Abstract
When challenged by DNA-damaging agents, Escherichia coli cells respond by inducing the SOS stress response, which leads to an increase in mutation frequency by two mechanisms: translesion replication, a process that causes mutations because of misinsertion opposite the lesions, and an inducible mutator activity, which acts at undamaged sites. Here we report that DNA polymerase V (pol V; UmuC), which previously has been shown to be a lesion-bypass DNA polymerase, was highly mutagenic during in vitro gap-filling replication of a gapped plasmid carrying the cro reporter gene. This reaction required, in addition to pol V, UmuD′, RecA, and single-stranded DNA (ssDNA)-binding protein. pol V produced point mutations at a frequency of 2.1 × 10−4 per nucleotide (2.1% per cro gene), 41-fold higher than DNA polymerase III holoenzyme. The mutational spectrum of pol V was dominated by transversions (53%), which were formed at a frequency of 1.3 × 10−4 per nucleotide (1.1% per cro gene), 74-fold higher than with pol III holoenzyme. The prevalence of transversions and the protein requirements of this system are similar to those of in vivo untargeted mutagenesis (SOS mutator activity). This finding suggests that replication by pol V, in the presence of UmuD′, RecA, and ssDNA-binding protein, is the basis of chromosomal SOS untargeted mutagenesis.
The SOS stress response is induced in Escherichia coli by single-stranded DNA (ssDNA) gaps formed when DNA lesions that have escaped repair block replication (1–3). Unable to remove the lesion from such gap/lesion structures, the cells activate a tolerance response, which involves filling in the DNA gaps without removal of the lesion, thereby restoring genome continuity. Thereafter, a second attempt to eliminate the lesion by DNA repair can be made. Filling in of the gap is done by patching of a homologous DNA segment from the fully replicated sister chromatid via recombinational repair (4, 5) or by translesion replication, which requires the SOS-inducible proteins UmuC, UmuD′, and RecA (6, 7). The latter process is mutagenic, because of the miscoding promoted by most DNA lesions. Recently, it was found that UmuC is a DNA polymerase, termed DNA polymerase V (pol V), with a remarkable capability to replicate through DNA lesions that severely block other DNA polymerases (8, 9).
In addition to this mutagenesis process, which is targeted to DNA lesions, a mutator activity is induced under SOS conditions, which produces mutations in the apparent absence of DNA damage (untargeted mutagenesis) (10–12). Chromosomal untargeted mutagenesis requires the SOS-inducible proteins RecA, UmuD′, and UmuC (1, 13, 14), the same proteins that are required for translesion replication. In addition, it exhibits a particular mutational specificity, namely, the selective generation of transversions (14–17). Another pathway of untargeted mutagenesis is observed by transfecting UV-irradiated E. coli cells with unirradiated phage λ (18). This phage untargeted mutagenesis requires the dinB, uvrA, and polA gene products (19–22) and produces frameshift mutations (23). Recently, dinB (a homologue of umuC) was shown to encode an error-prone DNA polymerase termed pol IV, which tends to produce frameshifts (24). The role of pol IV in E. coli cells is not clear, because dinB mutants are proficient both in untargeted and targeted SOS mutagenesis (20, 25).
Here we report that pol V (UmuC), in the presence of UmuD′, RecA, and ssDNA-binding protein (SSB), is highly mutagenic and exhibits a specificity for transversion mutations. These protein requirements and mutagenic specificity suggest that replication of ssDNA regions by pol V, in the presence of UmuD′, RecA, and SSB, is the mechanistic basis of SOS untargeted mutagenesis.
Materials and Methods
Materials.
The sources of materials used were as follows: nucleotides and DTT, Boehringer Mannheim; ethidium bromide, Sigma; and [α-32P]dTTP, Amersham.
Proteins.
The fusion maltose-binding protein (MBP)-UmuC protein and UmuD′ were purified as described previously (9, 26). pol III holoenzyme, SSB, and RecA were purified according to published procedures (refs. 27–29, respectively), except that a phosphocellulose purification step was added for RecA. DNA polymerase II was a gift from M. Goodman (University of Southern California, Los Angeles), and the E. coli MutM (Fpg) protein was a gift from J. Laval (Institute Gustave Roussy, Villejuif, France) and S. Boiteux (Commissariat Energie Atomique, Fontenay Aux Roses, France). Uracil DNA N-glycosylase was purchased from United States Biochemical; pol I, exonuclease III, BSA, and proteinase K were from Boehringer Mannheim; S1 nuclease was from Promega, and restriction nuclease AatII, dam methylase, and T4 DNA ligase were from New England Biolabs.
Gapped Plasmid.
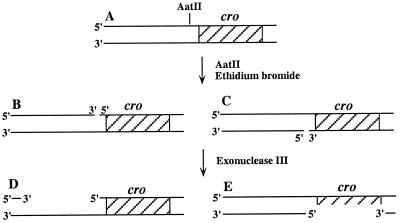
Plasmid pOC2 is a pBR322 derivative carrying the cro gene, which was used previously in our laboratory for mutagenesis studies (30–34). Treatment of pOC2 with the restriction nuclease AatII in the presence of ethidium bromide (35) produced two populations of plasmid, each nicked in one of the two complementary strands (Fig. 1). Subsequently, exonuclease III was added to extend the nicks into gaps. Notice that the cro region was single-stranded and, therefore, could be replicated in only half of the molecules (Fig. 1). This limitation, however, did not interfere with the assay, because the unreplicated DNA did not add a significant mutagenesis background (see below). The nicks were introduced upstream to the cro gene, using 0.025 unit/μl of the restriction nuclease AatII, in the presence of 0.11 μg/μl ethidium bromide, and 77 nM plasmid pOC2, at 37°C for 30 min. The DNA was precipitated with ethanol to remove the ethidium bromide, then extracted with phenol, and precipitated again. The gap was generated in a reaction mixture containing 30 nM nicked pOC2, 1 unit/μl exonuclease III, 66 nM Tris⋅HCl (pH 7.5), 0.66 mM MgCl2, 1 mM DTT, and 90 mM NaCl. The reaction was carried out at 37°C for 20 min to obtain a ssDNA region of approximately 350 nt. The size of the gap was deduced from the electrophoretic migration of the DNA after treatment with nuclease S1, which digested the single-stranded region in the plasmid.
Figure 1.
Preparation of the gapped plasmid. Plasmid pOC2 (A) was nicked upstream to the cro gene with restriction enzyme AatII in the presence of ethidium bromide. This generated two subpopulations of nicked plasmid (B and C). Addition of exonuclease III extended the nicks into gaps in the 3′ → 5′ direction. Half of the molecules contain the cro gene in the single-stranded region.
In Vitro Replication Fidelity Assay.
The standard gap-filling replication reaction mixture (50 μl) contained 20 mM Tris⋅HCl (pH 7.5), 8 μg/ml BSA, 5 mM DTT, 0.1 mM EDTA, 4% glycerol, 1 mM ATP, 10 mM MgCl2, 0.5 mM each of dATP, dGTP, dTTP, and dCTP, and 1 μg (6.2 nM) gapped pOC2. The replication was carried out with 0.5 μM UmuC fusion protein in the presence of 4.8 μM UmuD′, 0.6 μM SSB, 4.2 μM RecA, and 200 units of T4 DNA ligase. Control reactions were performed with 1.5 nM pol III holoenzyme, 11 nM DNA pol I, or 11 nM DNA pol II. Reactions were carried out at 37°C for 20 min, after which they were terminated by heat inactivation at 65°C for 10 min. The DNA then was methylated by adding 32 units of dam methylase, 80 μM S-adenosylmethionine, 50 mM Tris⋅HCl (pH 7.5), 10 mM EDTA, and 5 mM 2-mercaptoethanol, in a total volume of 100 μl, at 37°C for 1 hr. The reaction was terminated by adding SDS to 0.2% and EDTA to 15 mM, and the DNA was purified by digestion with 0.4 mg/ml proteinase K at 37°C for 1 hr, followed by phenol extraction and ethanol precipitation. DNA molecules carrying Cro− mutations that were formed during the in vitro replication stage were detected in a subsequent bioassay step (30), by transformation of an indicator strain, E. coli WBY11T (33), and plating on lactose-indicator plates containing kanamycin (70 μg/ml). Mutants were scored after an incubation period of 21 hr at 37°C. Under these conditions Cro− mutants yield dark-red colonies, whereas Cro+ plasmids yield white colonies. Typically, each transformation plate contained a total of 3–4 × 104 colonies, and for each DNA sample, eight plates were plated. The mutation frequencies of pol V and pol III holoenzyme were obtained as an average of 8 and 15 experiments, respectively. Parallel reactions were performed to determine the amount of DNA synthesis. This was done under the same conditions, except that [α-32P]dTTP was included at 50 μM. Reaction products were analyzed by agarose gel electrophoresis followed by phosphorimaging.
Calculation of Mutation Frequency.
The observed mutation frequency per gene was calculated by dividing the number of dark-red Cro− mutants by the total number of colonies on the plate. To obtain the actual mutation frequency, two corrections were made. (i) The subpopulation of the substrate that contained double-stranded cro transformed the indicator strain and led to the formation of kanR colonies, but did not contribute a significant number of Cro− mutants (see Table 1). To compensate for this, the mutation frequency obtained with each of the DNA polymerases was multiplied by 2. To check the accuracy of this correction factor, we pretreated the gapped plasmid with restriction nuclease EcoRI, which cuts within cro. Because ssDNA is resistant to EcoRI, all molecules in which cro is not in the gap are linearized, leaving only gapped circles with cro in the gap. When this DNA was used as a substrate to determine the frequencies of pol III holoenzyme and of pol V, we obtained mutation frequencies that were 2-fold higher than with substrate that was not pretreated. This validates the multiplication of mutations frequencies by 2. (ii) Whereas DNA synthesis by pol III holoenzyme led to essentially quantitative filling in of the single-stranded gap in the plasmid, the amount of DNA synthesis by pol V was 29.4% that of pol III holoenzyme (see Fig. 3). To correct for that, mutation frequencies obtained with pol V were divided further by 0.294. This correction was not required for reactions with pol I or pol II, which filled in the gaps essentially quantitatively (data not shown), similar to pol III holoenzyme. It should be noted that the use of these correction factors, although necessary, introduces some error in the final mutation frequencies. Therefore, although our conclusions are not affected, these numbers are not precise.
Table 1.
Frequency of Cro− mutations generated during in vitro gap-filling replication
| Protein composition | Mutation frequency × 10−5 |
|---|---|
| Pol V (MBP-UmuC), UmuD′, RecA, SSB | 2,325 ± 408 |
| Component omitted: | |
| UmuD′ | 27 ± 12 |
| MBP-UmuC | 19 ± 2.6 |
| RecA | 25 ± 3.6 |
| SSB | 51 ± 12 |
| MBP-UmuC + MBP | 18 ± 3.4 |
| All proteins | 8 ± 4 |
| Pol III holoenzyme | 98 ± 36 |
| Pol III holoenzyme, UmuD′, RecA, SSB | 81 ± 36 |
| Pol I | 138 ± 52 |
| Pol I, UmuD′, RecA, SSB | 78 ± 12 |
| Pol II | 148 ± 40 |
| Pol II, UmuD′, RecA, SSB | 65 ± 22 |
Gap-filling replication reactions were performed with the indicated proteins, after which the DNA products were introduced into an E. coli indicator strain and plated on lactose-eosin/methylene blue (EMB) plates. Mutant (dark-red) and wild-type (white) colonies were counted. Mutation frequency was calculated as described in Materials and Methods. Transformation of untreated intact pOC2 yielded a mutation frequency of 0.9 × 10−5.
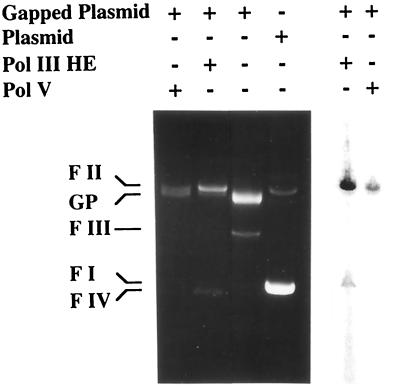
Figure 3.
Gap-filling DNA replication by pol V and pol III holoenzyme. Gap-filling replication was performed with pol V (MBP-UmuC) in the presence of UmuD′, RecA, and SSB, or with pol III holoenzyme, by using gapped pOC2 as a substrate. Reactions were performed in parallel in the presence and the absence of radiolabeled dTTP at 37°C for 20 min. Reaction products were fractionated by agarose gel electrophoresis and visualized by ethidium bromide staining (Left) or phosphorimaging (Right). Based on the analysis of the phosphorimage, the amount of radiolabel incorporated into DNA by pol V was 29.4% of the amount incorporated by pol III holoenzyme. FI, supercoiled plasmid; FII, open, circular plasmid; FIII, linearized plasmid; FIV, covalently closed and relaxed plasmid; GP, gapped plasmid.
The frequencies of specific types of mutations per gene were calculated by multiplying the fraction of that type of mutation out of all sequenced mutants (taken from Table 2) by the overall mutation frequency (presented in Table 1). To estimate base substitution mutation frequency per nucleotide, the mutation frequency per gene was divided by 87, the number of mutable sites in cro. This number is based on sequence data from 600 Cro− mutants that were sequenced in our laboratory over the years that showed that 96% of Cro− mutations mapped in the coding region, and they were distributed over 87 sites, representing 43% of the ORF. For frameshift, the number of mutable sites is the entire ORF (201 nt).
Table 2.
Mutations generated in the cro gene during in vitro gap-filling replication
| Mutation type | No. of mutations
|
||
|---|---|---|---|
| Pol III† | Pol V‡ | No polymerase§ | |
| Base substitution | 24 | 72 | 42 |
| Frameshift | 20 | 20 | 0 |
| Other¶ | 27 | 9 | 0 |
| All mutants | 71 | 101 | 42 |
| Transition | 13 | 23 | 27 |
| A → G | 0 | 0 | 1 |
| C → T | 11 | 5 | 24 |
| G → A | 2 | 2 | 1 |
| T → C | 0 | 16 | 1 |
| Transversion | 11 | 49 | 15 |
| A → C | 2 | 7 | 2 |
| A → T | 0 | 18 | 0 |
| C → A | 0 | 4 | 0 |
| C → G | 0 | 1 | 2 |
| G → C | 5 | 5 | 8 |
| G → T | 2 | 1 | 3 |
| T → A | 2 | 10 | 0 |
| T → G | 0 | 3 | 0 |
Gap-filling replication reactions were performed with the indicated DNA polymerases, after which the DNA products were introduced into an E. coli indicator strain and plated on lactose-EMB plates. Plasmids were extracted from dark-red mutant colonies, and the sequence of their cro gene was determined by DNA sequence analysis. The details are presented in Materials and Methods.
†Replication was performed with pol III holoenzyme.
‡Replication with pol V, in the presence of UmuD′, RecA, and SSB.
§Nonreplicated gapped plasmid was used to transform the indicator strain.
¶ Other mutations include big deletions and insertions as well as complex mutations.
DNA Sequence Analysis of the Mutants.
Mutant colonies were picked and their plasmid contents were extracted. The sequence of the cro gene in these plasmids was determined by the Biological Services Department in our institute by using automated DNA sequence analysis.
Treatment of the Gapped Plasmid with Uracil DNA N-Glycosylase and MutM Glycosylase/AP Lyase.
To eliminate from the gapped plasmid possible spontaneous lesions—i.e., abasic (AP) sites, uracils, and 8-oxoguanines—the gapped pOC2 was treated with uracil DNA N-glycosylase and MutM glycosylase/AP lyase before replication. The reaction mixture contained 26 nM gapped pOC2, 0.025 unit/μl uracil DNA N-glycosylase, 0.83 μM MutM, 0.1 M KCl, 1 μg/μl bovine gamma globulin, 20 mM Tris⋅HCl (pH 8.0), 10 mM NaCl, and 1 mM EDTA. The reaction was carried out at 37°C for 30 min, after which it was terminated by phenol extraction, and the DNA was precipitated with ethanol.
Results
Outline of the Experimental System.
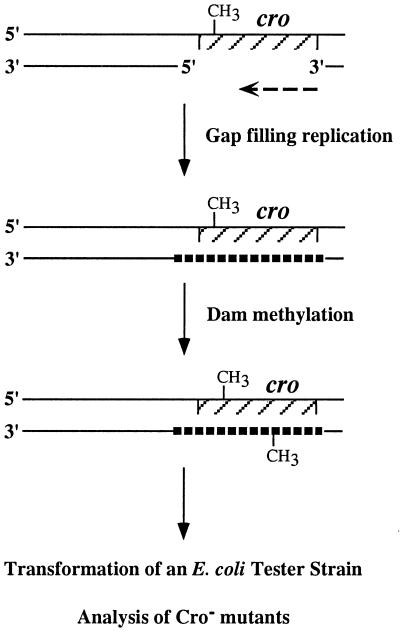
The replication fidelity of pol V was determined by using a fidelity assay (Fig. 2) in which mutations generated in a reporter gene during in vitro replication were analyzed by a subsequent bioassay. The substrate used was a gapped plasmid, carrying the phage λ cro reporter gene in the ssDNA region. The assay consisted of in vitro gap-filling replication with pol V (as a MBP-UmuC fusion protein), UmuD′, RecA, and SSB or with pol III holoenzyme as a control. This was followed by methylation of the plasmid with dam methylase to prevent removal of in vitro generated mutations by in vivo mismatch repair during the propagation in the tester strain. The gap-filled and methylated DNA then was used to transform an indicator E. coli strain, in which Cro− mutations were detected as dark-red colonies over a background of white Cro+ colonies (30, 32, 34).
Figure 2.
Outline of the cro replication fidelity assay.
Gap-Filling Replication by Pol V.
In vitro gap-filling replication of the gapped plasmid with pol III holoenzyme led to efficient filling-in of the 350-nt ssDNA gap. This is indicated by the strong reduction in the amount of the gapped-plasmid DNA band and the appearance of the nicked and covalently closed circular forms of the plasmid (Fig. 3 Left). Similarly, pol V in the presence of UmuD′, RecA, and SSB promoted gap-filling replication, but to a lesser extent, as indicated by the persistence of a fraction of the substrate (Fig. 3 Left). To quantify the differences in DNA synthesis promoted by the two DNA polymerases, the replication reactions were conducted in the presence of α-32P-radiolabeled dTTP. After replication, the substrates were fractionated by agarose gel electrophoresis, and the amount of radiolabel incorporated into the DNA was determined by phosphorimaging. It was found that DNA synthesis promoted by pol V, UmuD′, RecA, and SSB amounted to 29.4% of that of pol III holoenzyme (Fig. 3 Right).
Replication by Pol V Is Error-Prone.
The fidelity of the gap-filling replication reaction was determined by transforming an indicator strain with the replication products and scoring Cro− mutant colonies on lactose-EMB plates. The plasmid replicated by pol III holoenzyme yielded a mutation frequency of 98 × 10−5 per gene, 12-fold higher than the nonreplicated, gapped DNA (8 × 10−5; Table 1). The intact untreated plasmid pOC2 gave even a lower mutation frequency of 0.9 × 10−5. Experiments performed with pol I or pol II yielded mutation frequencies of 138 × 10−5 and 148 × 10−5, respectively, indicating a similar fidelity of all three DNA polymerases in this assay system (Table 1).
When replication was conducted with pol V (as an MBP-UmuC fusion protein) in the presence of UmuD′, SSB, and RecA, the overall Cro− mutation frequency was 2,325 × 10−5, 24-fold higher than pol III holoenzyme. This frequency reflects primarily errors made by pol V during the replication reaction, because its omission from the reaction led to a drastic 122-fold decrease in mutation frequency (19 × 10−5; Table 1). We have shown previously that gap-filling replication by pol V requires UmuD′, RecA, and SSB (9). Similarly, the formation of Cro− mutations during the in vitro reaction required all components: omission of each of pol V, UmuD′, RecA, or SSB abolished the increase of mutagenesis (Table 1). Using the MBP tag instead of the MBP-UmuC protein also abolished the mutagenic effect (Table 1). Addition of UmuD′, RecA, and SSB to pol I, pol II, or pol III holoenzyme did not decrease the fidelity of these DNA polymerases in the gap-filling assay (Table 1). This shows that pol V could not be replaced by any of these DNA polymerases, suggesting that the effect of UmuD′, RecA, and SSB is specific to pol V. These results on the error-prone nature of pol V are consistent with those of Tang et al. (36), who recently reported that a purified UmuD′2C complex promoted misinsertion during DNA synthesis.
Pol V-Generated Mutations Are Mainly Transversions.
DNA sequence analysis of 214 mutants was performed to examine the specificity of the in vitro generated mutations. It was found that pol III holoenzyme produced transitions, transversions, and frameshifts, together with more complex events, mostly deletions (Fig. 4; Table 2). The major class of mutation generated by pol III holoenzyme was frameshift mutation (45% of the point mutations; Tables 2 and 3), which was formed with an average frequency of 1.4 × 10−6 per nucleotide (Table 3). A dominance of frameshifts among mutations generated by pol III holoenzyme was observed recently in two other systems based on the lacI (37) and rpsL (38) reporter genes. The frequency of base-substitution mutations generated by pol III holoenzyme in our system was 33.1 × 10−5 per gene or approximately 3.8 × 10−6 per nucleotide. This is made up of transitions at 2.1 × 10−6 per nucleotide and transversions at 1.7 × 10−6 per nucleotide (Table 3). The mutagenic specificity of pol III holoenzyme remained essentially unchanged when replication was performed with pol III holoenzyme in the presence of SSB and RecA (data not shown).
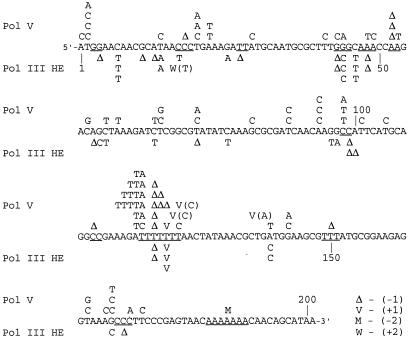
Figure 4.
Spectra of mutations generated in the cro gene during in vitro replication by pol V or by pol III holoenzyme. The 201 nt of the coding region of cro are shown. Mutations generated by pol V are shown above the cro sequence, whereas mutations generated by pol III holoenzyme are shown underneath the sequence. Δ, −1 deletion; M, −2 deletion; V, +1 insertion next to the marked nucleotide. The identity of the inserted nucleotide is shown in parentheses, unless it is identical to the template nucleotide after which it was inserted. W, +2 insertion. In addition to mutations in the coding region, eight mutations were found upstream to cro: for pol V, two C → A and one each of T → G, T → A, C → T, +T; for pol III holoenzyme, C → T and G → T, one each.
Table 3.
Frequency of major classes of mutations generated during in vitro gap-filling replication by pol III holoenzyme and by pol V
| Mutation | Mutation frequency × 10−5 per cro gene
|
Mutation frequency × 10−5 per nucleotide
|
Pol V/Pol III | ||
|---|---|---|---|---|---|
| Pol III | Pol V | Pol III | Pol V | ||
| Base substitution | 33.1 | 1,657 | 0.38 | 19.0 | 50 |
| Transition | 17.9 (30%) | 529 (25%) | 0.21 | 6.1 | 30 |
| Transversion | 15.2 (25%) | 1,128 (53%) | 0.17 | 13.0 | 74 |
| Frameshift | 27.6 (45%) | 460 (22%) | 0.14 | 2.3 | 17 |
| Total point mutations | 60.7 | 2,118 | 0.52 | 21.3 | 35/41* |
Mutation frequency per cro gene was calculated based on the data presented in Tables 1 and 2. Mutation frequency per nucleotide was calculated by dividing the mutation frequency per gene by the number of mutable sites (87 for base substitution and 201 for frameshift), as described in Materials and Methods.
The ratio is 35 for mutation frequency per gene and 41 for mutation frequency per nucleotide. The difference stems from the fact that the number of mutable sites is different for base substitutions and frameshifts.
When replication was performed with pol V in the presence of RecA, UmuD′, and SSB, the frequency of all types of mutations was much higher compared with pol III holoenzyme. The most dramatic difference was in transversion mutations, which were generated by pol V at a frequency 74-fold higher than pol III holoenzyme, reaching an average value of 1.13% per gene or 13.0 × 10−5 per nucleotide (Fig. 4; Tables 2 and 3). Also, other types of mutations were higher: transitions were 30-fold higher (0.53% per gene; 6.1 × 10−5 per nucleotide), and frameshifts were 17-fold higher (0.46% per gene; 2.3 × 10−5 per nucleotide) than with pol III holoenzyme. The control nonreplicated gapped pOC2 produced a distinct spectrum composed of exclusively base substitutions, of which 64% were transitions, mostly (89%) C → T (Table 2).
Analysis of the mutational spectra revealed that the most pronounced differences in specificity between the two polymerases were in the formation of A⋅A, T⋅G, T⋅T, C⋅T, A⋅G, and T⋅C mismatches (the template nucleotides are underlined), which were formed by pol V at frequencies 49- to 296-fold higher than by pol III holoenzyme (Table 4). The types of mismatches formed in DNA most frequently by pol V (≈0.1–0.4% per cro gene each) were A⋅A ≈ T⋅G > T⋅T > A⋅G > G⋅G ≈ C⋅A ≈ C⋅T (Table 4).
Table 4.
Frequency of specific types of base substitution mutations generated during in vitro gap-filling replication by pol V and by pol III holoenzyme
| Mutation | Mismatch* | Mutation frequency × 10−5 per gene
|
Pol V/Pol III | |
|---|---|---|---|---|
| Pol III | Pol V | |||
| Transition | ||||
| A → G | A⋅C | <1.4 | <23.0 | — |
| C → T | C⋅A | 15.2 | 115.1 | 8 |
| G → A | G⋅T | 2.8 | 46.0 | 16 |
| T → C | T⋅G | <1.4 | 368.3 | >263 |
| Transversion | ||||
| A → C | A⋅G | 2.8 | 161.1 | 58 |
| A → T | A⋅A | <1.4 | 414.4 | >296 |
| C → A | C⋅T | <1.4 | 92.1 | >66 |
| C → G | C⋅C | <1.4 | 23.0 | >16 |
| G → C | G⋅G | 6.9 | 115.1 | 17 |
| G → T | G⋅A | 2.8 | 23.0 | 8 |
| T → A | T⋅T | 2.8 | 230.2 | 82 |
| T → G | T⋅C | <1.4 | 69.1 | >49 |
Mutation frequency was calculated based on the data in Tables 1 and 2. The mismatches formed most frequently by pol V, and their frequencies are underlined. The largest differences between pol V and pol III holoenzyme are in boldface type.
The mismatches that gave rise to the observed mutations. The template nucleotide in each pair is shown first.
The gapped cro gene contains a run of seven T residues (Fig. 4; nucleotides 119–125), which was a mutational hot spot for both pol III holoenzyme and pol V. Eighteen of 101 mutations generated by pol V were located in this run, including 11 frameshifts and 7 base substitutions (Fig. 4). Five of the 71 mutations generated by pol III holoenzyme were located in this run, all of them frameshift mutations. The abundance of frameshift mutations in this T run is most likely due to slippage of the DNA polymerases (39, 40). A strong hot spot unique to pol V was located at the first nucleotide 5′ to the T run, where 12 A → T transversions were found. Interestingly, none of the 71 pol III holoenzyme mutations mapped in this site. Long T runs were shown previously to cause pauses during DNA synthesis with purified DNA polymerases (41), events that might facilitate slippage or misinsertion. In addition, long A:T runs form bent DNA (42), and that might have affected misincorporation.
The Common Spontaneous DNA Lesions Are Not the Cause of the in Vitro Generated Mutations.
A critical question is whether the observed mutations result from translesion replication of spontaneous DNA lesions in the DNA, rather than from the infidelity of pol V. The most common spontaneous DNA lesions currently known are apurinic sites, 8-oxoguanine, and uracil (43, 44). Based on their published rates of formation (44), no significant amount of spontaneous lesions was expected to accumulate. However, as a precaution, the gapped DNA was treated with purified uracil DNA N-glycosylase and the MutM glycosylase/AP lyase before replication. This combination of enzymes caused nicks in double-stranded DNA and in ssDNA at abasic sites, uracil, and 8-oxoguanine (44), as we have verified with substrates containing site-specific lesions (data not shown). Therefore, such a treatment of the gapped plasmid before replication was expected to cause linearization of plasmid molecules containing the most common spontaneous lesions. Because linear plasmid DNA transforms E. coli cells very poorly, the treatment was expected to eliminate substrate molecules carrying spontaneous lesions from the assay. It was found that the treatment did not reduce mutation frequency of the replicated DNA (data not shown), arguing against the involvement of the known spontaneous DNA lesions.
Another line of evidence against the involvement of the known spontaneous lesions in this system was the specificity of in vitro generated mutations. AP sites generate primarily G → T and A → T transversions, 8-oxoguanine produces primarily G → T transversions, and deamination of C produces C → T transitions (44). Of these three types of mutations, only A → T was observed at a significant frequency, but, even so, it comprised only 18% of all mutants, and most of them were located at a single hot spot (position 118). In addition, two of the major mutational events promoted by pol V were at T residues (T → A and T → C; Tables 2 and 4), where no significant spontaneous DNA lesion is known to be formed. Taken together, these results suggest that DNA lesions are not responsible for the mutations generated during in vitro replication by pol V.
Discussion
The gap-filling DNA replication by pol V, described above, is characterized by three elements: (i) it requires UmuD′, RecA, and SSB (Table 1); (ii) it is highly mutagenic, generating point mutations (base substitutions and frameshifts) at a frequency 35-fold higher than pol III holoenzyme (Table 3); and (iii) it has a distinct mutational specificity, namely, the tendency to form transversions (Tables 3 and 4). Whereas the spectrum of mutations generated by pol III holoenzyme was dominated by frameshifts (45%), the spectrum of pol V was dominated by transversion (53%). These features are similar to those of untargeted mutagenesis, a branch of SOS mutagenesis that occurs at undamaged DNA regions, also termed SOS mutator activity (6, 10, 12, 14, 44). Chromosomal untargeted mutagenesis was shown to require UmuC (pol V), UmuD′, and RecA (1, 10, 12–14) and produced preferentially transversion mutations (14–17). These similarities suggest that replication of undamaged DNA by pol V, UmuD′, RecA, and SSB is the mechanistic basis for SOS untargeted mutagenesis.
Under which circumstances might pol V produce mutations during SOS? It is possible that pol V acts in ssDNA gaps that are formed during DNA transactions in SOS-induced cells. Such gaps may be formed even in the absence of DNA damage, e.g., when replication is interrupted at some higher-order structures in DNA, or during the processes of recombination or transposition. In addition, pol V may produce mutations in the vicinity of lesions. Thus, when the replication fork is blocked at a DNA lesion, pol V, which is recruited to perform lesion bypass, might proceed well beyond the lesion, leading to an increased frequency of mutations downstream to the lesion (hitchhiking mutations; ref. 45).
The mutagenic DNA synthesis by pol V generates base pair mismatches, which might be substrates for the mismatch repair (MMR) system. Indeed, it was shown previously that in mutants defective in MMR, SOS untargeted mutations were higher than in MMR-proficient cells, indicating that untargeted mutations are subjected to mismatch correction (14, 46). Interestingly, this increase was mainly in transition, not transversion mutations (14). MMR is very effective in preventing transition mutations (i.e., correcting purine–pyrimidine mismatches) and frameshifts, but it is less efficient in preventing transversions (i.e., correcting purine–purine or pyrimidine–pyrimidine mismatches) (14, 47, 48). This specificity of MMR is well suited to correct replication errors, because the replicative polymerase, pol III holoenzyme, produces primarily frameshifts and transitions (75% of all point mutations; Table 3; see also refs. 37, 38, and 48). In addition to transversions, pol V also generates in vitro mismatches, which lead to transitions and frameshifts at high frequencies (Tables 3 and 4). When this occurs in the cell under in vivo SOS conditions, these mismatches are likely to be repaired by MMR. Thus, the net result of the activities of pol V and MMR would be to generate transversion mutations with a specificity higher than expected based solely on the fidelity of pol V. That pol V generates mutations that can escape mismatch repair is consistent with the notion that SOS has evolved as a means of increasing genetic diversity under stress, thereby accelerating adaptation of bacterial populations to hostile environments (12, 49, 50).
Acknowledgments
We thank Myron Goodman (University of Southern California, Los Angeles) for his generous gift of DNA polymerase II and J. Laval (Institute Gustave Roussy, Villejuif, France) and S. Boiteux (Commissariat Energie Atomique, Fontenay Aux Roses, France) for their generous gift of E. coli MutM DNA N-glycosylase. This research was supported by a grant from The U.S.–Israel Binational Science Foundation (96-00448). Z.L. is an incumbent of The Maxwell Ellis Professorial Chair in Biomedical Research.
Abbreviations
- ssDNA
single-stranded DNA
- SSB
ssDNA-binding protein
- pol III and V
DNA polymerases III and V
- MBP
maltose-binding protein
- EMB
eosin/methylene blue
- MMR
mismatch repair
References
- 1.Witkin E M. Bacteriol Rev. 1976;40:869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Little J W, Mount D W. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- 3.Sassanfar M, Roberts J F. J Mol Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 4.Eggleston A K, West S C. Trends Genet. 1996;12:20–26. doi: 10.1016/0168-9525(96)81384-9. [DOI] [PubMed] [Google Scholar]
- 5.Cox M M. Genes Cells. 1998;3:65–78. doi: 10.1046/j.1365-2443.1998.00175.x. [DOI] [PubMed] [Google Scholar]
- 6.Livneh Z, Cohen-Fix O, Skaliter R, Elizur T. CRC Crit Rev Biochem Mol Biol. 1993;28:465–513. doi: 10.3109/10409239309085136. [DOI] [PubMed] [Google Scholar]
- 7.Walker G C. Trends Biochem Sci. 1995;20:416–420. doi: 10.1016/s0968-0004(00)89091-x. [DOI] [PubMed] [Google Scholar]
- 8.Tang M, Shen X, Frank E G, O'Donnell M, Woodgate R, Goodman M F. Proc Natl Acad Sci USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reuven N B, Arad G, Maor-Shoshani A, Livneh Z. J Biol Chem. 1999;274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- 10.Witkin E M. Proc Natl Acad Sci USA. 1974;71:1930–1934. doi: 10.1073/pnas.71.5.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.George J, Castellazzi M, Buttin G. Mol Gen Genet. 1975;140:309–332. [PubMed] [Google Scholar]
- 12.Witkin E M, Wermundsen I E. Cold Spring Harbor Symp Quant Biol. 1979;43:881–886. doi: 10.1101/sqb.1979.043.01.095. [DOI] [PubMed] [Google Scholar]
- 13.Ciesla Z. Mol Gen Genet. 1982;186:289–300. doi: 10.1007/BF00331866. [DOI] [PubMed] [Google Scholar]
- 14.Fijalkowska I J, Dunn R L, Schaaper R M. J Bacteriol. 1997;179:7435–7445. doi: 10.1128/jb.179.23.7435-7445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller J H, Low K B. Cell. 1984;37:675–682. doi: 10.1016/0092-8674(84)90400-8. [DOI] [PubMed] [Google Scholar]
- 16.Yatagai F, Halliday J A, Glickman B W. Mol Gen Genet. 1991;230:75–80. doi: 10.1007/BF00290653. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe-Akanuma M, Woodgate R, Ohta T. Mutat Res. 1997;373:61–66. doi: 10.1016/s0027-5107(96)00189-3. [DOI] [PubMed] [Google Scholar]
- 18.Ichikawa-Ryo H, Kondo S. J Mol Biol. 1975;97:77–92. doi: 10.1016/s0022-2836(75)80023-4. [DOI] [PubMed] [Google Scholar]
- 19.Maenhaut-Michel G, Caillet-Fauquet P. J Mol Biol. 1984;177:181–187. doi: 10.1016/0022-2836(84)90064-0. [DOI] [PubMed] [Google Scholar]
- 20.Brotcorne-Lannoye A, Maenhaut-Michel G. Proc Natl Acad Sci USA. 1986;83:3904–3908. doi: 10.1073/pnas.83.11.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caillet-Fauquet P, Maenhaut-Michel G. Mol Gen Genet. 1988;213:491–498. doi: 10.1007/BF00339621. [DOI] [PubMed] [Google Scholar]
- 22.Kim S R, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H. Proc Natl Acad Sci USA. 1997;94:13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood R D, Hutchinson F. J Mol Biol. 1984;173:293–305. doi: 10.1016/0022-2836(84)90122-0. [DOI] [PubMed] [Google Scholar]
- 24.Wagner J, Gruz P, Kim S R, Yamada M, Matsui K, Fuchs R P P, Nohmi T. Mol Cell. 1999;4:281–286. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- 25.Kenyon C J, Walker G C. Proc Natl Acad Sci USA. 1980;77:2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reuven N B, Tomer G, Livneh Z. Mol Cell. 1998;2:191–199. doi: 10.1016/s1097-2765(00)80129-x. [DOI] [PubMed] [Google Scholar]
- 27.Cull M G, McHenry C S. Methods Enzymol. 1995;262:22–35. doi: 10.1016/0076-6879(95)62005-2. [DOI] [PubMed] [Google Scholar]
- 28.Lohman T M, Overman L B. J Biol Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- 29.Cox M M, McEntee K, Lehman I R. J Biol Chem. 1981;256:4676–4678. [PubMed] [Google Scholar]
- 30.Cohen-Fix O, Livneh Z. Proc Natl Acad Sci USA. 1992;89:3300–3304. doi: 10.1073/pnas.89.8.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen-Fix O, Livneh Z. J Biol Chem. 1994;269:4953–4958. [PubMed] [Google Scholar]
- 32.Skaliter R, Eichenbaum Z, Shwartz H, Ascarelli-Goell R, Livneh Z. Mutat Res. 1992;267:139–151. doi: 10.1016/0027-5107(92)90118-l. [DOI] [PubMed] [Google Scholar]
- 33.Barak Y, Cohen-Fix O, Livneh Z. J Biol Chem. 1995;270:24174–24179. doi: 10.1074/jbc.270.41.24174. [DOI] [PubMed] [Google Scholar]
- 34.Tomer G, Cohen-Fix O, O'Donnell M, Goodman M, Livneh Z. Proc Natl Acad Sci USA. 1996;93:1376–1380. doi: 10.1073/pnas.93.4.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barzilai R. J Mol Biol. 1973;74:739–742. doi: 10.1016/0022-2836(73)90062-4. [DOI] [PubMed] [Google Scholar]
- 36.Tang M, Bruck I, Eritja R, Turner J, Frank E G, Woodgate R, O'Donnell M, Goodman M F. Proc Natl Acad Sci USA. 1998;95:9755–9760. doi: 10.1073/pnas.95.17.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pham P T, Olson M W, McHenry C S, Schaaper R M. J Biol Chem. 1998;273:23575–23584. doi: 10.1074/jbc.273.36.23575. [DOI] [PubMed] [Google Scholar]
- 38.Fujii S, Akiyama M, Aoki K, Sugaya Y, Higuchi K, Hiraoka M, Miki Y, Saitoh N, Yoshiyama K, Ihara K, et al. J Mol Biol. 1999;289:835–850. doi: 10.1006/jmbi.1999.2802. [DOI] [PubMed] [Google Scholar]
- 39.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Cold Spring Harbor Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 40.Kunkel T A. Biochemistry. 1990;29:8003–8011. doi: 10.1021/bi00487a001. [DOI] [PubMed] [Google Scholar]
- 41.Weisman-Shomer P, Dube D K, Perrino F W, Stokes K, Loeb L A, Fry M. Biochem Biophys Res Commun. 1989;164:1149–1156. doi: 10.1016/0006-291x(89)91789-0. [DOI] [PubMed] [Google Scholar]
- 42.Koo H S, Wu H M, Crothers D M. Nature (London) 1986;320:501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- 43.Lindahl T. Nature (London) 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 44.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 45.Ruiz R M, Bridges B A. Mol Gen Genet. 1987;208:542–548. doi: 10.1007/BF00328153. [DOI] [PubMed] [Google Scholar]
- 46.Caillet-Fauquet P, Maenhaut-Michel G, Radman M. EMBO J. 1984;3:707–712. doi: 10.1002/j.1460-2075.1984.tb01873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaaper R M, Dunn R L. Proc Natl Acad Sci USA. 1987;84:6220–6224. doi: 10.1073/pnas.84.17.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaaper R M. J Biol Chem. 1993;268:23762–23765. [PubMed] [Google Scholar]
- 49.Radman M. In: Molecular Mechanisms for Repair of DNA. Hanawalt P, Setlow R B, editors. New York: Plenum; 1975. pp. 355–367. [Google Scholar]
- 50.Echols H. Cell. 1981;25:1–2. doi: 10.1016/0092-8674(81)90223-3. [DOI] [PubMed] [Google Scholar]