
viral TNF-binding protein
- TNFR
TNF receptor
- SPR
surface plasmon resonance
- Crm
cytokine response modifier
Footnotes
References
- 1.Locksley R M, Killeen N, Lenardo M J. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 2.Beutler B. Tumor Necrosis Factors: The Molecules and Their Emerging Role in Medicine. New York: Raven; 1992. [Google Scholar]
- 3.Chen G, Goeddel D V. Science. 2002;296:1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 4.Seet B T, Johnston J B, Brunetti C R, Barrett J W, Everett H, Cameron C, Sypula J, Nazarian S, Lucas A, McFadden G. Annu Rev Immunol. 2003;21:377–423. doi: 10.1146/annurev.immunol.21.120601.141049. [DOI] [PubMed] [Google Scholar]
- 5.Alcamí A, Koszinowski U H. Immunol Today. 2000;21:447–455. doi: 10.1016/S0167-5699(00)01699-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tortorella D, Gewurz B E, Furman M H, Schust D J, Ploegh H L. Annu Rev Immunol. 2000;18:861–926. doi: 10.1146/annurev.immunol.18.1.861. [DOI] [PubMed] [Google Scholar]
- 7.Moss B, Shisler J. Semin Immunol. 2001;13:59–66. doi: 10.1006/smim.2000.0296. [DOI] [PubMed] [Google Scholar]
- 8.Cunnion K M. Mol Genet Metab. 1999;67:278–282. doi: 10.1006/mgme.1999.2878. [DOI] [PubMed] [Google Scholar]
- 9.Smith C A, Davis T, Anderson D, Solam L, Beckman M P, Jerzy R, Dower S K, Cosman D, Goodwin R G. Science. 1990;248:1019–1023. doi: 10.1126/science.2160731. [DOI] [PubMed] [Google Scholar]
- 10.Smith C A, Davis T, Wignall J M, Din W S, Farrah T, Upton C, McFadden G, Goodwin R G. Biochem Biophys Res Commun. 1991;176:335–342. doi: 10.1016/0006-291x(91)90929-2. [DOI] [PubMed] [Google Scholar]
- 11.Upton C, Macen J L, Schreiber M, McFadden G. Virology. 1991;184:370–382. doi: 10.1016/0042-6822(91)90853-4. [DOI] [PubMed] [Google Scholar]
- 12.Xu X-M, Nash P, McFadden G. Virus Genes. 2000;21:97–109. [PubMed] [Google Scholar]
- 13.Hu F-Q, Smith C A, Pickup D J. Virology. 1994;204:343–356. doi: 10.1006/viro.1994.1539. [DOI] [PubMed] [Google Scholar]
- 14.Smith C A, Hu F-Q, Smith T D, Richards C L, Smolak P, Goodwin R G, Pickup D J. Virology. 1996;223:132–147. doi: 10.1006/viro.1996.0462. [DOI] [PubMed] [Google Scholar]
- 15.Loparev V N, Parsons J M, Knight J C, Panus J F, Ray C A, Buller R M L, Pickup D J, Esposito J J. Proc Natl Acad Sci USA. 1998;95:3786–3791. doi: 10.1073/pnas.95.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saraiva M, Alcami A. J Virol. 2001;75:226–233. doi: 10.1128/JVI.75.1.226-233.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Harven E, Yohn D S. Cancer Res. 1966;26:995–1008. [PubMed] [Google Scholar]
- 18.Espana C, Brayton M A, Ruebner B H. Exp Mol Pathol. 1971;15:34–42. doi: 10.1016/0014-4800(71)90017-7. [DOI] [PubMed] [Google Scholar]
- 19.Lee H-J, Essani K, Smith G L. Virology. 2001;281:170–192. doi: 10.1006/viro.2000.0761. [DOI] [PubMed] [Google Scholar]
- 20.Paulose M, Bennet B L, Manning A M, Essani K. Microb Pathog. 1998;25:33–41. doi: 10.1006/mpat.1998.0213. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Johnsson B, Lofas S, Lindquist G. Anal Biochem. 1991;198:268–277. doi: 10.1016/0003-2697(91)90424-r. [DOI] [PubMed] [Google Scholar]
- 23.Schreiber M, McFadden G. Virology. 1994;204:692–705. doi: 10.1006/viro.1994.1585. [DOI] [PubMed] [Google Scholar]
- 24.Massung R F, Jayarama V, Moyer R W. Virology. 1993;197:511–528. doi: 10.1006/viro.1993.1625. [DOI] [PubMed] [Google Scholar]
- 25.Afonso C L, Tulman E R, Lu Z, Zsak L, Osario F A, Balinsky C, Kutish G F, Rock D L. J Virol. 2002;76:783–790. doi: 10.1128/JVI.76.2.783-790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Essani K, Chalasani S, Eversole R, Beuving L, Birmingham L. Microb Pathog. 1994;17:347–353. doi: 10.1006/mpat.1994.1080. [DOI] [PubMed] [Google Scholar]
- 27.Mossman K, Upton C, McFadden G. J Biol Chem. 1995;270:3031–3038. doi: 10.1074/jbc.270.7.3031. [DOI] [PubMed] [Google Scholar]
- 28.Upton C, Mossman K, McFadden G. Science. 1992;258:1369–1372. doi: 10.1126/science.1455233. [DOI] [PubMed] [Google Scholar]
- 29.Williams A, Peh C A, Elliott T. Tissue Antigens. 2002;59:3–17. doi: 10.1034/j.1399-0039.2002.590103.x. [DOI] [PubMed] [Google Scholar]
- 30.Tulman E R, Afonso C L, Lu Z, Zsak L, Kutish G F, Rock D L. J Virol. 2001;75:7122–7130. doi: 10.1128/JVI.75.15.7122-7130.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tulman E R, Afonso C L, Lu Z, Zsak L, Sur J-H, Sandybaev N T, Kerembekova U Z, Zaitsev V L, Kutish G F, Rock D L. J Virol. 2002;76:6054–6061. doi: 10.1128/JVI.76.12.6054-6061.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Senkevich T G, Koonin E V, Bugert J J, Darai G, Moss B. Virology. 1997;233:19–42. doi: 10.1006/viro.1997.8607. [DOI] [PubMed] [Google Scholar]
- 33.Scallon B, Cai A, Solowski N, Rosenberg A, Song X-Y, Shealy D, Wagner C. J Pharmacol Exp Ther. 2002;301:418–426. doi: 10.1124/jpet.301.2.418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.