Abstract
Diverse members of the G protein-coupled receptor (GPCR) superfamily participate in a variety of physiological functions and are major targets of pharmaceutical drugs. Here we report that the repertoire of GPCRs for endogenous ligands consists of 367 receptors in humans and 392 in mice. Included here are 26 human and 83 mouse GPCRs not previously identified. A direct comparison of GPCRs in the two species reveals an unexpected level of orthology. The evolutionary preservation of these molecules argues against functional redundancy among highly related receptors. Phylogenetic analyses cluster 60% of GPCRs according to ligand preference, allowing prediction of ligand types for dozens of orphan receptors. Expression profiling of 100 GPCRs demonstrates that most are expressed in multiple tissues and that individual tissues express multiple GPCRs. Over 90% of GPCRs are expressed in the brain. Strikingly, however, the profiles of most GPCRs are unique, yielding thousands of tissue- and cell-specific receptor combinations for the modulation of physiological processes.
Mammalian G protein-coupled receptors (GPCRs) constitute a superfamily of diverse proteins with hundreds of members (1). All members have seven transmembrane domains but, on the basis of shared sequence motifs, they are grouped into four classes: A, B, C, and F/S (2).
GPCRs act as receptors for a multitude of different signals. One major group, referred to as chemosensory GPCRs (csGPCRs), are receptors for sensory signals of external origin that are sensed as odors, pheromones, or tastes (3–5). Most other GPCRs respond to endogenous signals, such as peptides, lipids, neurotransmitters, or nucleotides (6, 7). These GPCRs, the subject of this report, are involved in numerous physiological processes, including the regulation of neuronal excitability, metabolism, reproduction, development, hormonal homeostasis, and behavior. Considering that endogenous ligands are required for regulating these processes, in this report, we refer to this group of GPCRs as “endoGPCRs.”
A characteristic feature of endoGPCRs is that they are differentially expressed in many cell types in the body. This feature, together with their structural diversity, has proved important in medicinal chemistry. Of all currently marketed drugs, >30% are modulators of specific endoGPCRs (8). However, only 10% of endoGPCRs are targeted by these drugs, emphasizing the potential of the remaining 90% of the GPCR superfamily for the treatment of human disease.
Despite the importance of GPCRs in physiology and disease, the size of the endoGPCR superfamily is still uncertain. Celera's initial analysis of the human genome found 616 GPCRs, but not distinguish among endoGPCRs and csGPCRs (9), whereas the International Human Genome Sequencing Consortium reported a total of 569 “rhodopsin-like” (i.e., Class A) GPCRs (10), implying the inclusion of odorant receptors but the exclusion of the many endoGPCRs that are not “rhodopsin-like.” Focusing only on intronless genes, Takeda et al. (11) found 178 intronless nonchemosensory GPCRs. In addition, whereas most, if not all, csGPCRs are known to be selectively expressed in specific subsets of cells (11, 12), the expression patterns of most endoGPCRs are incomplete or unknown.
To address these issues, we conducted a comprehensive analysis that defined the endoGPCR superfamilies of two mammalian species, human and mouse.‡ Further analyses revealed evolutionary relationships indicative of a high level of nonredundancy in the endoGPCR superfamily as well as phylogenetic relationships that are predictive of ligands and/or functions for numerous endoGPCRs. Expression profiling showed that individual endoGPCRs are expressed in many tissues and that single tissues and brain regions express an unexpectedly large number of endoGPCRs. However, each tissue expresses a unique combination of endoGPCRs, indicating a combinatorial use of endoGPCRs for the regulation of diverse functions.
Materials and Methods
Database Mining and Phylogenetic Analyses.
The transmembrane domain regions of 254 known GPCRs (13) were each used as a query in tblastn searches of the National Center for Biotechnology Information human genome database; GPCR Class A, B, and C Hidden Markov Model models were also used as queries to search the International Protein Index proteome database (see Fig. 5 and Supporting Materials and Methods, which are published as supporting information on the PNAS web site, www.pnas.org). Neighbor-joining phylogenetic trees were built based on class-specific alignments (hmmalign). Bootstrap consensus trees were plotted by using treeview (14).
Expression Analyses.
RT-PCR/RNA was prepared (Totally RNA kit, Ambion, Austin, TX) from dissected tissues of adult mice and treated extensively with DNase. PCR was performed with cDNA prepared from 2, 20, or 200 ng of RNA in 25-μl reactions for 37 cycles. The alpha ease program (Alpha Innotech, San Leandro, CA) was used to estimate relative expression levels.
In Situ Hybridization.
In situ hybridization was performed as described (15) with minor modifications.
Results
The endoGPCR Repertoire.
To define the full complement of endoGPCRs in human and mouse, we embarked on a multistep process involving the identification of first known and then novel genes. Initially, we searched the available public literature and sequence databases for human and mouse endoGPCRs and then performed sequence comparisons. This identified a unique gene set for each species and defined the human and mouse orthologs. In total, 338 endoGPCRs were identified in human and 301 in mouse. Sequence alignments indicated that 260 of these molecules are common to both species (Fig. 5).
We then asked whether the remaining endoGPCR genes (78 human and 41 in mouse), which did not show a counterpart in the other species, might have undiscovered orthologs. Using the nonshared endoGPCRs as queries, the public human and mouse genome sequence databases were searched for orthologous genes using tblastn (16). These studies identified mouse orthologs for 56 of the human endoGPCRs, but no orthologs could be found for the remaining 22 (Fig. 5). No human orthologs were detected for 38 of the mouse genes. Eight additional mouse genes were discovered using 11 rat trace amine receptor sequences as queries. In total, 35 of these 46 mouse genes belonged to the trace amine receptor (17) and Mas1-related gene families (18, 19). These studies increased the number of endoGPCRs to 341 in human and 365 in mouse, with 319 endoGPCRs shared by the two species (Fig. 5).
We next undertook an exhaustive search for new human endoGPCR genes. Two different approaches were used. In the first, we used a homology-based strategy to search the human genome sequence database for genes encoding endoGPCRs. Two hundred fifty-four known endoGPCRs, representative of all classes, were each used as an independent query in tblastn searches of all human chromosomes. These searches yielded ≈500,000 matches, which were first reduced to ≈50,000 unique matches and then to 10,000 matches with homology to known GPCRs (see Materials and Methods). Among these, hits representing 315 of the 341 known endoGPCR genes were detected, consistent with the 90–95% coverage estimated for the human genome database. Approximately 1,000 hits were homologous to csGPCRs. Continued analysis of the remaining hits revealed 23 endoGPCR genes that were previously unknown.
In a second discovery method, the Hidden Markov Model profile-based approach (20) was used to search the human proteome. This method yielded 1,100 potential matches. Among these hits, 331 of the 341 known endoGPCRs were represented, confirming the validity of the search strategy. After elimination of known genes, three novel genes were identified. The combination of both genomic search strategies revealed 26 endoGPCR genes that were not previously described. These genes are referred to as PGR1 to PGR28 (Fig. 5). Searches of the mouse genome sequence database, together with RT-PCR analyses (see below), identified orthologs of 24 of the 26 genes in the mouse and three additional unique mouse genes.
Altogether, these searches identified a total of 367 endoGPCRs in human and 392 in mouse; 343 of the endoGPCRs were common to the two species.
Phylogeny and Ligand Prediction.
Phylogenetic and receptor–ligand relationships among the endoGPCRs were subsequently analyzed. Each human and mouse endoGPCR was first assigned to one of the four distinct classes of GPCRs (A, B, C, and F/S) on the basis of shared sequence motifs with a prototype of that class. All but five of the receptors (TPRA40, TM7SF1, TM7SF1L1, TM7SF1L2, and TM7SF3) could be assigned to one of the four classes by this method. These assignments indicate that of 367 human endoGPCRs, 284 belong to Class A, 50 to Class B, 17 to Class C, and 11 to Class F/S. Of 392 mouse endoGPCRs, 313, 47, 17, and 10 belong to Classes A, B, C, and F/S, respectively.
The endoGPCRs were next catalogued according to ligand specificities reported in the literature. This effort identified 224 human and 214 mouse endoGPCRs with known ligands. The remaining 143 human and 178 mouse endoGPCRs have no known ligands and are therefore orphan receptors. Among the orphan receptors, 98 human and 136 mouse receptors belong to Class A, 34 human and 31 mouse receptors to Class B, 6 human and 6 mouse receptors to Class C, and none to Class F/S (Fig. 5).
The endoGPCRs were subsequently divided into families of related receptors that either recognize the same/similar ligand(s) or are likely to do so. Sequence comparisons and phylogenetic analyses (see below) showed that endoGPCRs with highly related ligand specificities that are traditionally classified as belonging to the same “family” are at least 40% homologous in protein sequence. We therefore assigned endoGPCRs that showed at least 40% sequence homology to the same family. In this manner, 95 different families of endoGPCRs were identified, including 18 families of orphan receptors that have not been previously described (Fig. 5). These studies assigned 12 of 143 human and 49 of 178 mouse orphan endoGPCRs to seven different families of receptors that interact with known ligands and thus can be predicted to recognize ligands similar to those detected by other family members.
To further investigate sequence–ligand relationships among human endoGPCRs, we conducted a phylogenetic analysis. endoGPCRs were aligned to the class-specific Hidden Markov Model profile model by using hmmalign (21). These alignments were used for the construction of phylogenetic trees by clustal w (22). The phylogenetic trees were then overlaid with information on the ligand specificities of individual receptors, where available.
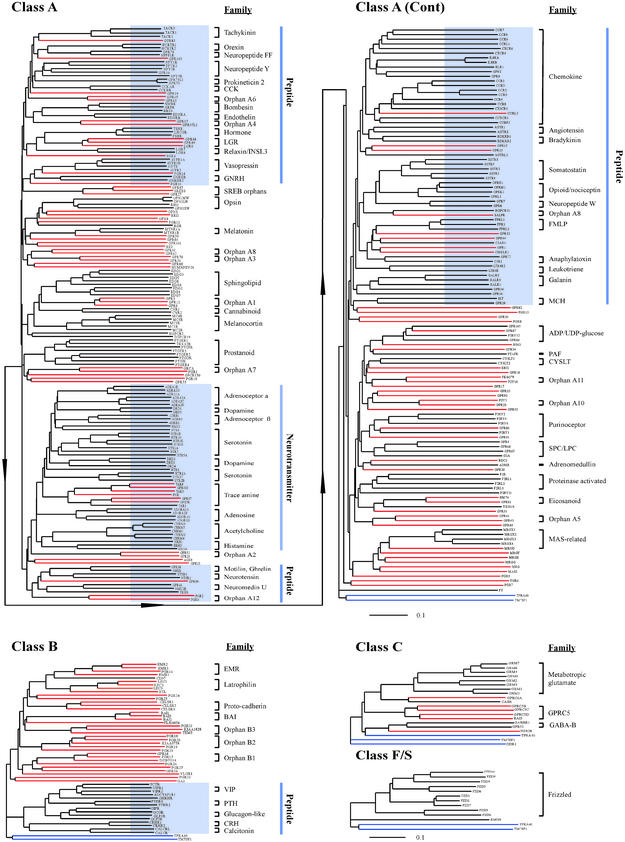
The combined phylogenetic/ligand analyses of human endoGPCRs are shown in Fig. 1. The phylogenetic tree of the Class A receptors, the largest set, was composed of a number of major branches that were progressively subdivided into smaller branches containing increasingly related endoGPCRs. The three smaller classes of receptors (Classes B, C, and F/S) exhibited a similar organization, but fewer branches. endoGPCRs that recognize the same ligand, such as receptors for the neurotransmitter acetylcholine or receptors that belong to the same family, were clustered together in small branches.
Figure 1.
Phylogenetic trees of human endoGPCRs. Lines corresponding to individual proteins are colored black for those with known ligands, red for orphan proteins, and blue for proteins with seven transmembrane domains but no homology to known endoGPCRs. The Class A tree was split in two parts due to size considerations (arrow line indicates the connection). Clusters of endoGPCRs with significant predictive value as to ligands are highlighted in blue on these bootstrap consensus trees (bootstrap values not shown). The ruler at the bottom of each tree indicates the horizontal distance equal to 10% sequence divergence.
The phylogenetic trees revealed a striking higher-order organization relevant to endoGPCR functions. Multiple receptor families with related functions or that recognize ligands of a particular chemical class were grouped in the same large branch. For example, the 40 neurotransmitter/neuromodulator receptors of the dopamine, serotonin, trace amine, adenosine, acetylcholine, histamine, and adrenoreceptor α and β families were all clustered phylogenetically. Moreover, the 106 endoGPCRs known to recognize peptide ligands were clustered in four large branches, three in the Class A tree and one in the Class B tree. This organization is of predictive value for numerous orphan endoGPCRs. For example, endoGPCRs such as PGR2, PGR3, PGR11, GPR19, GPR37, GPR39, GPR45, GPR63, and GPR103 could be predicted to have peptide ligands because they were grouped with other receptors activated by peptides. Other orphan receptors, such as GPR21 and GPR52, could conceivably be activated by amine neuromodulators, because they clustered phylogenetically with the large neurotransmitter branch of the Class A tree.
Combinatorial Expression of endoGPCRs.
To begin a dissection of the functions of individual endoGPCRs, we used RT-PCR to analyze the expression patterns of 100 mouse endoGPCRs randomly distributed throughout the phylogenetic trees in 17 peripheral tissues and 9 different brain regions (Figs. 2 and 3). All tissue samples were normalized according to their 18S rRNA content, and two different amounts of RNA from each tissue (2 and 20 ng) were used to facilitate estimation of relative expression levels. On the basis of observed amplification rates in the PCR reactions and the criterion used for scoring (a minimum of 60 ng of PCR product), it can be estimated that these experiments could detect roughly one mRNA molecule per 15 cells in a given tissue. This was important for analyses of brain regions, which can contain a large variety of cell types. The conditions used reliably reproduced expression profiles of several tissue-specific genes, including those encoding the GPCRs blue opsin (retina only) and MC1R (skin only) (data not shown). To exclude the possibility of false positives due to contaminating DNA, all RNA samples were pretreated at least twice with DNase and then analyzed extensively for DNA contamination (see Materials and Methods). Although we cannot absolutely exclude that some positive signals were derived from DNA, comparisons of RT-PCR results with those obtained by in situ hybridization argue against this possibility (see below).
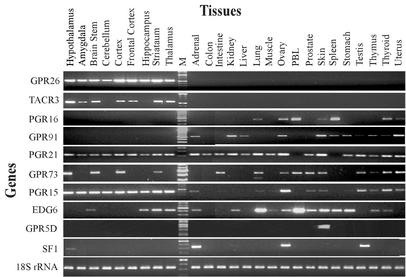
Figure 2.
Expression profiles of nine endoGPCRs by RT-PCR.
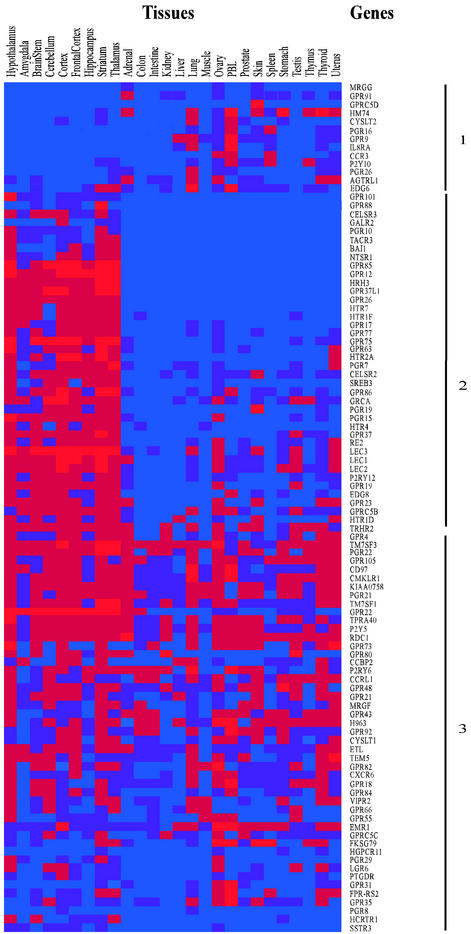
Figure 3.
Summary of tissue expression of 100 endoGPCR genes. Genes were analyzed individually by RT-PCR as shown and the intensity of the observed bands determined by scanning. Each gene is represented by a single row of colored boxes, with four different expression levels: no expression, blue; low expression, purple; moderate expression, dark red; strong expression, pure red. Gene and tissues, as well as groups of expression patterns, are indicated.
Several striking features of endoGPCR expression emerged in these studies (Figs. 2 and 3). These features are illustrated by the RT-PCR results shown in Fig. 2 for nine different endoGPCRs and a control gene, SF1 (23–25). First, most endoGPCRs were expressed in multiple tissues. Second, specific patterns of expression were clearly delineated. For example, GPR26 and TACR3 were exclusively expressed in the brain, whereas GPR91 and PGR16 were expressed solely in peripheral tissues. Four other genes, GPR73, EDG6, PGR15, and PGR21, were expressed in both brain and peripheral tissues. Also shown is GPRC5D, the only endoGPCR found to be expressed in just a single tissue, skin. Note that each endoGPCR has a different expression pattern.
A scattergram of RT-PCR results obtained for 100 different endoGPCRs in 26 mouse tissues is shown in Fig. 3. The most remarkable finding was that 93% of endoGPCRs were detected in the brain, generally in four to five distinct anatomical areas. The largest number of genes was detected in the hypothalamus (82 genes), a brain region of high cellular complexity. Individual peripheral tissues also showed expression of multiple different endoGPCRs, ranging from 18 genes in muscle to 72 genes in ovary. One concern was that blood present in some tissues might be the source of some GPCR cDNAs. This may be the case for lung, ovary, and thyroid, where most of the endoGPCRs detected in peripheral blood leukocytes (PBLs), were also seen. However, most tissues did not show expression of the same endoGPCRs as PBLs, arguing against this possibility.
Although individual endoGPCR genes were generally expressed in numerous tissues, most genes had unique expression profiles. Nonetheless, three groups of endoGPCRs with broadly related profiles were observed. In the first group were genes expressed primarily in peripheral tissues. Seven of these genes were expressed exclusively in peripheral tissues and not in the brain. The second group contained genes expressed primarily in brain. Of these 41 genes, 14 were solely expressed in brain and not in peripheral tissues. In the third group, the genes were broadly expressed in the brain and throughout the periphery.
Complex Patterns of endoGPCRs in the Brain.
The expression of 93% of endoGPCRs in the brain was unexpected. To further investigate endoGPCR expression in the brain, we used in situ hybridization (26). Individual endoGPCR cRNA probes were hybridized to tissue sections spanning the entire brain, except the olfactory bulb. Of the 44 endoGPCRs analyzed, mRNAs encoding 37 (84%) were detected in the brain. The concordance of the two methods, RT-PCR and in situ hybridization, was high. Of the endoGPCR genes expressed in the brain, 32 were also tested by RT-PCR. Comparisons of results obtained for these 32 genes in 9 brain regions (288 comparisons) showed that the RT-PCR results accurately predicted the in situ hybridization results in 94% of cases (270/288). Similarly, results obtained by in situ hybridization were echoed by RT-PCR in 87% of comparisons (251/288).
Fig. 4 presents expression patterns seen for endoGPCRs in the brain that are illustrative, but not totally inclusive, of those observed. One pattern is exemplified by PGR15, which was highly expressed in numerous subregions of the hypothalamus, with much less labeling in the adjacent thalamus or striatum (Fig. 4h). Other endoGPCRs, such as PGR7, were highly expressed in a single nucleus or region, with relatively little signal observed elsewhere (Fig. 4b). In contrast, several orphan receptors were widely distributed throughout the brain, but with highest levels noted in specific regions. For example, GPR63 was robustly expressed both in the pyramidal cells of the hippocampus (Fig. 4a) and in the Purkinje cell layer of the cerebellum (Fig. 4d).
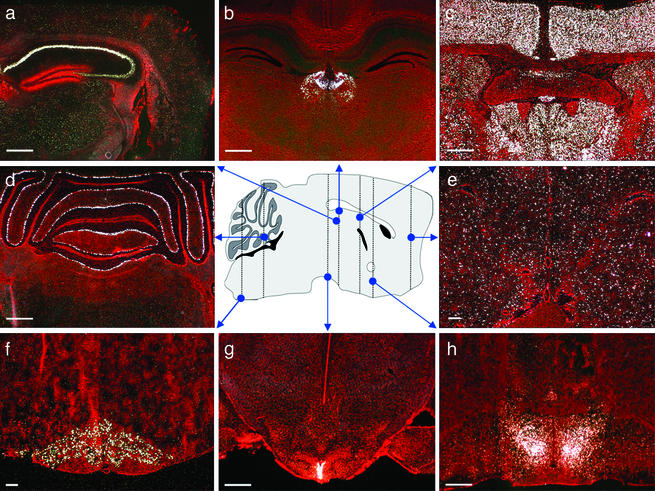
Figure 4.
Representative in situ hybridization photomicrographs of endoGPCR expression in the mouse brain. (a) GPR63 in the Ammons horn (CA) regions of the hippocampus; (b) PGR7 in the habenula; (c) GRCA in the cortex and thalamus; (d) GPR63 in the Purkinje cells of the cerebellum; (e) GPR37 in the frontal cortex; (f) GPR26 in the inferior olive; (g) GPR50 in the cells lining the third ventricle; and (h) PGR15 in the preoptic region of the hypothalamus. Vertical lines on sagittal mouse brain drawing represent approximate coronal plane of photomicrographs. (Bars = 500 μm.)
Other orphan receptors exhibited a nonlocalized profile. For instance, GRCA mRNA was detected in nearly every neuronal region in the entire brain, whereas the white matter regions were conspicuously devoid of GRCA mRNA (Fig. 4c). In contrast, the orphan gene GPR37 was diffusely expressed in scattered cells from the frontal cortex (Fig. 4e) to the medulla, in both white and gray matter, suggesting a glial cell distribution. A surprising number of endoGPCRs were prominently expressed in circumventricular organs, the choroid plexus, and the ependymal cells of the ventricles, areas involved in chemical communication between the brain and periphery. This pattern is exemplified by GPR50, which was found at very high levels in virtually all cells lining the ventral portion of the third ventricle (Fig. 4g).
The in situ hybridization analyses, in addition to confirming the results obtained by RT-PCR for different brain regions, reveal that endoGPCRs are expressed in diverse patterns, further highlighting the involvement of combinations of endoGPCRs in different functions.
Discussion
Here, we conducted a comprehensive analysis of the endoGPCR superfamily of two mammalian species, human and mouse, and profiled the expression patterns of 100 mouse endoGPCR genes. These studies identified a number of unknown endoGPCRs and uncovered phylogenetic relationships of predictive value for ligands of orphan receptors. They also revealed an unexpected complexity of endoGPCR expression patterns that is of high significance for studies of physiological processes and for the use of endoGPCRs as pharmaceutical targets in the treatment of human disease.
These studies identified a total of 367 endoGPCRs in human and 392 in mouse, of which 26 human and 83 mouse genes are reported here, to our knowledge for the first time. Given that there are ≈350 olfactory receptors and 30 other chemosensory receptors in human (27, 28), this places the total number of human GPCRs at 750, exceeding the estimate of 616 GPCRs based on human genome sequence annotation (9). The existence of additional human GPCRs cannot be excluded (29). However, given the near-complete status of the human genome sequence searched here, the exhaustive nature of the search, and the inability to detect additional GPCRs using a different search strategy (30), the 367 endoGPCRs identified are likely to represent the full repertoire.
Of the 367 human and 392 mouse endoGPCRs, 343 are common to the two species. The persistence of the vast majority of endoGPCRs over the 50–60 million years of evolutionary time separating the two species is significant in two regards. First, it suggests that the functions of most endoGPCRs are conserved in human and mouse, a finding important to the use of mouse as a model system for human. Second, it argues against the idea that highly related endoGPCRs with the same ligand have overlapping functions. In sharp contrast, the V1R family of csGPCRs for pheromones has >100 members in mouse, but only three in human (31). Considering the diverse expression patterns of endoGPCRs seen here, the maintenance of multiple receptors for the same ligand through evolution is likely to result from their expression in different cell types and, thereby, their involvement in different functions. A small number of human and mouse endoGPCR genes were found in only one of the two species. It may be that these genes actually exist in both species but are located in small regions of the human or mouse genome that have not yet been completely sequenced. Alternatively, some or all of these endoGPCR genes could have arisen by gene duplication events after divergence from a common ancestor and eventually come to serve a function unique to one species.
The phylogenetic analyses and sequence comparisons conducted here indicate that the human endoGPCR superfamily can be divided into 95 different families. Of 187 orphan endoGPCRs, 51 belong to families with known ligands. These studies also revealed a higher-level phylogenetic organization in which clusters of families with common ligand chemistry, or a shared function, are evident. The 106 endoGPCRs with known peptide/protein ligands are clustered into four large groups; the sole exception within these groups is a pair of leukotriene receptors. Similarly, all 40 known neurotransmitter receptors are members of yet another large cluster. With this degree of accuracy (>90%), one can predict that six of the orphan endoGPCRs are neurotransmitter receptors, and that another 27 are receptors for peptide or protein ligands.
The most surprising finding of these studies is the combinatorial expression of a multitude of different endoGPCRs in different tissues and cell types. In sharp contrast, csGPCR expression is typically restricted to a single tissue (32–34). This feature, along with the evolutionary conservation of endoGPCRs and their ligand source, clearly distinguishes endoGPCRs from csGPCRs. Although the two groups of receptors are structurally related, share sequence motifs, and transmit signals by similar mechanisms, they differ substantially in other aspects. In the case of csGPCRs, the signals are from an exogenous source, the receptors are not well conserved between human and mouse, and the genes are primarily expressed in the sensory organs. On the contrary, for endoGPCRs, the signals are from endogenous sources, the receptors are well conserved between human and mouse, and the genes are widely expressed throughout the body with a high preference for the brain.
On average, each of the 100 endoGPCRs analyzed is expressed in 14 different tissues. Even more remarkably, individual tissues express a large number of different endoGPCRs. Strikingly, however, different endoGPCRs are expressed in diverse combinations of tissues, and most exhibit a unique expression pattern. Moreover, different tissues use different combinations of endoGPCRs, indicating that complex sets of these receptors are involved in numerous physiological processes.
Given the expression of individual endoGPCRs in multiple tissues, one might predict that mice lacking a particular gene would exhibit multiple defects. However, this is not generally the case. In some cases, ablation of an endoGPCR has no apparent effect, whereas in many others, a selective defect is seen in a particular function (35). These findings have prompted suggestions that there is a degree of redundancy built into physiological processes that ensures their functions in the face of genetic polymorphisms and ongoing mutation. In this scenario, loss of an endoGPCR might be deleterious to one function in which it is involved but not to others. This model may also explain the unusual success of endoGPCRs as targets for pharmaceutical intervention in the treatment of diseases.
Supplementary Material
Acknowledgments
We thank Robert Nowinski and the rest of our colleagues at Primal for their suggestions and support during the course of this work. In particular, we mention Linda Madisen, Maria Pavlova, Alex Rohde, Jeanna Strout, and Laura Johnson for contributions in assembling the endoGPCR gene list. All mouse and human GPCR sequences are available at Primal's web site, www.primalinc.com.
Abbreviations
- GPCR
G protein-coupled receptor
- csGPCR
chemosensory GPCR
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY255527–AY255623).
The sequences can be accessed at www.primalinc.com.
References
- 1.Bockaert J, Pin J P. EMBO J. 1999;18:1723–1729. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horn F, Weare J, Beukers M W, Horsch S, Bairoch A, Chen W, Edvardsen O, Campagne F, Vriend G. Nucleic Acids Res. 1998;26:275–279. doi: 10.1093/nar/26.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buck L, Axel R. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 4.Mombaerts P. Science. 1999;286:707–711. doi: 10.1126/science.286.5440.707. [DOI] [PubMed] [Google Scholar]
- 5.Firestein S. Nature. 2000;404:552–553. doi: 10.1038/35007167. [DOI] [PubMed] [Google Scholar]
- 6.Howard A D, McAllister G, Feighner S D, Liu Q, Nargund R P, Van der Ploeg L H, Patchett A A. Trends Pharmacol Sci. 2001;22:132–140. doi: 10.1016/s0165-6147(00)01636-9. [DOI] [PubMed] [Google Scholar]
- 7.Lee D K, George S R, Evans J F, Lynch K R, O'Dowd B F. Curr Opin Pharmacol. 2001;1:31–39. doi: 10.1016/s1471-4892(01)00003-0. [DOI] [PubMed] [Google Scholar]
- 8.Wise A, Gearing K, Rees S. Drug Disc Today. 2002;7:235–246. doi: 10.1016/s1359-6446(01)02131-6. [DOI] [PubMed] [Google Scholar]
- 9.Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 10.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 11.Meneses A. Neurosci Biobehav Rev. 1999;23:1111–1125. doi: 10.1016/s0149-7634(99)00067-6. [DOI] [PubMed] [Google Scholar]
- 12.Csaba Z, Dournaud P. Neuropeptides. 2001;35:1–23. doi: 10.1054/npep.2001.0848. [DOI] [PubMed] [Google Scholar]
- 13.Tusnady G E, Simon I. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 14.Page R D. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 15.Hohmann J G, Jureus A, Teklemichael D N, Matsumoto A M, Clifton D K, Steiner R A. Neuroscience. 2002;117:105–117. doi: 10.1016/s0306-4522(02)00798-4. [DOI] [PubMed] [Google Scholar]
- 16.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 17.Borowsky B, Adham N, Jones K A, Raddatz R, Artymyshyn R, Ogozalek K L, Durkin M M, Lakhlani P P, Bonini J A, Pathirana S, et al. Proc Natl Acad Sci USA. 2001;98:8966–8971. doi: 10.1073/pnas.151105198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong X, Han S, Zylka M J, Simon M I, Anderson D J. Cell. 2001;106:619–632. doi: 10.1016/s0092-8674(01)00483-4. [DOI] [PubMed] [Google Scholar]
- 19.Lembo P M, Grazzini E, Groblewski T, O'Donnell D, Roy M O, Zhang J, Hoffert C, Cao J, Schmidt R, Pelletier M, et al. Nat Neurosci. 2002;5:201–209. doi: 10.1038/nn815. [DOI] [PubMed] [Google Scholar]
- 20.Krogh A, Brown M, Mian I S, Sjolander K, Haussler D. J Mol Biol. 1994;235:1501–1531. doi: 10.1006/jmbi.1994.1104. [DOI] [PubMed] [Google Scholar]
- 21.Eddy S R. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 22.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo X, Ikeda Y, Parker K L. Cell. 1994;77:481–490. doi: 10.1016/0092-8674(94)90211-9. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda Y, Luo X, Abbud R, Nilson J H, Parker K L. Mol Endocrinol. 1995;9:478–486. doi: 10.1210/mend.9.4.7659091. [DOI] [PubMed] [Google Scholar]
- 25.Lala D S, Rice D A, Parker K L. Mol Endocrinol. 1992;6:1249–1258. doi: 10.1210/mend.6.8.1406703. [DOI] [PubMed] [Google Scholar]
- 26.Pardue M L, Gall J G. Proc Natl Acad Sci USA. 1969;64:600–604. doi: 10.1073/pnas.64.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glusman G, Yanai I, Rubin I, Lancet D. Genome Res. 2001;11:685–702. doi: 10.1101/gr.171001. [DOI] [PubMed] [Google Scholar]
- 28.Zozulya S, Echeverri F, Nguyen T. Genome Biol. 2001;2:RESEARCH0018. doi: 10.1186/gb-2001-2-6-research0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foord S M. Curr Opin Pharmacol. 2002;2:561–566. doi: 10.1016/s1471-4892(02)00214-x. [DOI] [PubMed] [Google Scholar]
- 30.Takeda S, Kadowaki S, Haga T, Takaesu H, Mitaku S. FEBS Lett. 2002;520:97–101. doi: 10.1016/s0014-5793(02)02775-8. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez I, Punta K D, Rothman A, Ishii T, Mombaerts P. Nat Neurosci. 2002;5:134–140. doi: 10.1038/nn795. [DOI] [PubMed] [Google Scholar]
- 32.Ressler K J, Sullivan S L, Buck L B. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- 33.Vassar R, Ngai J, Axel R. Cell. 1993;74:309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- 34.Matsunami H, Montmayeur J P, Buck L B. Nature. 2000;404:601–604. doi: 10.1038/35007072. [DOI] [PubMed] [Google Scholar]
- 35.Rohrer D K, Kobilka B K. Physiol Rev. 1998;78:35–52. doi: 10.1152/physrev.1998.78.1.35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.