Abstract
l-Arginine is the only endogenous nitrogen-containing substrate of NO synthase (NOS), and it thus governs the production of NO during nervous system development as well as in disease states such as stroke, multiple sclerosis, Parkinson's disease, and HIV dementia. The “arginine paradox” refers to the dependence of cellular NO production on exogenous l-arginine concentration despite the theoretical saturation of NOS enzymes with intracellular l-arginine. Herein, we report that decreased availability of l-arginine blocked induction of NO production in cytokine-stimulated astrocytes, owing to inhibition of inducible NOS (iNOS) protein expression. However, activity of the promoter of the iNOS gene, induction of iNOS mRNA, and stability of iNOS protein were not inhibited under these conditions. Our results indicate that inhibition of iNOS activity by arginine depletion in stimulated astrocyte cultures occurs via inhibition of translation of iNOS mRNA. After stimulation by cytokines, uptake of l-arginine negatively regulates the phosphorylation status of the eukaryotic initiation factor (eIF2α), which, in turn, regulates translation of iNOS mRNA. eIF2α phosphorylation correlates with phosphorylation of the mammalian homolog of yeast GCN2 eIF2α kinase. As the kinase activity of GCN2 is activated by phosphorylation, these findings suggest that GCN2 activity represents a proximal step in the iNOS translational regulation by availability of l-arginine. These results provide an explanation for the arginine paradox for iNOS and define a distinct mechanism by which a substrate can regulate the activity of its associated enzyme.
Nitric oxide (NO) is a diffusible neuronal second messenger that can be synthesized in the nervous system by three distinct enzymes: neuronal NO synthase (NOS) (1), endothelial NOS (2–4), and inducible NO synthase (iNOS) (5). Neuronal NOS and endothelial NOS differ from iNOS in that they are tightly regulated by calcium-activated calmodulin, specific phosphorylation, interaction with plasma membrane ionotropic receptors, or compartmentalization in caveolae (6). This tight regulation makes neuronal or endothelial NOS ideal for generating NO as a signaling molecule that can regulate physiological processes such as differentiation and plasticity in the nervous system (6, 7). By contrast, iNOS enzyme is commonly up-regulated by inflammatory mediators, and it produces NO as long as the molecule is intact and its substrate arginine is available (8, 9). Indeed, in vitro, iNOS-expressing cells can produce large amounts of NO for days or longer. Whereas the high activity of iNOS enzyme makes it well suited as an effector of cellular defense against invading microorganisms or as a mediator of homeostatic responses to hypoxia, persistent activation of iNOS can lead to toxic levels of NO production that are deleterious for the host. Because NO is a free radical, it can oxidize protein, lipids, or DNA, and its toxic oxidative properties are enhanced if it reacts with a superoxide anion radical to form peroxynitrite (10). Indeed, excessive NO generated from iNOS has been implicated in a host of disease states including multiple sclerosis, HIV dementia, Huntington's disease, Alzheimer's disease, Parkinson's disease, stroke, and amyotrophic lateral sclerosis. In all of these diseases, iNOS protein levels are increased in microglia or astrocytes; molecular deletion or pharmacological inhibition of iNOS can reduce cell loss in rodent models of stroke, Parkinson's disease, and Huntington's disease (11–13).
Because of its strong association with neuropathology, the regulation of iNOS has become the subject of intense scrutiny. However, specific intracellular regulatory mechanisms that will turn off iNOS enzyme activity or, alternatively, prevent it from being activated, are not well understood. NO is synthesized by iNOS via a five-electron oxidation of a nitrogen atom from the L-arginine guanidinium group. As L-arginine is the only nitrogen-containing substrate capable of participating in the complex enzymatic mechanisms involved in NO synthesis, much attention has been focused on the transport, synthesis, and degradation of arginine as a strategy for regulating iNOS therapeutically.
Arginine levels in the cerebrospinal fluid and the CNS pericellular space have been estimated at 20–40 μM (14). In response to inflammatory stimuli, arginine is transported into microglia or astrocytes via specific transporters (15). Astrocytes have been found to transport L-arginine mainly by system y+, a high-affinity Na+-independent transporter of arginine, lysine, and ornithine. Transport of arginine into astrocytes is necessary for maximal iNOS activity and NO generation (16). However, it remains unclear why extracellular arginine is required for iNOS activity as basal, intracellular concentrations of arginine are well above the Km level and should be sufficient to saturate iNOS. This phenomenon, termed the “arginine paradox,” has yet to be adequately resolved (17).
Herein, we resolve the arginine paradox for iNOS. We demonstrate that in resting astrocyte cultures, iNOS is not expressed. In response to cytokines, iNOS transcription is activated, and uptake of arginine leads to derepression of the translational control apparatus and enhanced levels of iNOS protein. Extracellular arginine deprivation or intracellular arginase overexpression leads to decreased intracellular arginine and decreased translation of iNOS.
Materials and Methods
Reagents.
Recombinant murine IFN-γ, DMEM, FBS, penicillin, and streptomycin were from Life Technologies (Grand Island, NY). Dibutryl cAMP was from Sigma. L-Arginine was obtained from Calbiochem. L-[Guanido-14C]arginine and [α-32P]dCTP [3,000 Ci (1 Ci = 37 GBq)/nmol] were purchased from DuPont NEN. Antibodies against rat iNOS, β-actin, GFP, and glial fibrillary acidic protein (GFAP) were purchased from Santa Cruz Biotechnology.
Isolation and Culture of Rat Neonate Astrocytes.
Astrocyte precursors were isolated from 1- to 3-day-old rat pups as described (18). Cells were stimulated with cAMP and/or IFN-γ in DMEM culture.
Adenoviral Infection.
Astrocytes were plated at a density of 2 × 105 cells per 100-mm culture plates (Corning). Subconfluent astrocytes were infected with recombinant adenovirus containing GFP cDNA (19) and Arg I cDNA, eukaryotic initiation factor 2α (eIF2α) S51A cDNA, eIF2α S51D cDNA, or only GFP cDNA as control at designated multiplicities of infection (mois) as described (20).
Determination of Arginase Activity.
Arginase activity in cell lysates was measured by the conversion of [guanido-14C]arginine to 14C urea, using a derivation of the method of Russell and Ruegg (22). Activation of cell lysates and initiation of the arginase assay were exactly as described (22).
Measurement of Intracellular Arginine Level.
The neutralized extracts were used for determining amino acid (arginine) by HPLC methods, as described by Wu and Morris (15).
Results and Discussion
Induction of iNOS, but Not Arginase, in Astrocytes by cAMP/IFN-γ or Lipopolysaccharide (LPS)/IFN-γ.
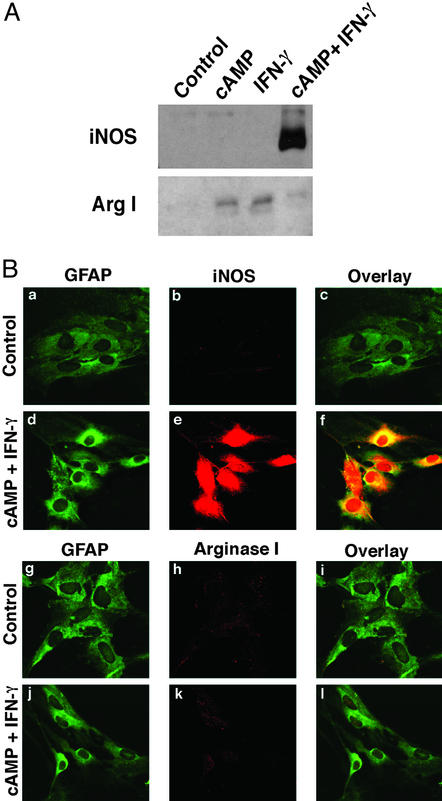
To evaluate the arginine paradox in astrocyte cultures, we identified stimuli that induced iNOS expression but did not induce the arginine-degrading enzyme arginase I. Astrocytes were stimulated with dbcAMP (1 mM)/IFN-γ (100 units/ml) or LPS (1 μg/ml)/IFN-γ (100 units/ml) for 24 h, and iNOS protein levels were monitored by immunoblot analysis. cAMP/IFN-γ (Fig. 1A) and LPS/IFN-γ (not shown) robustly induced iNOS protein levels (24). Interestingly, when cAMP or IFN-γ were added separately to astrocytes, induction of iNOS was not observed. Rather, arginase I was strongly induced by the separate addition of each agent (Fig. 1A). Confocal microscopy verified that iNOS expression colocalized with astrocytic, GFAP-positive cells (Fig. 1B) and that concomitant addition of cAMP/IFN-γ to astrocytes did not affect arginase I expression (Fig. 1B). The induction of arginase I by IFN-γ seen in primary astrocytes herein is in contrast to previous observations that IFN-γ decreases arginase levels in cells whose origins are outside of the nervous system (25–27). Although the precise reasons for these differences are not clear, they may reflect cell type-specific regulation of arginases.
Figure 1.
Regulation of iNOS and arginase I in primary cultured rat astrocytes. (A) Immunoblot analyses of iNOS and arginase I expression. Rat astrocytes were treated with cAMP (1 mM), IFN-γ (100 units/ml), and cAMP plus IFN-γ for 24 h. Whole-cell lysates were subjected to immunoblotting with antibodies specific for iNOS or arginase I. (B) cAMP and IFN-γ induce iNOS but not arginase I in astrocytes. Rat astrocytes were treated with vehicle (control) or cAMP plus IFN-γ for 24 h. Cells were fixed with 4% paraformaldehyde and incubated with mouse anti-GFAP (a, d, g, and j), rabbit anti-iNOS (b and e), or rabbit anti-arginase I (h and k) antibody. Fluorescein-conjugated secondary antibodies were incubated, and indirect fluorescence was detected by confocal microscopy.
Overexpression of Arginase I in Astrocytes Decreases Cytokine-Induced NO Production by Decreasing iNOS Protein Levels.
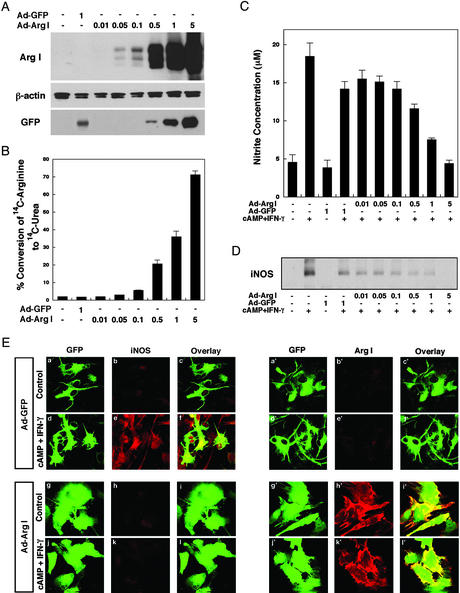
To increase intracellular arginase levels and evaluate the effects of decreasing arginine on iNOS activity, we generated a replication-deficient, adenoviral vector containing the rat arginase I cDNA (Ad-ArgI). Arginase I is a 35- to 38-kDa cytoplasmic protein that cleaves arginine into urea and ornithine (15). The enzyme is highly expressed in liver and functions as part of the urea cycle (15). Expression of arginase I has been observed in other organs in addition to liver including brain, but the function of arginase in brain remains unclear (15). Infection of astrocytes with the Ad-ArgI vector resulted in an moi-dependent increase in arginase I expression and activity (Fig. 2 A and B). An adenoviral vector expressing GFP alone did not affect arginase I levels or activity (Fig. 2 A and B). We evaluated the effect of enforced expression of arginase I on iNOS activity; as expected, we found that as intracellular arginase I levels were increased, iNOS activity (measured by nitrite in the medium) decreased (Fig. 2C). Indeed, the highest moi used resulted in sufficient expression of arginase I to completely suppress cAMP/IFN-γ-induced iNOS activity. Previous studies using stable nonneuronal cell lines have demonstrated that enforced expression of arginase I can inhibit NOS activity, and these studies have reasonably assumed that inhibition results from the ability of arginase to steal arginine substrate from iNOS (28). However, we found that suppression of iNOS activity by arginase I was associated with decreased iNOS protein expression as monitored by immunoblot analysis (Fig. 2D) or confocal microscopy (Fig. 2E). Measurement of intracellular amino acid levels verified that infection of astrocytes with Ad-ArgI (moi = 5) resulted in a 60% decrease in intracellular arginine but did not affect other amino acids such as asparagine and glycine (data not shown). These results indicate that arginase activity leads to specific decreases in arginine levels, and reduced intracellular arginine regulates iNOS activity by regulating iNOS expression.
Figure 2.
(A) Increased expression of arginase I in astrocytes by using an adenoviral vector. Rat astrocytes were infected with an adenoviral vector containing arginase I (Ad-Arg I) or vector control (Ad-GFP) for 48 h. Whole-cell lysates were subjected to immunoblotting with antibodies specific for arginase I, β-actin, or GFP. (B) Increased arginase activity in virally transfected astrocytes. Rat astrocytes were infected with the indicated moi of Ad-Arg I or Ad-GFP for 48 h. Arginase activity reflects percent conversion of [14C]arginine added to the culture medium to 14C urea. Results are mean ± SD of triplicate experiments. (C) Overexpression of arginase I in astrocytes can decrease cytokine-induced NO levels in astrocytes. Rat astrocytes were infected with the indicated moi of Ad-Arg I or Ad-GFP for 24 h and then further treated with cAMP (1 mM) and IFN-γ (100 units/ml) to induce NO product. After 16 h, NO levels were quantified by measuring the accumulation of nitrite in the culture medium of astrocytes with the Griess reagent. (D) Overexpression of arginase I in astrocytes can decrease cytokine-induced iNOS protein levels. Rat astrocytes were infected with Ad-Arg I or Ad-GFP (vector control) for 24 h and subsequently treated with cAMP (1 mM) and IFN-γ (100 units/ml) for 16 h. Immunoblot analysis of iNOS in whole-cell lysates is shown. (E) For confocal microscopy cells were fixed with 4% paraformaldehyde and incubated with rabbit anti-iNOS (b, e, h, and k) or rabbit anti-arginase I (b′, e′, h′, and k′) antibody. The adenovirus transfer vector used in these studies has a double expression cassette, and GFP has been subcloned into Ad-GFP (a, a′, d, and d′) or Ad-Arg I (g, g′, j, and j′) to monitor vector-derived expression.
To determine whether changes in iNOS protein expression induced by intracellular arginase I could be attributed merely to global suppression of protein synthesis, we measured incorporation of radioactive cysteine and methionine into perchloric acid-precipitable protein fractions. Infection of astrocytes with Ad-GFP at an moi of 1 reduced protein synthesis by ≈50%, comparable to the magnitude of the decrease of iNOS protein observed with the vector control as compared with uninfected cells. Interestingly, protein synthesis rates in astrocytes infected with varying mois (0.1–5) of Ad-ArgI were comparable to the vector control. These results suggest that suppression of iNOS expression by overexpressing arginase I cannot be attributed simply to a decrease in global protein synthesis.
In addition to depleting arginine, arginase I produces ornithine and urea and thus can provide substrate for the ornithine decarboxylase-dependent generation of polyamines. A suicide inhibitor of ornithine decarboxylase, α-difluoromethylornithine, had no effect on iNOS protein levels in astrocytes overexpressing arginase, suggesting that the polyamine pathway was unlikely to be involved in the regulation of iNOS by arginase.
Extracellular Arginine Availability Regulates Levels of iNOS Induced by Cytokines.
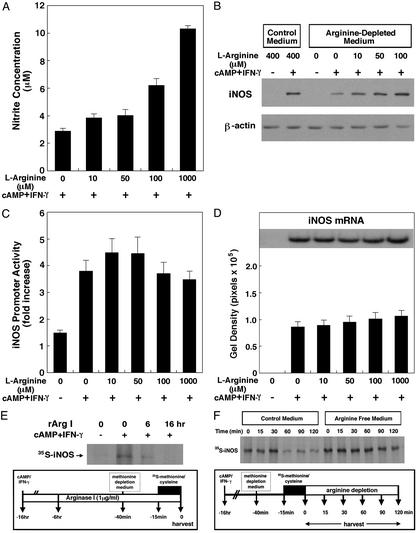
Induction of iNOS activity by inflammatory mediators in astrocytes depends on the concentration of arginine in the culture medium (Fig. 3A). These observations confirm previous studies that demonstrated that as the concentration of extracellular arginine increased, iNOS activity in astrocytes also increased (29). To determine whether manipulating extracellular arginine affects iNOS protein levels, astrocytes were switched to medium containing varying concentrations of arginine (0–100 μM) at the time of cytokine treatment, and protein lysates were harvested for immunoblotting 24 h later. As expected from our findings with overexpressed arginase, we found that the level of induction of iNOS protein depends on the level of extracellular arginine (Fig. 3B). As concentrations of arginine in cerebrospinal fluid have been estimated at 20–40 μM (14), these results suggest that physiological concentrations of extracellular arginine regulate iNOS activity by affecting iNOS protein levels.
Figure 3.
Regulation of iNOS expression by extracellular arginine. (A) Extracellular arginine regulates intracellular production of NO by iNOS. Astrocytes were cultured in L-arginine-free media containing the designated concentration of L-arginine supplementation. Parallel cultures were also treated with cAMP/IFN-γ to induce iNOS expression, and NO levels were quantitated as described. (B) Extracellular levels of L-arginine regulate intracellular iNOS expression induced by cAMP and IFN-γ in astrocytes. (C) Manipulating extracellular arginine does not affect iNOS promoter activity. iNOS promoter (firefly luciferase) activity was normalized to Renilla luciferase activity. The data are mean ± SE obtained from three separate experiments. (D) Manipulating extracellular arginine does not affect iNOS mRNA levels. Northern blot analysis data were obtained from seven separate experiments. (E) Manipulation of extracellular arginine using recombinant arginase I diminishes de novo synthesis of iNOS. Rat astrocytes were treated with cAMP (1 mM) and IFN-γ (100 units/ml) for 16 h before cell harvest. Recombinant arginase I (1 μg/ml) was added 6 or 16 h before cell harvest. Forty minutes before cell harvest, cells were placed in methionine/cysteine-free medium. [35S]methionine/cysteine was added for 15 min before harvest. Harvested cell lysates were immunoprecipitated with an anti-iNOS antibody. Immunoprecipitated samples were resolved by SDS/PAGE, and radiolabeled proteins were detected by autoradiography. (F) Extracellular manipulation of arginine does not trigger the degradation of iNOS protein. Rat astrocytes were treated with cAMP (1 mM) and IFN-γ (100 units/ml) for 16 h before the chase with nonradiolabeled amino acids. Subsequently, cells were placed in methionine/cysteine-free medium 40 min before the chase addition; [35S]methionine/cysteine was added for 15 min before the chase addition. Cells were chased in L-arginine-depleted medium or L-arginine-supplemented medium (control) for the indicated time period. At the designated times, cell lysates were harvested, and levels of radiolabeled iNOS were detected as described above.
Recent studies have documented the ability of specific amino acids to control mammalian gene transcription. Indeed, arginine depletion in HeLa cells can lead to activation of the transcription factor, ATF-2, and expression of the CHOP (gadd153) gene (30). To determine whether arginine depletion can inhibit iNOS expression by inhibiting iNOS transcription, we looked at whether varying concentrations of arginine affected activation of the iNOS promoter. We used a promoter–reporter gene construct that contains 1.5 kb of noncoding sequence 5′ to the ATG start site of the iNOS gene coupled to a luciferase reporter gene (31). iNOS promoter activity in astrocytes cultured in the absence of arginine and presence of cAMP/IFN-γ did not differ significantly from cytokine-exposed astrocytes cultured in the presence of gradually increasing concentrations (0–1,000 μM) of arginine (Fig. 3C).
Arginine depletion has been shown to decrease expression of the CD3 ζ chain of the T cell receptor by significantly reducing the half-life of the CD3 ζ chain mRNA (32). However, Northern blot analysis using a radiolabeled iNOS cDNA probe showed that varying the concentration of extracellular arginine also did not affect iNOS mRNA levels (Fig. 3D). Taken together, these observations indicate that changes in iNOS protein resulting from manipulation of extracellular arginine concentration or intracellular arginase expression cannot be attributed to changes in iNOS transcription or mRNA stability.
Arginine Determines iNOS Protein Levels via Translational Control Mechanisms.
After eliminating the possibility that changes in iNOS transcription or mRNA stability accounted for changes in iNOS protein levels in response to varying extracellular arginine concentrations, we turned our attention to the hypothesis that changes in extracellular arginine may regulate translation control mechanisms to alter expression of iNOS protein. To directly test whether arginine manipulation alters the translation of iNOS protein from its mRNA, we immunoprecipitated iNOS from lysates of astrocytes metabolically labeled with 35S-methionine/cysteine. When the radiolabeled immunoprecipitate was subjected to SDS/PAGE and autoradiography, we found incorporation of radiolabel into a band that migrated at the predicted molecular weight of iNOS. As expected, we found decreased radiolabeled iNOS in astrocytes cultured in arginine-deficient medium as compared with those plated in the presence of normal extracellular arginine (Fig. 3E). Subsequent pulse–chase experiments confirmed that decreased iNOS expression under conditions of arginine deprivation is due to decreased synthesis rather than increased degradation. Indeed, we found that although arginine depletion led to a decrease in cytokine-induced iNOS protein levels, the half-life of the synthesized protein was increased (Fig. 3F). These findings support the notion that arginine regulates translational control mechanisms to modulate iNOS protein levels.
How does arginine regulate the translation of iNOS? Amino acid deprivation in yeast and eukaryotes has been shown to activate the GCN2 kinase and result in the phosphorylation of eIF2α (33, 34). eIF2α is a translational initiation factor that is involved in the first regulated step of translation. Recent evidence has underscored the role that amino acids such as arginine and leucine play in the initiation phase of mRNA translation. The first of two regulated steps in translational control involves the binding of methionyl-tRNA to the 40S ribosomal subunit to form the 43S preinitiation complex. For this to occur, eIF2α must form a ternary complex with methionyl tRNA and GTP. This complex then binds to the 40S ribosomal subunit. Translation is initiated when specific mRNAs bind to the 43S preinitiation complex. eIF2α phosphorylation at serine 51 leads to inhibition of guanine nucleotide exchange, inhibition of ternary complex formation (eIF2-GTP-Met-tRNA), and cessation of cap-dependent translational initiation. The phosphorylation of eIF2α is a reversible and highly dynamic process that is regulated by a small family of kinases, including the double-stranded RNA-activated protein kinase PKR, heme regulated inhibitor protein kinase HRI, unfolded protein response kinase PERK, and the amino acid control kinase GCN2. These kinases are activated as part of a physiological response to stresses such as heat shock, iron deficiency, viral infection, oxidative stress, cerebral ischemia, and amino acid starvation (33–36). By phosphorylating eIF2α, these kinases act to suppress the translation of some cellular mRNAs. In the specific case of amino acid starvation, GCN2 is activated, eIF2α is phosphorylated, and translation is suppressed, thus liberating amino acids and alleviating the cellular debt. The ability of amino acids to regulate eIF2α phosphorylation and protein translation led us to postulate the following model. Cytokine-stimulated transport of arginine from the extracellular milieu into astrocytes results in increases in iNOS activity by diminishing eIF2α phosphorylation and increasing iNOS translation. As cytoplasmic arginine is also regulated by arginase I, we also postulated that enforced expression of the intracellular arginine-degrading enzyme, arginase, could negatively regulate iNOS activity through increases in eIF2α phosphorylation.
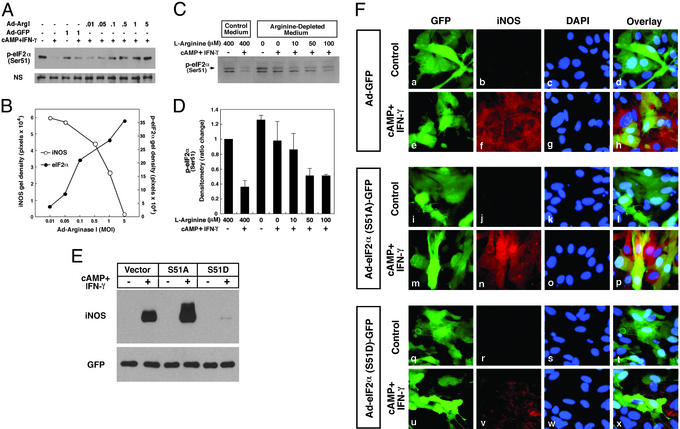
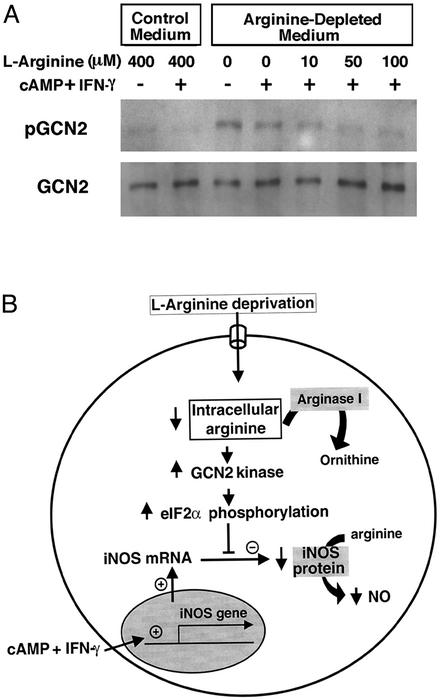
To test the model, we first examined whether overexpression of arginase I leads to an increase in eIF2α phosphorylation at serine 51, using an antibody that recognizes the phospho-form of eIF2α. Under basal conditions, astrocytes unexpectedly possess significant eIF2α phosphorylation (Fig. 4A). Treatment of astrocytes with cAMP and IFN-γ, which induces iNOS expression, decreased eIF2α phosphorylation (Fig. 4A). Viral transfer of arginase I to astrocytes led to an moi-dependent increase in arginase I activity and a concomitant decrease in cAMP/IFN-γ-induced iNOS expression (Fig. 2D). The decrease in iNOS expression correlated directly with an increase in eIF2α phosphorylation (Fig. 4B). As expected from these observations, decreasing concentrations of extracellular arginine that decrease iNOS protein expression correlate directly with increased eIF2α phosphorylation (Fig. 4 C and D). To test directly whether the phosphorylation of eIF2α is sufficient to decrease iNOS expression, we constructed an adenoviral vector containing a cDNA for a phosphomimetic form of eIF2α in which serine 51 has been mutated to aspartic acid (37). As expected, we found that overexpression of this form of eIF2α significantly decreased expression of iNOS as compared with the vector control (Fig. 4 E and F). By contrast, overexpression of a form of eIF2α that cannot be phosphorylated (serine 51 is replaced with alanine; ref. 38), results in increased expression of iNOS as compared with the vector control or the S51D mutant (Fig. 4 E and F). Together, these results are consistent with the hypothesis that intracellular arginine regulates iNOS activity by regulating iNOS expression through translational control pathways involving the amino acid control kinase GCN2 and eIF2α phosphorylation. Accordingly, we also found that experimental manipulations that decrease intracellular arginine during cytokine treatment in astrocytes also result in increased GCN2 phosphorylation, the activated form of this eIF2α kinase (Fig. 5A).
Figure 4.
Extracellular or intracellular manipulation of intracellular arginine decreases iNOS expression by translational control mechanisms involving eIF2α phosphorylation. (A) Arginase I-induced decreases in iNOS protein levels are associated with the phosphorylation of eIF2α. Rat astrocytes were infected with Ad-Arg I or Ad-GFP for 24 h. Cells were then stimulated by cAMP (1 mM) and IFN-γ (100 units/ml) for 16 h. Whole-cell lysates were subjected to immunoblotting with antibodies specific for the form of eIF2α that is phosphorylated at serine 51. (B) An inverse correlation is observed between the level of iNOS protein and the amount of phospho-eIF2α protein. The densitometric values were calculated from iNOS and phospho-eIF2α immunoblots shown in A and Fig. 2D. (C) Increased levels of extracellular arginine are correlated with a decrease in the level of intracellular phospho-eIF2α. (D) The densitometric value (mean ± SD) of phospho-eIF2α from three separate experiments. Rat astrocytes were cultured in l-arginine-depleted medium or l-arginine-supplemented medium for 24 h. Cells were then stimulated by cAMP (1 mM) and IFN-γ (100 units/ml) for 16 h. (E) Rat astrocytes were infected with Ad-GFP (vector control), Ad-S51A (serine to alanine, nonphosphorylatable mutant), or Ad-S51D (serine to aspartate, phosphomimetic mutant) at an moi of 5 for 24 h and subsequently treated with cAMP (1 mM) and IFN-γ (100 units/ml) for 16 h. Immunoblot analysis of iNOS in whole-cell lysates is shown. (F) For immunofluorescence microscopy, cells were fixed with 4% paraformaldehyde and incubated with rabbit anti-iNOS antibody (b, f, j, n, r, and v) or the nuclear stain 4′,6-diamidino-2-phenylindole (DAPI) (c, g, k, o, s, and w). The adenovirus transfer vector used in these studies has a double expression cassette, and GFP has been subcloned into Ad-GFP (a and e), Ad-S51A (i and m), or Ad-S51D (q and u) to monitor vector-derived expression.
Figure 5.
(A) Increased extracellular arginine levels are also associated with decreased phosphorylation of the GCN2 kinase. Cell lysates were immunoprecipitated with GCN2 antibody and immunoblotted with a phospho-GCN2 or a GCN2 antibody. (B) A model for regulation of iNOS activity by arginine. Cytokine stimulation of astrocytes leads to increased transcription of iNOS message. The translation of this message into iNOS protein is governed, in part, by the transport of extracellular arginine into the cell. Increases in intracellular arginine lead to charging of tRNAs, decreased GCN2 phosphorylation and activity, and subsequent decreased phosphorylation of eIF2α. Decreased eIF2α phosphorylation results in increased translational initiation of iNOS. This model resolves the arginine paradox for iNOS and suggests that extracellular arginine regulates the activity of intracellular iNOS by regulating its translation.
Recent studies have highlighted the importance that eIF2α phosphorylation plays in adaptive cell stress responses. These responses, which are highly conserved from yeast to humans, involve eIF2α phosphorylation-mediated translational attenuation, transcriptional induction, and cell survival or death (33–35). Such homeostatic responses have been best characterized under conditions such as amino acid deprivation, viral infection, and endoplasmic reticulum stress. Our studies suggest eIF2α phosphorylation may also regulate inflammatory responses under basal conditions by inhibiting the translation of iNOS. After cytokine treatment, arginine is transported intracellularly, eIF2α phosphorylation decreases, and iNOS translation can ensue.
Taken together, these data are consistent with a model in which cytokines induce the transcriptional up-regulation of iNOS mRNA. The amount of this mRNA that is translated into protein depends on the transport of arginine into the cell, likely via the specific form of the y+ transporter known as CAT-2 (29). The resultant increased intracellular arginine leads to decreased phosphorylation of GCN2 and decreased phosphorylation of eIF2α. Decreased phosphorylation of eIF2α allows increased translation of iNOS mRNA and increases in iNOS protein (Fig. 5B). Regulators of GCN2 activity or eIF2α phosphorylation may therefore be propitious targets for neurological diseases such as stroke, Parkinson's disease, Huntington's disease, and HIV dementia where aberrant iNOS expression has been causally associated with neuronal loss and neuropathology.
Translational Control of iNOS by Arginine Can Explain the Arginine Paradox.
In addition to defining a molecular target for iNOS regulation in neurological disease states, our studies suggest an explanation for the arginine paradox for iNOS. This term has been used to refer to situations in which exogenous L-arginine concentrations apparently determine the level of NO production even when intracellular levels of L-arginine are available in excess. Data supporting other explanations for this paradox have been put forward by other investigators (17, 21, 36, 39). For example, Giugliano et al. (39) demonstrated that i.v. infusions of L-arginine stimulate insulin release and that this insulin release, rather than increased endothelial NOS activity and NO formation, is responsible for vasodilation, decreased platelet aggregation, and decrease in blood viscosity. Moreover, an endogenous competitive inhibitor of NOS, asymmetric dimethylarginine, accumulates in renal failure, preeclampsia, and the serum of cholesterol-fed rabbits (36). Thus, increasing the concentration of extracellular arginine would overcome the effect of the competitive inhibitor and thereby increase NOS activity. However, a role for asymmetric dimethylarginine in vivo has yet to be definitively established.
Our studies, using the experimental leverage of a defined in vitro system, provide another explanation for the arginine paradox in the case of iNOS, and the scenario outlined herein may have specific adaptive functions. It has been shown previously that arginine starvation can lead to NOS-driven superoxide production in cells engineered to overexpress neuronal NOS (23). By coupling arginine levels to iNOS protein synthesis, the cell provides a mechanism for ensuring that iNOS is not expressed in arginine-depleted cells and that toxic superoxide cannot be produced.
In summary, we demonstrate that, as expected, iNOS activity in astrocytes is governed by arginine transported into the cell from the extracellular medium. Unexpectedly, however, we found that arginine concentration not only regulates NO production by limiting availability of substrate for iNOS, it also regulates iNOS expression via translational control of iNOS mRNA.
Acknowledgments
We thank H. Harding and D. Ron for advice, the GCN2 antibody, and the eIF2α constructs; D. Ash for recombinant arginase; C. Lowenstein for the iNOS promoter–reporter construct; and M. Waters for the iNOS cDNA. J.L. is an awardee of the Korea Science and Engineering Foundation. This work was generously supported by National Institutes of Health and Veterans Adminstration of America grants (to R.R.R., S.M.M., and R.J.F.).
Abbreviations
- NOS
NO synthase
- iNOS
inducible NOS
- GFAP
glial fibrillary acidic protein
- moi
multiplicity of infection
- eIF
eukaryotic initiation factor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bredt D S, Hwang P M, Glatt C E, Lowenstein C, Reed R R, Snyder S H. Nature. 1991;351:714–718. doi: 10.1038/351714a0. [DOI] [PubMed] [Google Scholar]
- 2.Furchgott R F. Biosci Rep. 1999;19:235–251. doi: 10.1023/a:1020537506008. [DOI] [PubMed] [Google Scholar]
- 3.Ignarro L J. Biosci Rep. 1999;19:51–71. doi: 10.1023/a:1020150124721. [DOI] [PubMed] [Google Scholar]
- 4.Palmer R M, Ashton D S, Moncada S. Nature. 1988;333:664–666. doi: 10.1038/333664a0. [DOI] [PubMed] [Google Scholar]
- 5.Davis K L, Martin E, Turko I V, Murad F. Annu Rev Pharmacol Toxicol. 2001;41:203–236. doi: 10.1146/annurev.pharmtox.41.1.203. [DOI] [PubMed] [Google Scholar]
- 6.Peunova N, Enikolopov G. Nature. 1995;375:68–73. doi: 10.1038/375068a0. [DOI] [PubMed] [Google Scholar]
- 7.Lu Y F, Kandel E R, Hawkins R D. J Neurosci. 1999;19:10250–10261. doi: 10.1523/JNEUROSCI.19-23-10250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kroncke K D, Fehsel K, Suschek C, Kolb-Bachofen V. Int Immunopharmacol. 2001;1:1407–1420. doi: 10.1016/s1567-5769(01)00087-x. [DOI] [PubMed] [Google Scholar]
- 9.Heneka M T, Feinstein D L. J Neuroimmunol. 2001;114:8–18. doi: 10.1016/s0165-5728(01)00246-6. [DOI] [PubMed] [Google Scholar]
- 10.Estevez A G, Spear N, Manuel S M, Barbeito L, Radi R, Beckman J S. Prog Brain Res. 1998;118:269–280. doi: 10.1016/s0079-6123(08)63214-8. [DOI] [PubMed] [Google Scholar]
- 11.Dawson V L, Dawson T M. Prog Brain Res. 1998;118:215–229. doi: 10.1016/s0079-6123(08)63210-0. [DOI] [PubMed] [Google Scholar]
- 12.Zhao X, Haensel C, Araki E, Ross M E, Iadecola C. Brain Res. 2000;872:215–218. doi: 10.1016/s0006-8993(00)02459-8. [DOI] [PubMed] [Google Scholar]
- 13.Hewett S J, Csernansky C A, Choi D W. Neuron. 1994;13:487–494. doi: 10.1016/0896-6273(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 14.Kuiper M A, Teerlink T, Visser J J, Bergmans P L M, Scheltens P, Wolters E Ch. J Neural Transmem. 2000;107:183–189. doi: 10.1007/s007020050016. [DOI] [PubMed] [Google Scholar]
- 15.Wu G, Morris S M., Jr Biochem J. 1998;336:1–17. doi: 10.1042/bj3360001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiesinger H. Prog Neurobiol. 2001;64:365–391. doi: 10.1016/s0301-0082(00)00056-3. [DOI] [PubMed] [Google Scholar]
- 17.Kurz S, Harrison D G. J Clin Invest. 1997;99:369–370. doi: 10.1172/JCI119166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer G, Kettenmann H. Exp Cell Res. 1985;159:273–279. doi: 10.1016/s0014-4827(85)80001-x. [DOI] [PubMed] [Google Scholar]
- 19.He T C, Zhou S, da Costa L T, Yu J, Kinzler K W, Vogelstein B. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai D, Deng K, Mellado W, Lee J, Ratan R, Filbin M T. Neuron. 2002;35:711–719. doi: 10.1016/s0896-6273(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 21.McDonald K K, Zharikov S, Block E R, Kilberg M S. J Biol Chem. 1997;272:31213–31216. doi: 10.1074/jbc.272.50.31213. [DOI] [PubMed] [Google Scholar]
- 22.Russell A S, Ruegg U T. J Immunol Methods. 1980;32:375–382. doi: 10.1016/0022-1759(80)90029-0. [DOI] [PubMed] [Google Scholar]
- 23.Xia Y, Dawson V L, Dawson T M, Snyder S H, Zweier J L. Proc Natl Acad Sci USA. 1996;93:6770–6774. doi: 10.1073/pnas.93.13.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burgher K L, Heroux J A, Ringheim G E. Neurochem Int. 1997;30:483–489. doi: 10.1016/s0197-0186(96)00085-x. [DOI] [PubMed] [Google Scholar]
- 25.Wang W W, Jenkinson C P, Griscavage J M, Kern R M, Arabolos N S, Byrns R E, Cederbaum S D, Ignarro L J. Biochem Biophys Res Commun. 1995;210:1009–1016. doi: 10.1006/bbrc.1995.1757. [DOI] [PubMed] [Google Scholar]
- 26.Buga G M, Singh R, Pervin S, Rogers N E, Schmitz D A, Jenkinson C P, Cederbaum S D, Ignarro L J. Am J Physiol. 1996;271:H1988–H1998. doi: 10.1152/ajpheart.1996.271.5.H1988. [DOI] [PubMed] [Google Scholar]
- 27.Wei L H, Jacobs A T, Morris S M, Jr, Ignarro L J. Am J Physiol. 2000;279:C248–C256. doi: 10.1152/ajpcell.2000.279.1.C248. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Meininger C J, Hawker J R, Jr, Haynes T E, Kepka-Lenhart D, Mistry S K, Morris S M, Jr, Wu G. Am J Physiol. 2001;280:E75–E82. doi: 10.1152/ajpendo.2001.280.1.E75. [DOI] [PubMed] [Google Scholar]
- 29.Stevens B R, Kakuda D K, Yu K, Waters M, Vo C B, Raizada M K. J Biol Chem. 1996;271:24017–24022. doi: 10.1074/jbc.271.39.24017. [DOI] [PubMed] [Google Scholar]
- 30.Jousse C, Bruhat A, Harding H P, Ferrara M, Ron D, Fafournoux P. FEBS Lett. 1999;448:211–216. doi: 10.1016/s0014-5793(99)00373-7. [DOI] [PubMed] [Google Scholar]
- 31.Lowenstein C J, Alley E W, Raval P, Snowman A M, Snyder S H, Russell S W, Murphy W J. Proc Natl Acad Sci USA. 1993;90:9730–9734. doi: 10.1073/pnas.90.20.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez P C, Zea A H, Culotta K S, Zabaleta J, Ochoa J B, Ochoa A C. J Biol Chem. 2002;277:21123–21129. doi: 10.1074/jbc.M110675200. [DOI] [PubMed] [Google Scholar]
- 33.Sattlegger E, Hinnebusch A G. EMBO J. 2000;19:6622–6633. doi: 10.1093/emboj/19.23.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harding H P, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini D D, Ron D. Mol Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 35.Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman R J. Mol Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 36.Tsikas D, Boger R H, Sandmann J, Bode-Boger S M, Frolich J C. FEBS Lett. 2000;478:1–3. doi: 10.1016/s0014-5793(00)01686-0. [DOI] [PubMed] [Google Scholar]
- 37.Kaufman R J, Davies M V, Pathak V K, Hershey J W. Mol Cell Biol. 1989;9:946–958. doi: 10.1128/mcb.9.3.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pathak V K, Nielsen P J, Traschel H, Hershey J W. Cell. 1988;54:633–639. doi: 10.1016/s0092-8674(88)80007-2. [DOI] [PubMed] [Google Scholar]
- 39.Giugliano D, Marfella R, Verrazzo G, Acampora R, Coppola L, Cozzolino D, D'Onofrio F. J Clin Invest. 1997;99:433–438. doi: 10.1172/JCI119177. [DOI] [PMC free article] [PubMed] [Google Scholar]