Abstract
Current cell-free protein synthesis systems can synthesize proteins with high speed and accuracy, but produce only a low yield because of their instability over time. Here we describe the preparation of a highly efficient but also robust cell-free system from wheat embryos. We first investigated the source of the instability of existing systems in light of endogenous ribosome-inactivating proteins and found that ribosome inactivation by tritin occurs already during extract preparation and continues during incubation for protein synthesis. Therefore, we prepared our system from extensively washed embryos that are devoid of contamination by endosperm, the source of tritin and possibly other inhibitors. In a batch system, we observed continuous translation for 4 h, and sucrose density gradient analysis showed formation of large polysomes, indicating high protein synthesis activity. When the reaction was performed in a dialysis bag, enabling the continuous supply of substrates together with the continuous removal of small byproducts, translation proceeded for >60 h, yielding 1–4 mg of enzymatically active proteins, and 0.6 mg of a 126-kDa tobacco mosaic virus protein, per milliliter of reaction volume. Our results demonstrate that plants contain endogenous inhibitors of translation and that after their elimination the translational apparatus is very stable. This contrasts with the common belief that cell-free translation systems are inherently unstable, even fragile. Our method is useful for the preparation of large amounts of active protein as well as for the study of protein synthesis itself.
The development of a system capable of synthesizing any desired protein on a preparative scale is one of the most important endeavors in biotechnology today. Three strategies are currently being used: chemical synthesis, in vivo expression, and cell-free protein synthesis. The first two methods have severe limitations: chemical synthesis is not feasible for the synthesis of long peptides because of low yield, and in vivo expression can produce only those proteins that do not affect the physiology of the host cell (1–3). Cell-free translation systems, in contrast, can synthesize proteins with high speed and accuracy, approaching in vivo rates (4–5), and they can express proteins that would interfere with cell physiology. However, they are relatively inefficient because of their instability (6).
Because cell free systems nonetheless have great potential for large scale protein synthesis, many efforts have been made to increase their efficiency. Spirin et al. (7) proposed a continuous flow cell-free translation system, in which a solution containing amino acids and energy sources is supplied to the reaction chamber through a filtration membrane. This design is significantly more efficient than conventional batch systems: The reaction works for tens of hours and produces hundreds of micrograms per milliliter of reaction volume (7–9). Recently, several modified versions of the Spirin system have been reported (10–13). Kigawa et al. showed that, by using a dialysis membrane to facilitate the continuous supply of substrates and removal of byproducts, an Escherichia coli-coupled transcription–translation system yields as much as 6 mg of protein per milliliter of reaction volume (12). This high productivity can, however, only be expected with fairly small proteins such as Ras protein (21 kDa) or chloramphenicol acetyltransferase (26 kDa). The problem with larger proteins is that with the increasing molecular weight of the mRNAs their degradation by endogenous E. coli ribonuclease(s) also increases. Kawarasaki et al. showed that in a wheat germ cell-free system translational efficiency increases after neutralization of endogenous ribonucleases and phosphatases with copper ions and antiphosphatase antibodies (13). For their improvements, these groups focused on modifying the reaction chamber and/or optimizing the reaction conditions while using conventional extracts. We used a different approach, instead focusing on clarifying the nature of the instability of the extracts.
We concentrated on wheat germ cell-free systems because they have numerous advantages such as low cost, easy availability in large amounts, low endogenous incorporation, and the capacity to synthesize high-molecular-weight proteins. Moreover they are eukaryotic systems and hence more suitable for the expression of eukaryotic proteins. After we discovered that the mechanism of action of the ricin toxin is ribosome inactivation (14–16), many other ribosome-inactivating proteins (RIPs) with identical mechanism of action have been found in higher plants (17). Most commonly these toxins are single-chain proteins, and they inhibit protein synthesis by removing a single adenine residue in a universally conserved stem-loop structure of 28S ribosomal RNA (14–17). Although the biological function of the RIPs is not known, it is generally believed that they are important for cell defense (17). The most widely studied example is an antiviral effect during infection by several plant viruses (18). As originally proposed by Ready et al. (19), the explanation for the antiviral activity of RIPs is that, when a cell wall is damaged, the RIP is released into the cytosol, where it inactivates ribosomes, thereby preventing virus replication. Tritin, found in wheat seeds and thought to be localized mainly in the endosperm, is such a single-chain RIP (20). Initially, it was reported that wheat embryonic ribosomes are resistant to this protein (20–22), which would render any contamination with tritin inconsequential.
To improve protein synthesis in wheat germ cell-free systems, we started with the hypothesis that the embryonic ribosomes are in fact susceptible to tritin. In this case, contamination of wheat germ preparations with tritin-containing endosperm fragments would be fatal. Accordingly, we prepared our cell-free system from extensively washed embryos and indeed found that the system became far more active.
In addition to the benefit of a better protein synthesis system, these results shed new light on the translational apparatus itself: Although it is usually seen as a rather fragile apparatus, it appears instead to be very stable: so stable, in fact, that plants seem to have developed a suicide mechanism (the RIPs) directed against the translational apparatus, further emphasizing its crucial role in cell physiology. We believe that the strategy we followed to improve the wheat cell-free system—elimination of endogenous translational inhibitors—is equally applicable for other systems.
Materials and Methods
General.
The following procedures were either described or cited previously (9, 14–15, 23–24): determination of RNA N-glycosidase activity, analysis of cell-free protein synthesis, sucrose density gradient analysis of polysomes, determination of proteins, the sources of m7GpppG, ribonucleotide triphosphates, SP6 RNA polymerase, T7 RNA polymerase, human placental ribonuclease inhibitor (133 units/ml), l-[U-14C]leucine, MTX immobilized on agarose, creatine kinase, spermidine, and the 20 amino acids. Dialysis membrane (molecular weight cutoff 12,000–14,000, regenerated cellulose, Viskase Sales, Chicago), the nonionic detergent Nonidet P-40, and proteinase inhibitor E64 were purchased from Nakarai Tesque (Kyoto). The luciferase assay kit (PiccaGene) was from Wako Pure Chemical (Osaka). Low and high molecular weight marker kits (Rainbow marker) were from Amersham Pharmacia. Recombinant forms of luciferase and green fluorescent protein (GFP) (S65T) that were used as standards were purchased from Promega and CLONTECH, respectively. Plasmid pCaMV35S-sGFP(S65T)-NOS3′(25) carrying the GFP gene was kindly provided by Y. Niwa (School of Food and Nutritional Sciences, University of Shizuoka, Japan), and plasmid pSP-Luc+ carrying luciferase was obtained from Promega. Plasmid pTLW3 (26), covering the tobacco mosaic virus (TMV) genome, was a generous gift from Y. Watanabe (University of Tokyo).
Purification of Wheat Embryos and Extract Preparation.
Wheat seeds were ground in a mill (Roter Speed Mill model pulverisette 14, Fritsh, Germany), then were sieved through a 710- to 850-mm mesh. Embryos were selected with the solvent flotation method of Erickson and Blobel (27) by using a solvent containing cyclohexane and carbon tetrachloride (240:600, vol/vol). Damaged embryos and contaminants were discarded, and intact embryos were dried overnight in a fume hood. To remove contaminating endosperm, the embryos were washed three times with 10 vol of water under vigorous stirring, and then were sonicated for 3 min in a 0.5% solution of Nonidet P-40 by using a Bronson model 2210 sonicator (Yamato, Japan). Finally, the embryos were washed once more in the sonicator with sterile water.
Preparation of the Cell-Free Extract.
The method used is a slight modification of the procedure described by Erickson and Blobel (27). Washed embryos were ground to a fine powder in liquid nitrogen. Five grams of the powder were added to 5 ml of 2 × buffer A (40 mM Hepes, pH 7.6/100 mM potassium acetate/5 mM magnesium acetate/2 mM calcium chloride/4 mM DTT/0.3 mM of each of the 20 amino acids). The mixture was briefly vortexed and then was centrifuged at 30,000 × g for 30 min. The resulting supernatant was subjected to gel-filtration on a G-25 (fine) column, equilibrated with two volumes of buffer A. The void volume was collected and centrifuged at 30,000 × g for 10 min. The final supernatant was adjusted to 200 A260/ml with buffer A, was divided into small aliquots, and was stored in liquid nitrogen until use.
Cell-Free Translation.
In the batch system, 50 μl of reaction mixture contained 12.5 μl of extract (thus 24%); final concentrations of the various ingredients are 24 mM Hepes/KOH (pH 7.8), 1.2 mM ATP, 0.25 mM GTP, 16 mM creatine phosphate, 0.45 mg/ml creatine kinase, 2 mM DTT, 0.4 mM spermidine, 0.3 mM of each of the 20 amino acids including [14C]leucine (2 μCi/ml), 2.5 mM magnesium acetate, 100 mM potassium acetate, 50 μg/ml of deacylated tRNA prepared from wheat embryos, 0.05% Nonidet P-40, 1 μM E-64 as proteinase inhibitor, 0.005% NaN3, and 7.2 μg (0.02 nmol) of dihydrofolate reductase (DHFR) mRNA. The extract was not treated with micrococcal nuclease because we did not observe any positive effect of this treatment. Incubation was done at 26°C.
For the dialysis system, 500 μl of reaction mixture contained 300 μl of the extract and the same ingredients as described above. The dialysis bag was immersed in 5 ml of a solution containing all described ingredients except for creatine kinase. The reaction was done at 23°C, and, every 24 h, 0.05 nmol of DHFR mRNA (or equivalent moles of the other mRNAs) and 50 μg of creatine kinase were supplemented. The dialysis solution was also replaced every 24 h. To confirm the longevity of the system, [14C]leucine (the same concentration as above) was added into both reaction mixture and dialysis buffer at 52 h, then was incubated until 72 h (Fig. 4C). The autoradiogram of the gel was obtained by using a BAS-2000 phosphoimager (Fuji).
Figure 4.
Synthesis of luciferase (A), GFP (B), and 126-kDa TMV protein (C) in the dialysis system. Samples were analyzed as described in Materials and Methods. The standard samples were prepared by mixing a reaction mixture without mRNA with known amounts of luciferase or GFP before loading onto the gel. For the autoradiogram in C, [14C]leucine was added at 52 h, and samples were withdrawn after an additional 8 h (60 h total) or 20 h (72 h). Authentic GFP migrates slower than the cell-free product on the native gel, which is attributable to different amino acid compositions because both proteins work as a monomer form. Products and supplemented creatine kinase are marked with arrows and asterisks, respectively.
Preparation of mRNA.
Capped mRNA encoding DHFR was synthesized by in vitro transcription of linearized plasmid pSP65 carrying the gene under SP6 RNA polymerase promoter control (9). The transcript is 1,079 nucleotides long and consists of the sequence m7GpppGAAUACACGGAAUUCGAGCUCGCCCGGGAAAUCUCAAUG (the italicized sequence is the initiation codon) at its 5′ end, a 477-nt coding sequence, and a 3′ noncoding region of 565 nucleotides with a poly(A) tail of 100 adenosines (9). Coding sequences for GFP (717 nucleotides) (25) and luciferase (1,650 nucleotides) were cloned into the above plasmid in such a way that the 5′- and 3′-untranslated regions of DHFR were preserved. Capped TMV RNA (6,388 nucleotides) was transcribed from linearized plasmid pTLW3 carrying the genome under T7 RNA polymerase promoter control (26).
Analysis of Products and Their Enzymatic Activities.
The amount of protein synthesized was determined as follows: Aliquots were withdrawn, and samples containing 1 μl of reaction mixture were separated on 12.5% SDS polyacrylamide gels (8% gels for TMV protein) or 12.5% native polyacrylamide gels (for GFP), then were stained with Coomassie brilliant blue. The product amount was estimated by densitometric scanning of the bands and comparison to standards. The standard samples were prepared by mixing a reaction mixture without mRNA with known amounts of standard proteins (DHFR, GFP, or luciferase) before loading onto the gel. Because pure, authentic, 126-kDa TMV protein is not available, the amount of this protein was estimated with less accuracy by calculating its relative amount compared with molecular markers included as internal standards by using average 105- and 160-kDa band intensities. The amount of DHFR was confirmed by determining the amount of methotrexate-agarose column purified protein, and its activity was measured colorimetrically as described (9). Luciferase activity was determined by using a commercial kit and a liquid scintillation counter as described (28). The specific activities of recombinant luciferase and the synthesized protein were 3.4 × 105 and 5.1 × 106 cpm/pg, respectively. Semiquantitative measurement (28) of GFP activity on the native gel was carried out by using a UV-illuminator (Dark Reader, Clare Chemical Research, Denver) with a wavelength of 400–500 nm. Subsequent scanning of photographs of the UV images and comparison of the intensities of the bands to those of the recombinant protein revealed that the translation product had more activity than the standard by a factor of 1.4.
Results and Discussion
Removal of Contaminants such as Tritin from Wheat Embryos Leads to a More Active Cell-Free Protein Synthesis System.
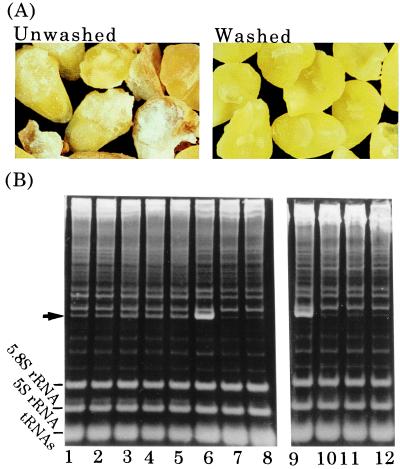
Since the first report of solvent flotation for the enrichment of viable, intact embryos from wheat seeds by Johnston and Stern (29), this method has commonly been used for the preparation of wheat embryos. We first addressed the possibility of a tritin contamination originating from endosperm as the reason for the instability of wheat germ cell-free systems. If wheat germs are isolated from dry wheat seeds by conventional procedures (27), microscopic examination reveals that the sample contains embryos as well as some white material and a number of white and brownish granules (Fig. 1A). Analysis of ribosomal RNAs from a protein synthesis reaction prepared from such a sample showed that depurination of ribosomes occurs, contradicting earlier reports (20–22) (Fig. 1B). After 4 h of incubation, 24% of the ribosome population had been depurinated, as judged by the aniline-dependent formation of a specific RNA fragment (Fig. 1B, arrow). Furthermore, even at the start of the incubation, 7% of the population had already been depurinated. The site of depurination was confirmed by direct sequencing of the fragment to be in the universally conserved sarcin/ricin domain of 28S rRNA (data not shown). When RNA was extracted directly from embryos by guanidine isothiocyanate-phenol, little formation of the aniline-induced fragment was observed (Fig. 1B, lanes 7 and 8). Thus, depurination must have occurred during the extract preparation and then continued during the protein synthesis reaction.
Figure 1.
Removal of tritin from embryos. Extracts were prepared from unwashed or washed embryos (A), and the depurination assay was performed (B). Translation mixtures prepared with the extract from unwashed embryos were incubated for 0, 1, 2, 3, and 4 h (B, lanes 1–5 respectively); mixtures with washed embryos were incubated for 0, 2, and 4 h (lanes 10–12, respectively). Isolated RNA was treated with acid/aniline, then was separated on 4.5% polyacrylamide gels. Additionally, RNA was directly extracted from embryos with guanidine isothiocyanate-phenol and was analyzed as above before (B, lane7) and after (B, lane 8) treatment with acid/aniline. For the fragment marker (B, lanes 6 and 9), incubation was carried out in the presence of gypsophilin (40), a highly active RIP from Gypsophila elegance; the arrow indicates the aniline-induced fragment.
The observed extent of depurination constitutes a considerable damage to protein synthesis because inactivation of any one ribosome among the actively translating ribosomes on an mRNA results in blockage of the respective polyribosome and cessation of translation (16). Attempts were made to neutralize the depurinating enzyme with synthetic RNA aptamers that tightly bind to the RIP (30), but these attempts failed. Instead, careful selection and subsequent extensive washing of the embryos yielded better results. These embryos had few contaminants (Fig. 1A Right), and when the depurination assay was performed, no aniline-induced cleavage was detectable (Fig. 1B, lanes 10–12), indicating minimal, if any, depurination during preparation as well as incubation.
As shown in Fig. 2, the cell-free system prepared from washed embryos has much higher translational activity than the conventional system (compare Fig. 2 A and B). When programmed with mRNA coding for DHFR, it has almost linear kinetics in DHFR synthesis over 4 h in a system containing 24% extract, as opposed to the regular system, which ceased to function after 1.5 h. When the content of washed extract in the reaction volume was increased to 48%, amino acid incorporation occurred initially at a rate twice that with 24% extract, but then stopped after 1 h. However, this halting was caused by a shortage of substrates rather than an irreversible inactivation of ribosomes or factors necessary for translation: Addition of amino acids, ATP, and GTP after cessation of the reaction (Fig. 2 A and B, arrows) restarted translation with kinetics similar to the initial rate. In contrast, if conventional extract was added to 48%, protein synthesis actually decreased compared with the 24% extract reaction. Furthermore, the halting of protein synthesis in the reaction with 24% extract could not be reversed by the addition of more substrate, indicating an irreversible damage by contaminants from endosperm (Fig. 2B).
Figure 2.
Protein synthesis with an extract prepared from washed embryos. The batch system contains either 12 μl (24%) or 24 μl (48%) of extracts from washed (A) or unwashed (B) wheat embryos. Protein synthesis was measured as hot trichloroacetic acid insoluble radioactivity. Arrows show addition of substrates. C shows the polysome profiles of 15 μl of reaction mixture aliquots loaded onto a linear 10% to 45% sucrose gradient in 25 mM Tris⋅HCl (pH 7.6), 100 mM KCl, and 5 mM MgCl2. After centrifugation, fractions were collected from the bottom of the tubes and were measured at 260 nm as described (24). Incubation times were 0 h (open circles in a), 1 h (closed circles in a), and 2 h (b) in the absence (open circles in b) or presence (closed circles in b) of 0.4 μM cycloheximide. In c, the translation system prepared from unwashed embryos was incubated for 2 h. In d and e, aliquots from the dialysis system were withdrawn after 48 and 60 h and were incubated in the presence of 0.4 μM cycloheximide for another 60 min at 26°C (closed circles). Similar analyses of the samples were carried out in the absence of mRNA (d and e, open circles) as negative controls.
High protein synthesis activity of the system with washed embryos can also be demonstrated by sucrose density gradient analysis (Fig. 2C). Significant formation of polysomes was observed after 1 h of incubation, and at 2 h a shift to heavier polysomes with a concomitant decrease of 80S monosomes was seen (Fig. 2C a and b). In the presence of low concentrations of cycloheximide polysome formation is a measure of translational initiation (31). A concentration of cycloheximide of 0.4 μM reduced the incorporation of [14C]leucine to 21% of the control (data not shown) and resulted in an accumulation of large polysomes, with 78% of ribosomes in polysomes (open circle in Fig. 2C b). A similar analysis of cell-free reactions prepared with regular extracts (27), but done in the absence of cycloheximide, did not show significant polysome formation (Fig. 2C c). The high efficiency of our system, therefore, can be attributed to at least two factors: first, high initiation, elongation, and termination rates (efficient usage and recycling of ribosomes); and second, low endogenous ribonuclease activity (retention of heavy polysomes for prolonged time).
There is an additional explanation for the dramatic improvement of protein synthesis after washing of the embryos. Thionins are a group of small basic and cysteine-rich proteins, originally purified as antifungal proteins from a variety of plants, including wheat seeds (32). Wheat γ-thionin is known to be in the endosperm of seeds (33), and, recently, Brummer et al. have shown in a wheat germ translation system that α- and β-thionin from barley endosperm are potent inhibitors of protein synthesis initiation (34). In addition, several ribonucleases have been reported in the endosperm of the seeds (35). Thus, it is possible that the washing of the embryos resulted in elimination of thionin and ribonucleases as well as tritin.
The Continuous-Flow Cell-Free System on a Preparative Scale.
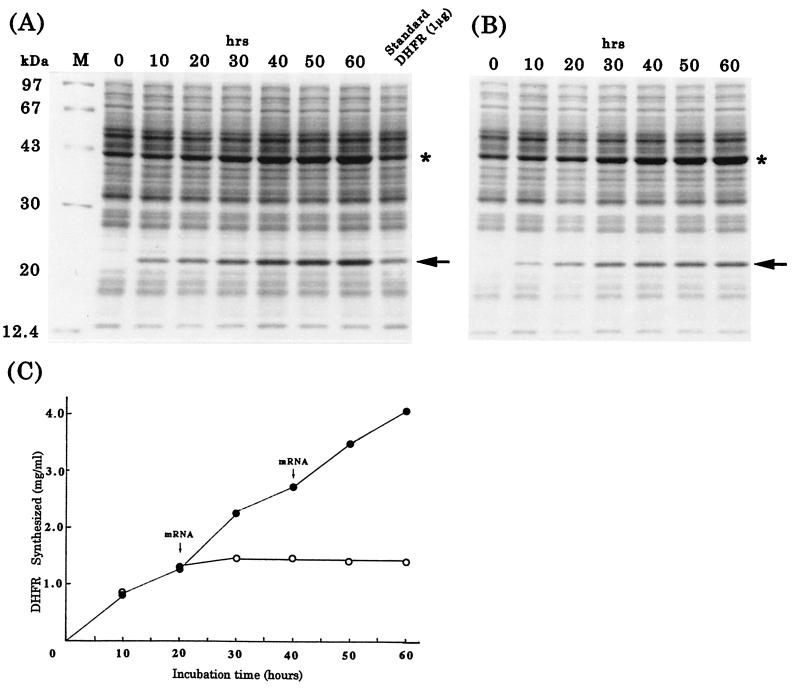
After establishing a procedure for the preparation of highly active wheat embryo extract, we addressed its possible application for the large scale production of protein. For this purpose, we chose a dialysis system because of its continuous supply of substrates and continuous removal of small byproducts (12). With DHFR mRNA as template, protein synthesis worked efficiently, as demonstrated by a Coomassie blue stained gel (Fig. 3A, arrow). Densitometric quantitation as well as a direct determination of purified DHFR revealed that the reaction proceeded up to 60 h, yielding 4 mg of enzyme in a 1-ml reaction (Fig. 3C). This yield was achieved when the system was supplemented with fresh mRNA every 24 h; without the addition of fresh mRNA, the reaction ceased after 24 h and yielded 1 mg of DHFR (Fig. 3 B and C, open circles). When aliquots of the reaction mixtures were withdrawn after 48 and 60 h and then were incubated in the presence of a low dose of cycloheximide for an additional 1 h, sucrose gradient centrifugation revealed polysome formation (Fig. 2C d and e). This is a direct indication of a robust system with high translational activity. The product has a similar specific activity as the authentic enzyme, 15.3 vs. 19.1 units/mg (9).
Figure 3.
Protein synthesis in the dialysis system. (A and B) Coomassie blue-stained SDS polyacrylamide gels showing DHFR synthesis with (A) or without (B) addition of new mRNA. Arrows and asterisks mark DHFR and creatine kinase, respectively. The standard sample was prepared by mixing a reaction mixture without mRNA with known amounts of DHFR before loading onto the gel. (C) Amounts of DHFR synthesized as determined from densitometric scans of the gels in A (closed circles) and B (open circles).
As shown in Fig. 4, the system also synthesized proteins of higher molecular weight in a preparative scale: 1.1 mg of luciferase (65 kDa), 1.2 mg of GFP (45 kDa). These proteins had the same or even higher specific activity compared with commercially available recombinant forms (Fig. 4 A and B; see Materials and Methods). Furthermore, the 126-kDa replicase of TMV, a major genome product (36) during infection, was produced with a yield of as much as 0.6 mg (Fig. 4). The synthesis proceeded for up to 72 h, as shown by the increase in intensity of the Coomassie brilliant blue-stained bands. This point was confirmed by autoradiography and analysis of amino acid incorporation: [14C]leucine was added after 52 h, samples were withdrawn at 60 and 72 h, and the samples were analyzed by SDS gel electrophoresis and autoradiography (Fig. 4). Densitometric quantitation of the bands showed linear synthesis: The photostimulated luminescence of the sample after 8 h of synthesis (at the 60-h time point) was 186, and after 20 h (at the 72-h point) it was 465, even though the rate of protein synthesis as measured by leucine incorporation was 21% of the rate at the beginning of incubation. This is another direct evidence of the robustness of the system and its efficiency in synthesizing even a 126-kDa protein for 3 days.
The structures of 5′- and 3′-untranslated regions are important for the efficiency of initiation and termination and also for the stability of mRNA (37). The mRNA constructs used here were not optimized in this respect, and we believe that the yields in our experiments do not, therefore, necessarily reflect maximum capacity. Efficient mRNA translation and its regulation requires a series of protein–mRNA and protein–protein interactions (37), and Wells et al. have recently shown the circularization of mRNA in vitro (38). Our method provides, in addition to its protein synthesis capacity, the opportunity to study translation itself, including the phenomenon of circular mRNA or the characterization of untranslated regions of mRNA in terms of efficient initiation or stability.
We show here that removal of endosperm contaminants, which contain protein synthesis inhibitor(s), from the embryo fraction improves protein synthesis in a wheat germ cell-free system. The improvement likely is caused by increased translational activity resulting from elimination of inhibitors of initiation (e. g. the thionins) and ribonucleases, as well as elimination of the RIP tritin. It is generally believed that cell-free translation systems are inherently unstable, but our results demonstrate the opposite: The translational apparatus appears to be very stable, in vitro and presumably also in vivo. We believe that our results shed light on the biological function of the nearly ubiquitous plant RIPs. We propose that plants acquired during evolution a suicide system useful to prevent larger damage and that because of its stability the translational machinery is the most important target of a suicide system. Viral attack would be one instance in which this suicide mechanism is employed. Ribosomes are a popular target of antibiotics also, emphasizing their central role in cell metabolism. The observed high stability of the translational apparatus might be an essential requirement for the evolution of life: Certain basic physiological processes such as protein synthesis might be required to function even in adverse conditions.
It is likely that the strategy that we followed to improve the wheat cell-free system, i.e., the inactivation of the translational suicide system, is successful with other systems as well. For instance, the widely used cell-free system from E. coli contains high ribonuclease activity and is hampered by a low efficiency in the translation of large mRNAs. Because of significant levels of template degradation, E. coli systems are limited when selecting large polypeptides for polysome display.
Our protein synthesis system has several advantages compared with existing systems in addition to its high efficiency: As a eukaryotic system, it is more amenable to the production of eukaryotic proteins from their natural mRNAs: i.e. no cDNA modification is needed; the system can produce high molecular weight proteins; because of little template degradation, it is useful for polysome display (39); and proteins that would normally interfere with cell physiology can be synthesized. Additionally, it should be a useful tool in the study of translation itself.
Acknowledgments
This work was supported by The Japan Society for the Promotion of Science Grant JSPS-RFTF 96100305 (to Y.E.).
Abbreviations
- RIP
ribosome-inactivating protein
- TMV
tobacco mosaic virus
- GFP
green fluorescent protein
- DHFR
dihydrofolate reductase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Golf S A, Goldberg A L. J Biol Chem. 1987;262:4508–4515. [PubMed] [Google Scholar]
- 2.Chrunyk B A, Evans J, Lillquist J, Young P, Wetzel R. J Biol Chem. 1993;268:18053–18061. [PubMed] [Google Scholar]
- 3.Henrich B, Lubitz W, Plapp R. Mol Gen Genet. 1982;185:493–497. doi: 10.1007/BF00334146. [DOI] [PubMed] [Google Scholar]
- 4.Kurland C G. Cell. 1982;28:201–202. doi: 10.1016/0092-8674(82)90336-1. [DOI] [PubMed] [Google Scholar]
- 5.Pavlov M Y, Ehrenberg M. Arch Biochem Biophys. 1996;328:9–16. doi: 10.1006/abbi.1996.0136. [DOI] [PubMed] [Google Scholar]
- 6.Roberts B E, Paterson B M. Proc Natl Acad Sci USA. 1973;70:2330–2334. doi: 10.1073/pnas.70.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spirin A S, Baranov V I, Ryabova L A, Ovodov S, Yu, Alakhov Yu B, Alakhov Yu B. Science. 1988;242:1162–1164. doi: 10.1126/science.3055301. [DOI] [PubMed] [Google Scholar]
- 8.Baranov V I, Morozov I, Yu, Ortlepp S A, Spirin A S. Gene. 1989;84:463–466. doi: 10.1016/0378-1119(89)90521-0. [DOI] [PubMed] [Google Scholar]
- 9.Endo Y, Otsuzuki S, Ito K, Miura K. J Biotechnol. 1992;25:221–230. doi: 10.1016/0168-1656(92)90157-5. [DOI] [PubMed] [Google Scholar]
- 10.Kigawa T, Yokoyama S. J Biochem. 1991;110:166–168. doi: 10.1093/oxfordjournals.jbchem.a123551. [DOI] [PubMed] [Google Scholar]
- 11.Kim D-M, Kigawa T, Choi C-Y, Yokoyama S. Eur J Biochem. 1996;239:881–886. doi: 10.1111/j.1432-1033.1996.0881u.x. [DOI] [PubMed] [Google Scholar]
- 12.Kigawa T, Yabuki T, Yoshida Y, Tsutsui M, Ito Y, Shibata T, Yokoyama S. FEBS Lett. 1999;442:15–19. doi: 10.1016/s0014-5793(98)01620-2. [DOI] [PubMed] [Google Scholar]
- 13.Kawarasaki Y, Kawai T, Nakano H, Yamane T. Anal Biochem. 1995;226:320–324. doi: 10.1006/abio.1995.1231. [DOI] [PubMed] [Google Scholar]
- 14.Endo Y, Mitsui K, Motizuki M, Tsurugi K. J Biol Chem. 1987;262:5908–5912. [PubMed] [Google Scholar]
- 15.Endo Y, Tsurugi K. J Biol Chem. 1987;262:8128–8130. [PubMed] [Google Scholar]
- 16.Wool I G, Glück A, Endo Y. Trends Biochem Sci. 1992;17:266–269. doi: 10.1016/0968-0004(92)90407-z. [DOI] [PubMed] [Google Scholar]
- 17.Barbieri L, Battelli M G, Stirpe F. Biochim Biophys Acta. 1993;1154:237–282. doi: 10.1016/0304-4157(93)90002-6. [DOI] [PubMed] [Google Scholar]
- 18.Taylor S, Massiah A, Lomonossoff G, Robert L M, Lord J M, Hartely M R. Plant J. 1994;5:827–853. doi: 10.1046/j.1365-313x.1994.5060827.x. [DOI] [PubMed] [Google Scholar]
- 19.Ready M P, Brown D T, Robertus J D. Proc Natl Acad Sci USA. 1986;83:5053–5056. doi: 10.1073/pnas.83.14.5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massiah A J, Hartely M R. Planta. 1995;197:633–640. doi: 10.1007/BF00191571. [DOI] [PubMed] [Google Scholar]
- 21.Stewart T S, Hruby D E, Sharma O K, Robert W K. Biochim Bipphys Acta. 1977;479:31–38. doi: 10.1016/0005-2787(77)90123-x. [DOI] [PubMed] [Google Scholar]
- 22.Taylor B E, Irvin J D. FEBS Lett. 1990;273:144–146. doi: 10.1016/0014-5793(90)81070-5. [DOI] [PubMed] [Google Scholar]
- 23.Yoshinari S, Koresawa S, Yokota S, Sawamoto H, Tamura M, Endo Y. Biosci Biotechnol Biochem. 1997;61:324–331. doi: 10.1271/bbb.61.324. [DOI] [PubMed] [Google Scholar]
- 24.Hase M, Endo Y, Natori Y. J Biochem. 1982;91:1457–1465. doi: 10.1093/oxfordjournals.jbchem.a133837. [DOI] [PubMed] [Google Scholar]
- 25.Chiu W-L, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheet J. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- 26.Hamamoto H, Sugiyama K, Nakagawa N, Hashida E, Matsunaga Y, Takemoto S, Watanabe Y, Okada Y. Bio/Technology. 1999;11:930–932. doi: 10.1038/nbt0893-930. [DOI] [PubMed] [Google Scholar]
- 27.Erickson A H, Blobel G. Methods Enzymol. 1983;96:38–50. doi: 10.1016/s0076-6879(83)96007-x. [DOI] [PubMed] [Google Scholar]
- 28.Aleen R C. Methods Enzymol. 1986;133:449–493. doi: 10.1016/0076-6879(86)33085-4. [DOI] [PubMed] [Google Scholar]
- 29.Johnston F B, Stern H. Nature (London) 1957;179:160–161. doi: 10.1038/179160b0. [DOI] [PubMed] [Google Scholar]
- 30.Hirao I, Yoshinari S, Yokoyama S, Endo Y, Ellington A D. Nucleic Acids Symp Ser. 1997;37:283–284. [PubMed] [Google Scholar]
- 31.Lodish H F, Housman D, Jacobsen M. Biochemistry. 1971;10:2348–2356. doi: 10.1021/bi00788a027. [DOI] [PubMed] [Google Scholar]
- 32.Bohlmann H. Crit Rev Plant Sci. 1994;13:1–16. [Google Scholar]
- 33.Colilla F J, Rocher A, Mendez E. FEBS Lett. 1990;270:191–194. doi: 10.1016/0014-5793(90)81265-p. [DOI] [PubMed] [Google Scholar]
- 34.Brummer J, Thole H, Kloppstech K. Eur J Biochem. 1994;219:425–433. doi: 10.1111/j.1432-1033.1994.tb19955.x. [DOI] [PubMed] [Google Scholar]
- 35.Matsushita S. Memoirs Res Inst Food Sci, Kyoto Univ. 1959;19:1–4. [Google Scholar]
- 36.Dawson W O. Virology. 1992;186:359–367. doi: 10.1016/0042-6822(92)90001-6. [DOI] [PubMed] [Google Scholar]
- 37.Sachs A B, Sarnow P, Hentze M W. Cell. 1997;89:831–835. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 38.Wells S E, Hillner P E, Vale R D, Sachs A B. Mol Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 39.Mattheakis L C, Bhatt R R, Dower W J. Proc Natl Acad Sci USA. 1994;91:9022–9026. doi: 10.1073/pnas.91.19.9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshinari S, Koresawa S, Yokota S, Sawamoto H, Tamura M, Endo Y. Biosci Biotechnol Biochem. 1997;61:324–331. doi: 10.1271/bbb.61.324. [DOI] [PubMed] [Google Scholar]