Abstract
We used a systematic approach to build a network of genes associated with developmental and stress responses in rice by identifying interaction domains for 200 proteins from stressed and developing tissues, by measuring the associated gene expression changes in different tissues exposed to a variety of environmental, biological, and chemical stress treatments, and by localizing the cognate genes to regions of stress-tolerance trait genetic loci. The integrated data set suggests that similar genes respond to environmental cues and stresses, and some may also regulate development. We demonstrate that the data can be used to correctly predict gene function in monocots and dicots. As a result, we have identified five genes that contribute to disease resistance in Arabidopsis.
Plant disease response often mimics certain normal developmental processes. For example, plants respond to fungal gibberellic acid and fusicoccin toxin similarly to the way they respectively respond to plant-produced gibberellin and auxin (1, 2). The same can be said for abiotic stress responses and certain stages of plant development. Leaf cells undergoing dehydration stress express some of the same genes that embryonic cells express during development or seed desiccation (3, 4). Because systematic regulation of gene expression drives developmental processes and stress responses (5, 6), it is likely that there is a broader overlapping set of genes and their cognate proteins involved in such responses.
Only a few reports describe large-scale integrated approaches used to characterize sets of genes responsible for phenotypes (7, 8). Now that a draft of the rice genome is available (9), we can apply similar functional genomics approaches in this system. We have studied protein–protein interactions, measured gene expression under a variety of conditions, localized rice genes to regions within quantitative trait loci (QTL), and evaluated mutant plants. Our goal was to discover sets of genes that may be modulated by stressful and developmental stimuli and to identify cognate proteins governed by associations with other proteins involved in similar stress and developmental responses. With this integrated set of genomics data, we show that we can ascribe function to >200 rice genes and verify our models by evaluating the phenotypes of plants with mutations in these genes.
Materials and Methods
Yeast Two-Hybrid (Y2H).
A Y2H assay performed by Myriad Genetics (Salt Lake City) was used to discover protein–protein interactions (10, 11). Constructs were designed around characteristic protein domains. PCR oligonucleotide primers were made to amplify segments from cDNA libraries, and products were mobilized into protein expression vectors in yeast and fused to the DNA-binding domain of the yeast GAL4 transcription factor (TF). Each nonself-activating segment was screened against two libraries made up of random primed cDNA gene fragments fused to GAL4 TF activation domain DNA. cDNA from one library (input trait) was generated from 6-wk-old untreated leaves, stems, and roots and 3- to 4-wk-old leaves of drought, high-salt, cold, and phytohormone-treated Oryza sativa ssp. japonica cv. Nipponbare, whereas the other cDNA library (output trait) was generated from seeds of different stages (milk, dough, hard dough, and germinating), callus, and panicles late in development (10–20 cm). More than five million bait/prey pairs were tested in reactions with each library. The inserted cDNA in each DNA-binding and activation domain vector that allowed for selective yeast growth was sequenced. The amino acid coordinates of interacting baits and prey can be found in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. The protein sequences (Table 2, which is published as supporting information on the PNAS web site), and the DNA sequences (Table 3, which is published as supporting information on the PNAS web site), were compared by blast (12) to sequences in GenBank. Protein sequences for which blast could not provide a functional annotation were searched with the SAM-T99 software package (13) against a library of 6,296 hidden Markov models representing all protein chains in the Protein Data Bank (14) to find remote homologs at the Structural Classification of Proteins superfamily level (15, 16). Interactions were confirmed by transforming paired constructs into naïve yeast cells and performing liquid culture β-galactosidase assays.
Gene Expression.
We compared the gene sequences of the baits and preys with the gene fragments represented on our GeneChip Rice Genome Array (Affymetrix, Santa Clara, CA) and experimentally determined expression as previously described (17, 18). The rice genome array contains 25-mer oligonucleotide probes with sequences corresponding to the 3′ ends of 21,000 predicted ORFs found in ≈42,000 contigs that make up the rice genome map (9, 17). Sixteen different probes were used to measure the expression level of each gene (the probe sets can be found at http://tmri.org/gene_exp_web/). An expression value was derived from the 72nd percentile of the ascending ordered intensity values for the probes. The fifth percentile was used as background. The expression level was calculated from the expression value minus the noise background associated for each probe set. Any expression level below background was floored to zero. Each chip was globally scaled by setting the average intensity of all probe sets to 100. Experiments included evaluating the differential gene expression from various plant tissues comprising seed (2 days postanthesis), 10-wk-old root, leaf and stem, panicle (4–7 cm), and pollen (mixed age). We also measured gene expression in plants exposed to environmental cold (14°C), osmotic pressure (media supplemented with 260 mM mannitol), drought (media supplemented with 25% polyethylene glycol 8000), salt (media supplemented with 150 mM NaCl) and abscisic acid (ABA)-inducible stresses (media supplemented to 50 μM) as described (6), infection by the fungal pathogen Magnaporthe grisea (rice blast), and supplemented media treatment with plant hormones jasmonate (JA, 100 μM), gibberellin (GA3, 50 μM), brassinolide (BL2, 10 μM), cytokinin (BAP, 10 μM), and auxin (2,4D, 10 μM) (Table 4, which is published as supporting information on the PNAS web site). Plants that were infected or from which untreated tissues were collected were grown in a greenhouse at 85°C, 65% relative humidity, and 12–14 h daylight. Environmentally and chemically conditioned plants were grown in sand for 6 weeks in a growth chamber at 28°C, 50% relative humidity, light intensity of 300 μE (1 E = 1 mol of photons), and 12-h light/dark cycles. The plants were fertilized three times per week with one-half strength Hoagland Solution (Sigma) containing 25 μM KH2PO4.
Genes Localized to QTLs.
Markers identifying the drought tolerance (osmotic adjustment) QTLs oa3.1 (between RZ313 and RG369) and oa8.1 (between RG978 and RG1) on rice chromosomes 3 and 8 (19), respectively, were searched for in the rice genome sequence contigs (9) and were used to align sequence contigs to chromosomal physical maps. Genes discovered in the two-hybrid analysis were compared by blast to genes identified in each QTL interval.
Phenotype Analysis.
Arabidopsis thaliana with T-DNA insertions in genes At1g63220 (line SAIL_320_D02), At2g36950 (line SAIL_779_E11), At1g02130 (line SAIL_680_D03), At5g05010 (SAIL_84_C10), and At4g22240 (SAIL_691_B11) were identified from a random insertion seed library (20). DNA regions surrounding the insertions were sequenced. Plants were self-pollinated, and plants homozygous for the T-DNA insertion were identified by PCR (20). Homozygous mutants, pad4–1 disease-susceptible mutants (21), and wild-type Columbia were challenged with Pseudomonas syringae pv. maculicola ES4326 at a concentration of 103 colony-forming units per leaf cm2, and plants were assayed for bacterial titer 3 days postinoculation (22). Data are reported as means and standard deviations of the log of colony-forming units per leaf cm2 for at least six replicates. The experiment was repeated, and similar results were obtained.
Results and Discussion
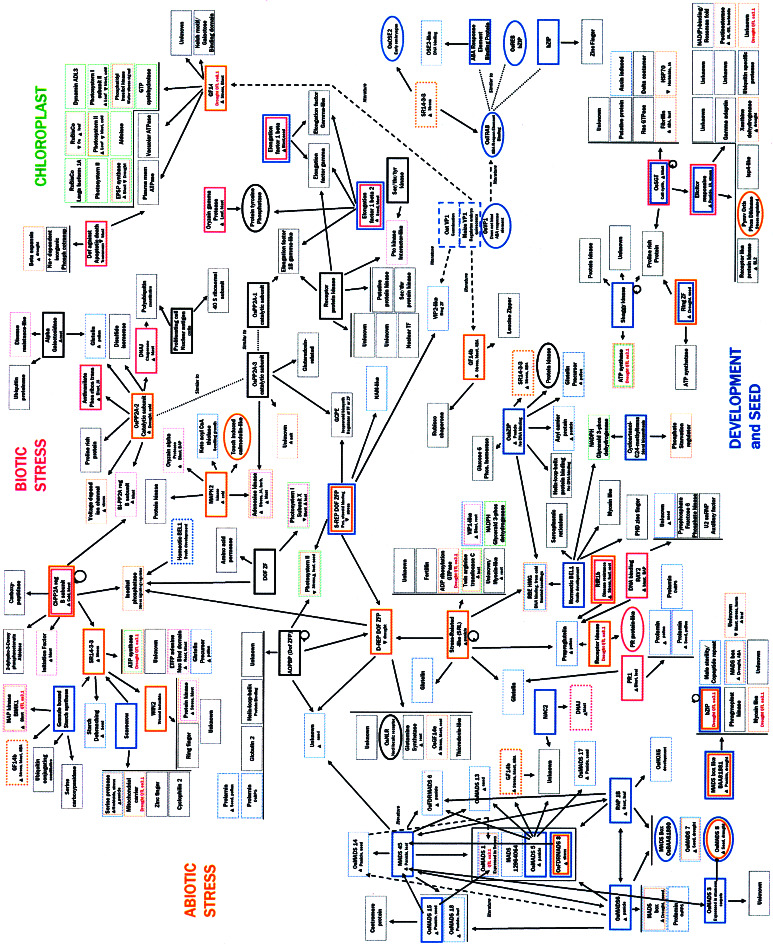
We performed a suite of genomics experiments and assembled the data to ascribe function to >200 rice genes. Experiments included determining protein–protein interactions by using Y2H, measuring gene expression data by using oligonucleotide arrays, localizing rice genes within QTLs, characterizing mutant plants, and performing bioinformatic analyses. Experimental data are published as supporting information on the PNAS web site and consist of protein domains for Y2H bait and interacting prey peptides; amino acid and DNA sequences for proteins; gene expression levels for 127 proteins in five different plant tissues, in leaves treated with the fungal pathogen M. grisea measured at 48 h postinoculation, in plants treated with five phytohormones sampled at 3 h postapplication (hpa), and in plants subjected to five stresses at 3, 27, and 75 hpa. Fig. 3, which is published as supporting information on the PNAS web site, is a key to Tables 1–4. For brevity, this report describes only some of these data (Fig. 1).
Figure 1.
Protein–protein interactions detected among rice proteins. Arrows indicate interaction direction between DNA-binding domain fused proteins (thick lined boxes or ovals) and activation domain fused proteins. Dotted boxes indicate that the protein occurs at more than one place on the map. Ovals rather than boxes indicate that a protein fused to the DNA-binding domain did not interact with other proteins. Circular arrows indicate self interactions. Dashed arrows indicate interactions described in published literature but not found in our study. Dotted lines refer to amino acid similarity between proteins. QTL and gene expression data, where available, are noted in each box. Box colors denote functional classification: blue, development; rose, biotic stress; orange, abiotic stress; green, chloroplast; black, undefined role.
Protein–Phosphatase Interactions.
Our quest to find a set of stress-response genes began with protein phosphatases of type 2A (PP2A) that participate in signal transduction cascades necessary for cell cycling, development, and disease resistance. PP2A specificity is due to the combination of different regulatory and catalytic subunits and differential expression and splicing of transcripts (23). One such PP2A regulatory B subunit is annotated in GenBank (GenBank accession no. embCAA90866) as a chilling-inducible protein (CI-PP2AregB). Our gene expression data confirmed that this gene is induced after cold treatment. We suspected that proteins that interact with CI-PP2AregB also are involved in stress-related responses. Y2H analysis revealed that CI-PP2AregB interacted with itself, a PP2A regulatory B subunit whose gene expression is induced by rice blast fungal infection (BI-PP2AregB), a carboxypeptidase (GenBank accession no. gb AAG46136), an ortholog of a wheat translation initiation factor, an inositol phosphatase-like protein (IPP), and a stress-regulated 14-3-3 protein (GenBank accession no. gb AAK38492, designated SR14-3-3) (Fig. 1). These associated proteins appear to be involved in stress responses. For example, initiation factors are regulated by phosphorylation under stressful situations and are implicated in degradation of nonsense mRNA that may form under stress conditions (24, 25), whereas a carboxypeptidase can regulate brassinosteroid signaling (26). IPP can be a negative regulator of ABA and stress signaling (27). This protein interacted with a drought-repressible zinc-finger protein (D-REP ZFP), thereby extending the interaction network.
In addition to the regulatory subunit, three catalytic subunits of PP2A were examined. The first, OsPP2A-2 (GenBank accession no. gi 11134218), interacted with BI-PP2AregB, as well as with a stress-induced voltage-dependent ion channel, a homolog of a maize DNAJ chaperone with a stress-related role, and a putative anthranilate phosphoribosyl transferase that is induced by rice blast. Similar genes are induced by viruses as part of resistance responses in other plants (28–30). The second catalytic subunit, OsPP2A-3 (GenBank accession no. gi 11134222), interacted with an unknown protein whose gene expression is induced under saline conditions, a homolog to a maize adenosine kinase whose gene expression is induced under a wide variety of stressful conditions, and a GcpE-like protein. The adenosine kinase, along with a protein with 54% similarity to a touch-induced calmodulin from A. thaliana, was found to interact with mitogen-activated protein kinase 2 (GenBank accession no. gb AAF61238), which is chilling-induced and may therefore participate in signaling events that are stress related. The third catalytic subunit OsPP2A-1 (gi 11134285) was shown to be part of a protein interaction network that included five translation elongation factors (EF; GenBank accession nos. gi 13626515, 3894214, and 6166140, and two previously undescribed EFs), a putative protein tyrosine phosphatase, and four putative protein kinases. This network of interacting proteins is likely to be stress regulated; translation is tightly controlled by phosphorylation and is regulated under stress conditions to prevent production of peptides from aberrant transcripts (31).
On the basis of these interactions, associated gene expression patterns, and similarities to proteins involved in stress responses, we hypothesize that many proteins that coordinate abiotic and biotic stress responses interact with protein phosphatases. BI-PP2AregB may be involved in both biotic and abiotic stress response pathways. We speculate that the two pathways are linked through protein phosphatases and regulated by dephosphorylation. These results support the notion that combined gene expression and protein interaction analyses can identify a network of genes involved in stress responses.
Transcription Factors.
Similar experimental approaches were used to study TFs to build a network of genes involved in development. MADS box TFs exist across kingdoms, and the plant homologs including APETELA and AGAMOUS have functions in meristem, flower, and seed development. Rice homologs such as OsMADS45 (GenBank accession no. gi 7446519) act similarly (32) and possibly control development by interaction with other MADS box TFs (33). OsMADS45 interacted with OsMADS6 (GenBank accession no. gi 7446517) and RAP1B (GenBank accession no. gi 7592642), which all cointeracted, and all three proteins interacted with a set of four other MADS box TFs. Our studies confirmed known interactions between OsMADS6 and OsMADS1, 5, 7, 8, 15, and 18 (33) and revealed additional interactions between OsMADS6 and MADS45, RAP1B, MADS 12964064, OsFDRMADS8, OsBAA81880, and a previously uncharacterized MADS box protein. All interacting MADS box peptides contained the protein-binding K domain (33). OsMADS5, 6, 7, 8, 13, 14, 15, 17, 45, and OsFDRMADS8 were expressed in seed tissue and/or panicles, consistent with more precise in situ tissue-specific expression data (34), whereas some appeared to be drought-induced. RAP1B was highly expressed in roots and leaves. Given its apparent tissue-general expression levels and the interaction between RAP1B and a HOX-like protein, which is similar to HOX TFs that are important for development in mammals, it is plausible that RAP1B is a general regulator of other MADS box TFs that may have more specific developmental functions.
MADS box TFs may act in conjunction with other unrelated TFs for regulating development. For example, OsMADS45 interacted with an undefined protein whose gene expression is induced in seeds. This undefined protein interacted with two ZFPs with a defined promoter-binding element (35) and with DOF domains resembling the Cys-2/Cys-2 zinc finger DNA-binding domains of steroid receptors (36). The first DOF ZFP interacted with a protein with a basic helix-loop-helix domain shared across kingdoms by a class of TFs responsible for developmental transcription (37), supporting the notion of an extended TF interaction network for developmental control. However, the interaction with the second similar D-REP DOF ZFP suggests an overlap between sets of genes involved in seed development and stress response. D-REP DOF ZFP is drought-repressible and appears to have a role in stress responses because of its interactions with a number of proteins, including the potential stress signal regulator inositol phosphatase-like protein that was shown to interact with CI-PP2AregB, a stress-repressible (S-REP) DOF ZFP, and a Stress-Related-like (SRL) protein, which is homologous to a protein found in frost-treated grape leaves.
The interactions between D-REP DOF ZFP, the SRL protein, and its interactors further support a hypothetical connection between proteins involved in stress responses and development. The SRL protein, whose gene expression is elevated in panicles, interacted with a high-mobility group protein (HMG) that accumulates in cold-treated rice seedlings (GenBank accession no. gb AAC78104 data). The HMG contains a DNA-binding domain and is part of a family of proteins that can enhance the binding of monocot TFs to transcriptional elements (38). HMG interacted with the homeotic BEL1-like protein (GenBank accession no. gb AAL58126) that may coordinate ovule and flower meristem development and OsbZIP (GenBank accession no. gb AAK01315), a basic/leucine zipper protein thought to have a role in plant development (39) whose gene expression was high in panicles. OsbZIP lacks a DNA-binding domain and possibly utilizes the DNA-binding domain of the HMG to affect gene transcription. A helix-loop-helix protein (GenBank accession no. gb AAK55467) may assist in protein binding. The interactions between SRL, HMG, and OsbZIP may mediate some coordinated transcriptional events for seed development and stress responses.
The interaction and transcriptional data for S-REP DOF ZFP (GenBank accession no. gi 4996640) and D-REP DOF ZFP also suggest that these proteins have dual functions in stress and development responses. GenBank annotation states the S-REP DOF ZFP transcript is present in germinated aleurone cells. S-REP DOF ZFP interacted with NAM-like (GenBank accession no. gi 18396807) and VIP2-like (GenBank accession no. gb AAK70903) TFs with possible meristematic or embryo dormancy roles (40, 41). It also interacted with a GcpE-like protein, which may be involved in isoprenoid biosynthesis and possibly produces a molecular activator of S-REP DOF ZFP (42). Whether separate molecular activators are necessary to control S-REP DOF ZFP during developmental or stressful stimuli remains to be determined.
14-3-3 Proteins.
Another class of proteins that may play dual roles in stress responses and development are 14-3-3 proteins that affect various biological processes through protein interactions. For example, 14-3-3 proteins, like rice GF14c, have been shown to be targeted to thylakoid, plasma, and vacuolar membranes and associated ATPase synthase complexes and to be involved in stress responses (2, 30, 43). Our analysis localizes stress and rice blast-inducible GF14c within the interval of drought tolerance QTL oa8.1 (19), and our interaction studies associated it with at least 10 proteins known to accumulate in the thylakoid. It remains to be seen whether GF14c or any other gene that localizes to similar QTL intervals is responsible for a specific drought tolerance trait. However, several genes encoding chloroplast proteins that interact with GF14c are also stress regulated, supporting the notion for a coordinated stress tolerance mechanism in chloroplast membranes.
Despite their membrane associations, 14-3-3 proteins interact with TFs involved in both stress and development responses. Rice 14-3-3 homologs GF14c (GenBank accession no. gi 7435022) and GF14b (GenBank accession no. gi 7435021) have been shown to interact with maize VP1 (44). We did not find similar interactions with a rice VP1 ortholog, but we did find other 14-3-3 protein/TF interactions. GF14b interacted with a leucine zipper protein (GenBank accession no. gb AAD37699) and OsMADS5 (GenBank accession no. gi 7446542). GF14b may also interact with a NAC2-like protein through an undefined intermediate (GenBank accession no. gi 14209582). GF14e interacted with D-REP DOF ZF, whereas SR14-3-3 interacted with a SCARECROW-like protein, OsbZIP, OsTRAB (GenBank accession no. gi 5821255), and OsOSE2 (GenBank accession no. gb AAF65459), TFs responsible for embryogenesis and other forms of development (45). It is possible that 14-3-3 proteins function by potentiating protein–protein interactions among TFs or by compartmentalizing TFs (46).
As signal regulators, 14-3-3 proteins can modulate nitrate reductase under limiting CO2 and nitrogen conditions and serve as targets for fungal toxin that results in the modulation of ATPase synthase and accelerated plasma membrane expansion (2, 47). Our results suggest similar stress signal regulation roles. SR14-3-3 interacted with an ATPase synthase that localizes to the region of drought tolerance QTL oa3.1 (19). SR14-3-3 interacted with CI-PP2AregB and a rice homolog to a wheat wound-inducible WIN2 protein. It is likely that CI-PP2AregB can regulate SR14-3-3, which, depending on its phosphorylation state or interaction with fungal toxins, regulates ATPase activity and controls membrane stability during stress. GF14e may have stress-related roles in its interaction with D-REP DOF ZFP and inositol phosphatase-like protein. The transcripts for GF14b, GF14e, and SR14-3-3 are all induced by a wide variety of stresses including ABA treatment, supporting the notion that 14-3-3 proteins act in stress response regulation. Some of the TFs that interacted with 14-3-3 proteins are ABA response element-binding proteins or DOF ZFPs that may respond to signals during development. Through a granule-bound starch synthase (GenBank accession no. gb AAC61675), GF14b and SR14-3-3 may interact with mitogen-activated protein (MAP) kinase BIMK1 (GenBank accession no. gb AAK01710). BIMK1 was induced by rice blast infection, and it lies within the interval for drought tolerance QTL oa3.1 (19). GenBank annotation states that BIMK1 participates in systemic acquired disease resistance, so these interactions may be important for this signaling process.
Disease Resistance Proteins.
Several other proteins associated with disease resistance may have roles in development. The rice homolog to PR1 (GenBank accession no. gi 7442184) was induced by rice blast and interacted with several different seed and pollen prolamins and glutelin (GenBank accession nos. gi 130946, 4126695, 4126691, and 82473). Two of the prolamins were also found to associate with OsMADS6 and a DOF ZFP. PR genes are induced by pathogens in a wide variety of plants; however, PR1 mRNA has also been found in tobacco stamens, which suggests that PR1 may have roles not directly related to disease resistance (48). The function of PR1 remains elusive, but these interactions with seed storage proteins portend a chaperone activity or an ability to provide osmotic stability under stress. Rir1b (GenBank accession no. gi 7489592) is a pathogen-inducible gene that can confer resistance to rice blast (49). It was induced by a variety of stresses but repressed by ABA. Rir1b interacted with a glutelin precursor (GenBank accession no. gi 121477) that also interacted with the SRL protein and a receptor kinase-like protein that localizes within the drought tolerance QTL oa8.1 (19). We hypothesize that globulins as well as PR1 homologs could serve as stabilizers for proteins involved in stress responses.
Plant homologs of the yeast SGT1 are required for resistance to several pathogens. AtSGT1b displays dual roles in development and disease resistance by complementing G1 and G2 arrest in yeast and by participating in RAR1-mediated R-gene defense responses (50, 51). In plants, SGT1 homologs likely bridge the two pathways through their activity in ubiquitin-dependent protein degradation and their interactions with RAR1, ubiquitin ligase complexes, COP signalosomes, and kinetichore subunits (50, 51). The rice homolog, OsSGT1, (GenBank accession no. gb AAF18438) may also participate in pathogen defense and development. Indeed, our studies indicate that OsSGT1 is inducible by rice blast infection. OsSGT1 interacted with several undefined and known proteins, including a fibrillin-like protein, a RAS GTPase-like protein (GenBank accession no. gi 730510), a δ coatamer (GenBank accession no. gi 2506139), and an elicitor-response protein (OsERP; GenBank accession no. gb AF090698) whose transcript is induced on treatment with a rice rice blast fungal elicitor according to GenBank and induced by stress in our hands. As a bait, OsERP interacted with other undefined proteins and a ubiquitin protease-related protein, implicating OsERP in SGT1-mediated protein degradation. Gene expression results reveal that OsERP is active in panicle as well as stressed tissues and thus may have a role in development. On the basis of the interactions and expression patterns, we suspect this set of rice proteins has roles in stress and disease resistance.
Verification of Predicted Functions.
At one level, the data presented in Table 1 state which bait peptides interacted with which prey peptides. However, more detailed information can be deduced. For example, two bait peptides comprising amino acids 1–227 and 1–150 for EF-1B2 interacted with EF-1γ prey peptides comprising amino acids 26–154, 54–270, and 26–288. This implies that the important domains for the interaction between EF-1B2 and EF-1γ are the 1–150 and 54–154 amino acid regions, respectively. These data also imply which regions are less important for interactions between proteins. These data could provide a basis for further research. Nevertheless, the redundancy of certain interactions suggests that they are real. For example, the bait for CI-PP2AregB (amino acids 100–250) interacted with two different-sized peptides of BI-PP2AregB. More than 30 interaction pairs were detected more than once, and we take this as a confirmation of these findings.
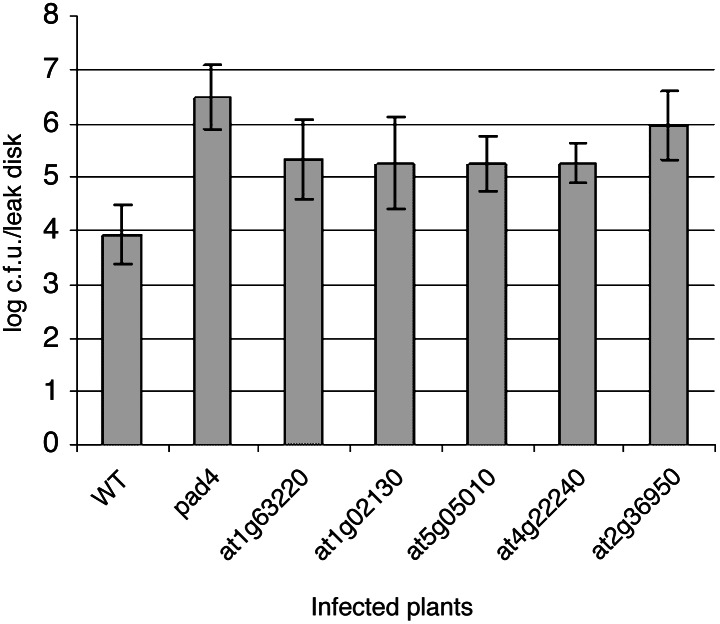
Alternative methods that detect protein interactions or gene expression could be used to support our results, but this may not be necessary to support gene function models. Instead, a combination of different genomics datasets, such as the one presented here, can be used to predict gene function in an organism (8) or across species (52). Thus, this genomics dataset may be useful for predicting gene function in rice or other plants. For example, we suspected that plants deficient in proteins that are potential interactors with AtSGT1 may have impaired disease defense responses. To test this hypothesis, we identified A. thaliana proteins homologous to proteins that interacted with OsSGT1. A. thaliana homologs to OsERP (At1g63220), the RAS GTPase-like protein (At1g02130), the fibrillin-like protein (At4g22240), and the δ coatamer (At5g05010) shared 75%, 90%, 79%, and 77% amino acid sequence similarity, respectively, with their rice counterparts. An undefined protein interacting with OsERP was also chosen for study and an A. thaliana homolog (At2g36950) with 52% amino acid sequence similarity identified. Next, we isolated A. thaliana mutants homozygous for insertion mutations in these genes (20). Sequencing the insertion sites revealed that the T-DNA insertion elements were located within exon 5 (1,362 nt) of At1g63220, exon 3 (1,508 nt) of At2g36950, exon 1 (504 nt) of At4g22240, and in the promoters of At1g02130 and At5g05010 (−992 and −191 nt). These plants were then challenged with P. syringae and assayed for disease susceptibility. By 3 days after inoculation, the mutant plants accumulated more than 10 times as many bacteria as wild-type plants (Fig. 2). Some mutants were nearly as susceptible as pad4-1, a mutant with extreme disease susceptibility and compromised in key defense signaling responses (53). Hence, these genes contribute to disease resistance in A. thaliana and may be associated with SGT1-mediated activation of defense responses. Other than increased susceptibility, none of the mutants had obvious developmental or morphological abnormalities.
Figure 2.
Titer of P. syringae in A. thaliana leaves 3 days postinoculation. Error bars are standard deviation.
In plants, homologs to RAS GTPases, δ coatamers, and fibrillins may respectively be involved in signaling during development (54), membrane or vesicle transport (55), and stabilizing membranes during osmotic stress (56). Obviously, each one of these functions could be necessary for contributing to disease resistance. More studies, however, will have to be performed to show any relationship to SGT1 in Arabidopsis. Notwithstanding, our systems approach to biology quickly revealed that these five uncharacterized proteins, including the homolog of the unknown protein that interacted with OsERP, play roles in disease resistance. Had we based our conclusions on our gene expression data alone, we may have incorrectly concluded that OsERP was not responsible for disease resistance or associated with OsSGT. We believe that combined datasets are more reliable for predicting gene functions in monocots and dicots, and that these predictions can be verified by examining phenotypes of mutants.
Supplementary Material
Acknowledgments
We thank Kim Campbell at Torrey Mesa Research Institute (TMRI) for growing plants; Todd Moughamer, Chris Martin, Tong Zhu, Xun Wang, Darrel Ricke, and Sherman Chang at TMRI and Nan Zhou at Syngenta Biotechnology, Incorporated, for contributions to the generation of gene expression data; and Terrece Pearman, Brad Swedlund, and Karen Heichman at Myriad Genetics for contributions to the Y2H project.
Abbreviations
- QTL
quantitative trait loci
- Y2H
yeast two-hybrid
- TF
transcription factor
- PP2A
protein phosphatase type 2A
- CI-PP2AregB
chilling-inducible PP2A regulatory B subunit
- SR14-3-3
stress-related 14-3-3 protein
- ZFP
zinc-finger protein
- D-REP
drought-repressible
- BI-PP2AregB
blast-inducible protein phosphatase type 2A regulatory B subunit
- EF
translation elongation factors
- S-REP
stress-repressible
- SRL
Stress-Related-like protein
- HMG
high-mobility group protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in GenBank database (accession nos. AY224421–224510).
References
- 1.Hedden P, Kamiya Y. Annu Rev Plant Physiol Plant Mol Biol. 1977;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- 2.Baunsgaard L, Fuglsang A T, Jahn T, Korthout H A, de Boer A H, Palmgren M G. Plant J. 1998;13:661–671. doi: 10.1046/j.1365-313x.1998.00083.x. [DOI] [PubMed] [Google Scholar]
- 3.Medina J, Catala R, Salinas J. Plant Physiol. 2001;125:1655–1666. doi: 10.1104/pp.125.4.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sivamani E, Bahieldin A, Wraith J M, Al-Niemi T, Dyer W E, Ho T D, Qu R. Plant Sci. 2000;155:1–9. doi: 10.1016/s0168-9452(99)00247-2. [DOI] [PubMed] [Google Scholar]
- 5.Harmer S L, Hogenesch J B, Straume M, Chang H S, Han B, Zhu T, Wang X, Kreps J A, Kay S A. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 6.Chen W, Provart N J, Glazebrook J, Katagiri F, Chang H S, Eulgem T, Mauch F, Luan S, Zou G, Whitham S A, et al. Plant Cell. 2002;14:559–574. doi: 10.1105/tpc.010410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ideker T, Thorsson V, Ranish J A, Christmas R, Buhler J, Eng J K, Bumgarner R, Goodlett D R, Aebersold R, Hood L. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 8.Boulton S J, Gartner A, Reboul J, Vaglio P, Dyson N, Hill D E, Vidal M. Science. 2002;295:127–131. doi: 10.1126/science.1065986. [DOI] [PubMed] [Google Scholar]
- 9.Goff S A, Ricke D, Lan T H, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 10.Garrus J E, von Schwedler U K, Pornillos O W, Morham S G, Zavitz K H, Wang H E, Wettstein D A, Stray K M, Cote M, Rich R L, et al. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 11.Bartel P L, Fields S. The Yeast Two Hybrid Screen. New York: Oxford Univ. Press; 1997. [Google Scholar]
- 12.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karplus K, Barrett C, Hughey R. Bioinformatics. 1998;14:846–856. doi: 10.1093/bioinformatics/14.10.846. [DOI] [PubMed] [Google Scholar]
- 14.Berman H M, Westbrook J, Feng Z, Gilliland G, Bhat T N, Weissig H, Shindyalov I N, Bourne P E. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murzin A G, Brenner S E, Hubbard T, Chothia C. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 16.Lo Conte L, Brenner S E, Hubbard T J, Chothia C, Murzin A G. Nucleic Acids Res. 2002;30:264–267. doi: 10.1093/nar/30.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu T, Budworth P, Chen W, Provart N, Chang H S, Guimil S, Su W, Estes B, Zou G, Wang X. Plant Biotechnol J. 2003;1:59–70. doi: 10.1046/j.1467-7652.2003.00006.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhu T, Budworth P, Han B, Brown B, Chang H S, Zou G, Wang X. Plant Physiol Biochem. 2001;39:221–242. [Google Scholar]
- 19.Zhang J, Zheng H G, Aarti A, Pantuwan G, Nguyen T T, Tripathy J N, Sarial A K, Robin S, Babu R C, Nguyen B D, et al. Theor Appl Genet. 2001;103:19–29. [Google Scholar]
- 20.Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jirage D, Tootle T L, Reuber T L, Frost L N, Feys B J, Parker J E, Ausubel F M, Glazebrook J. Proc Natl Acad Sci USA. 1999;96:13583–13588. doi: 10.1073/pnas.96.23.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glazebrook J, Rogers E E, Ausubel F M. Genetics. 1996;143:973–982. doi: 10.1093/genetics/143.2.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janssens V, Goris J. Biochem J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez J, Yaman I, Merrick W C, Koromilas A, Wek R C, Sood R, Hensold J, Hatzoglou M. J Biol Chem. 2002;277:2050–2058. doi: 10.1074/jbc.M109199200. [DOI] [PubMed] [Google Scholar]
- 25.Ishigaki Y, Li X, Serin G, Maquat L E. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Lease K A, Tax F E, Walker J C. Proc Natl Acad Sci USA. 2001;98:5916–5921. doi: 10.1073/pnas.091065998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong L, Lee B, Ishitani M, Lee H, Zhang C, Zhu J K. Genes Dev. 2001;15:1971–1984. doi: 10.1101/gad.891901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clough S J, Fengler K A, Yu I C, Lippok B, Smith R K, Jr, Bent A F. Proc Natl Acad Sci USA. 2000;97:9323–9328. doi: 10.1073/pnas.150005697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitham S A, Anderberg R J, Chisholm S T, Carrington J C. Plant Cell. 2000;12:569–582. doi: 10.1105/tpc.12.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper B. Plant J. 2001;26:339–349. doi: 10.1046/j.1365-313x.2001.01030.x. [DOI] [PubMed] [Google Scholar]
- 31.Smailov S K, Lee A V, Iskakov B K. FEBS Lett. 1993;321:219–223. doi: 10.1016/0014-5793(93)80112-8. [DOI] [PubMed] [Google Scholar]
- 32.Greco R, Stagi L, Colombo L, Angenent G C, Sari-Gorla M, Pe M E. Mol Gen Genet. 1997;253:615–623. doi: 10.1007/s004380050364. [DOI] [PubMed] [Google Scholar]
- 33.Moon Y H, Kang H G, Jung J Y, Jeon J S, Sung S K, An G. Plant Physiol. 1999;120:1193–1204. doi: 10.1104/pp.120.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kyozuka J, Kobayashi T, Morita M, Shimamoto K. Plant Cell Physiol. 2000;41:710–718. doi: 10.1093/pcp/41.6.710. [DOI] [PubMed] [Google Scholar]
- 35.Kisu Y, Ono T, Shimofurutani N, Suzuki M, Esaka M. Plant Cell Physiol. 1998;39:1054–1064. doi: 10.1093/oxfordjournals.pcp.a029302. [DOI] [PubMed] [Google Scholar]
- 36.Shimofurutani N, Kisu Y, Suzuki M, Esaka M. FEBS Lett. 1998;430:251–256. doi: 10.1016/s0014-5793(98)00670-x. [DOI] [PubMed] [Google Scholar]
- 37.Ledent V, Vervoort M. Genome Res. 2001;11:754–770. doi: 10.1101/gr.177001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez-Garcia J F, Quail P H. Plant J. 1999;18:173–183. doi: 10.1046/j.1365-313x.1999.00440.x. [DOI] [PubMed] [Google Scholar]
- 39.Izawa T, Foster R, Nakajima M, Shimamoto K, Chua N H. Plant Cell. 1994;6:1277–1287. doi: 10.1105/tpc.6.9.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Souer E, van Houwelingen A, Kloos D, Mol J, Koes R. Cell. 1996;85:159–170. doi: 10.1016/s0092-8674(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 41.Jones H D, Kurup S, Peters N C, Holdsworth M J. Plant J. 2000;21:133–142. doi: 10.1046/j.1365-313x.2000.00662.x. [DOI] [PubMed] [Google Scholar]
- 42.Campos N, Rodriguez-Concepcion M, Seemann M, Rohmer M, Boronat A. FEBS Lett. 2001;488:170–173. doi: 10.1016/s0014-5793(00)02420-0. [DOI] [PubMed] [Google Scholar]
- 43.Sehnke P C, Henry R, Cline K, Ferl R J. Plant Physiol. 2000;122:235–242. doi: 10.1104/pp.122.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schultz T F, Medina J, Hill A, Quatrano R S. Plant Cell. 1998;10:837–847. doi: 10.1105/tpc.10.5.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim J, Helariutta Y, Specht C D, Jung J, Sims L, Bruce W B, Diehn S, Benfey P N. Plant Cell. 2000;12:1307–1318. doi: 10.1105/tpc.12.8.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Igarashi D, Ishida S, Fukazawa J, Takahashi Y. Plant Cell. 2001;13:2483–2497. doi: 10.1105/tpc.010188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bachmann M, Huber J L, Athwal G S, Wu K, Ferl R J, Huber S C. FEBS Lett. 1996;398:26–30. doi: 10.1016/s0014-5793(96)01188-x. [DOI] [PubMed] [Google Scholar]
- 48.Lotan T, Ori N, Fluhr R. Plant Cell. 1989;1:881–887. doi: 10.1105/tpc.1.9.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaffrath U, Mauch F, Freydl E, Schweizer P, Dudler R. Plant Mol Biol. 2000;43:59–66. doi: 10.1023/a:1006423232753. [DOI] [PubMed] [Google Scholar]
- 50.Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P. Science. 2002;295:2073–2076. doi: 10.1126/science.1067554. [DOI] [PubMed] [Google Scholar]
- 51.Austin M J, Muskett P, Kahn K, Feys B J, Jones J D, Parker J E. Science. 2002;295:2077–2080. doi: 10.1126/science.1067747. [DOI] [PubMed] [Google Scholar]
- 52.Matthews L R, Vaglio P, Reboul J, Ge H, Davis B P, Garrels J, Vincent S, Vidal M. Genome Res. 2001;11:2120–2126. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou N, Tootle T L, Tsui F, Klessig D F, Glazebrook J. Plant Cell. 1998;10:1021–1030. doi: 10.1105/tpc.10.6.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winge P, Brembu T, Bones A M. Plant Mol Biol. 1997;35:483–495. doi: 10.1023/a:1005804508902. [DOI] [PubMed] [Google Scholar]
- 55.Presley J F, Ward T H, Pfeifer A C, Siggia E D, Phair R D, Lippincott-Schwartz J. Nature. 2002;417:187–193. doi: 10.1038/417187a. [DOI] [PubMed] [Google Scholar]
- 56.Gillet B, Beyly A, Peltier G, Rey P. Plant J. 1998;16:257–262. doi: 10.1046/j.1365-313x.1998.00292.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.