Abstract
The COP9 signalosome (CSN) is involved in multiple developmental processes. It interacts with SCF ubiquitin ligases and deconjugates Nedd8/Rub1 from cullins (deneddylation). CSN is highly expressed in Arabidopsis floral tissues. To investigate the role of CSN in flower development, we examined the expression pattern of CSN in developing flowers. We report here that two csn1 partially deficient Arabidopsis strains exhibit aberrant development of floral organs, decline of APETALA3 (AP3) expression, and low fertility in addition to defects in shoot and inflorescence meristems. We show that UNUSUAL FLORAL ORGANS (UFO) forms a SCFUFO complex, which is associated with CSN in vivo. Genetic interaction analysis indicates that CSN is necessary for the gain-of-function activity of the F-box protein UFO in AP3 activation and in floral organ transformation. Compared with the previously reported csn5 antisense and csn1 null mutants, partial deficiency of CSN1 causes a reduction in the level of CUL1 in the mutant flowers without an obvious defect in CUL1 deneddylation. We conclude that CSN is an essential regulator of Arabidopsis flower development and suggest that CSN regulates Arabidopsis flower development in part by modulating SCFUFO-mediated AP3 activation.
INTRODUCTION
The COP9 signalosome (CSN), previously known as the COP9 complex, was identified genetically as a negative regulator of photomorphogenesis. Null mutations in the CSN subunits lead to a constitutively photomorphogenic/deetiolated/fusca (cop/det/fus) phenotype, which also is associated with seedling lethality (for reviews, see Wei and Deng, 1996, 1999). CSN consists of eight subunits, CSN1 to CSN8, six of which have been reported to correspond to six of the cop/det/fus loci of Arabidopsis (Schwechheimer and Deng, 2001; Serino et al., 2003). Homologs of the Arabidopsis CSN have been found in humans, flies, and fission yeast (Deng et al., 2000).
CSN has been shown to interact with the SCF (Skp1–cullin–F-box protein) family of E3 ubiquitin ligase complexes (Lyapina et al., 2001; Schwechheimer et al., 2001). A typical SCF complex consists of CULLIN1 (CUL1), a small RING-H2 protein, Rbx1/Roc1/Hrt1, Skp1, and an F-box protein that recruits the substrates for ubiquitination (for review, see Deshaies, 1999). The ubiquitinated proteins, in most cases, are targeted for degradation by the 26S proteasome. Cullins are covalently modified with a ubiquitin-like protein, Nedd8/Rub1, in a conjugation reaction referred to as neddylation. CSN is responsible for cullin deneddylation (Cope et al., 2002). csn null mutants lack deneddylation activity and accumulate hyperneddylated CUL1 (Lyapina et al., 2001; Schwechheimer et al., 2001; Zhou et al., 2001; Mundt et al., 2002; Wang et al., 2002). In addition, CSN has been found to associate with protein kinase activities and to interact with numerous transcription factors and other important cellular regulators (Schwechheimer and Deng, 2001; Bech-Otschir et al., 2002). Through these activities, CSN participates in a wide range of signal transduction pathways and developmental processes of eukaryotic organisms (Schwechheimer et al., 2001; Mundt et al., 2002; Suh et al., 2002; Wang et al., 2002; Yang et al., 2002).
As a result of the postseedling lethality of the original Arabidopsis CSN mutants, studies of its role in Arabidopsis development have focused on the seedling stage. More recently, partial loss-of-function csn lines generated by antisense suppression or cosuppression, or by transgenic rescue of null mutants, have revealed that CSN is involved in multifaceted developmental processes throughout the plant life cycle, including flower development (Peng et al., 2001a, 2001b; Schwechheimer et al., 2001; Wang et al., 2002; Serino et al., 2003). In this study, in an effort to dissect the function of CSN1, we focused our investigation on the specific role of CSN during flower development using two weak csn1 mutant lines that we generated previously (Wang et al., 2002).
Through a sophisticated interplay of numerous flower genes, Arabidopsis floral meristem cells follow defined developmental steps to form a flower (Smyth et al., 1990; Irish, 1999). The development of the four floral organ types—sepals, petals, stamens, and carpels—is governed largely by the overlapping activities of three classes of regulatory genes, known as the A, B, and C genes (Coen and Meyerowitz, 1991). In particular, the B-class genes, APETALA3 (AP3) and PISTILLATA (PI), are necessary to specify petals and stamens (Krizek and Meyerowitz, 1996). Expression of the B-class genes requires the function of LEAFY (LFY), APETALA1 (AP1), and UNUSUAL FLORAL ORGANS (UFO) (Ingram et al., 1995; Levin and Meyerowitz, 1995; Wilkinson and Haughn, 1995; Lee et al., 1997; Samach et al., 1999; Ng and Yanofsky, 2001; Lamb et al., 2002). Unlike LFY and AP1, UFO is not a DNA binding transcription factor; instead, it is one of ∼700 F-box proteins in Arabidopsis that are believed to be components of the SCF-type E3 ubiquitin ligases (Gagne et al., 2002). At present, two F-box proteins in Arabidopsis, TIR1 and COI1, have been demonstrated experimentally to form SCFTIR1 (Gray et al., 1999) and SCFCOI1 (Xu et al., 2002) complexes, respectively, and to associate directly with CSN in vivo (Schwechheimer et al., 2001; Feng et al., 2003). Similarly, UFO has been shown to interact genetically and in yeast two-hybrid assays with the SKP1 homolog ASK1 (Samach et al., 1999; Zhao et al., 2001), although the mechanisms of UFO in the activation of class-B genes are unclear.
We report here the in situ expression pattern of CSN in developing flowers. Furthermore, we show that UFO forms an SCFUFO complex that interacts with CSN and that CSN1 is required for UFO-dependent AP3 expression. Thus, we substantiate the working model that CSN regulates development by modulating specific SCF function.
RESULTS
Expression Pattern of CSN during Flower Development
CSN is expressed widely and is highly enriched in flowers based on protein (Staub et al., 1996; Kwok et al., 1999) and RNA gel blot analyses (data not shown). However, the spatial and temporal expression patterns of CSN during flower development have not been reported. Thus, we determined the expression pattern of CSN in developing Arabidopsis flowers by immunohistochemical staining using antibodies against CSN4 and CSN6.
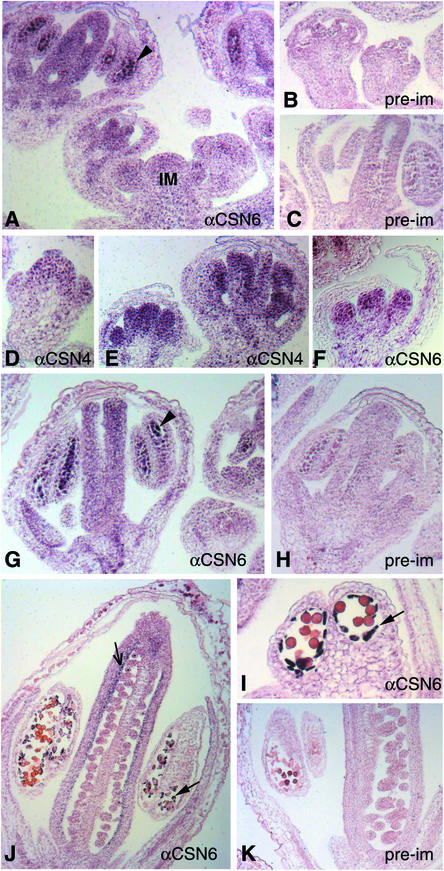
At early stages, CSN expression appeared widespread, with considerable staining in the inflorescence and floral meristems (Figures 1A and 1D). The staining was particularly prominent in the primordia of the inner whorls (whorls 2 to 4) after stage 5 (Figures 1E and 1F) compared with that in the preimmune control (Figures 1B, 1C, 1H, and 1K). We noticed that the nuclei were stained most intensely in these cells, consistent with CSN being a nucleus-enriched complex (Chamovitz et al., 1996; Staub et al., 1996). After stage 9 (Smyth et al., 1990), CSN accumulated in the pollen mother cells of the stamens (Figures 1A and 1G, arrowheads) and in the carpels. During later stages of flower development, CSN was particularly enriched in the inner carpel wall (Figure 1J, lined arrow) and in the tapetal cells of anthers (Figures 1J and 1I, solid arrows). Greater accumulation of CSN in these cell types likely corresponds to a function(s) of CSN in the corresponding stages of flower development.
Figure 1.
CSN Expression Pattern in Developing Arabidopsis Flowers.
Sections of flowers at various stages were immunostained with antibodies against CSN4 (αCSN4) ([D] and [E]) and CSN6 (αCSN6) ([A], [F], [G], [I], and [J]). Preimmune antiserum was used in an identical procedure as a background control ([B], [C], [H], and [K]). Arrowheads indicate pollen mother cells. The thin arrow in (J) indicates the carpel inner wall staining. Solid arrows indicate tapetal cells. IM, inflorescence meristem.
A CSN1 Deficiency Results in Aberrant Development of the Shoot Apex and Flowers
We previously described the characterization of a series of transgenic plants carrying CSN1 and its mutant derivatives in a csn1 null background, fus6-1 (Wang et al., 2002). Here, we focus on two lines, fus6/CSN1-11 (fus6-1/fus6-1; CSN1-11) and fus6/N270 (fus6-1/fus6-1; N270).
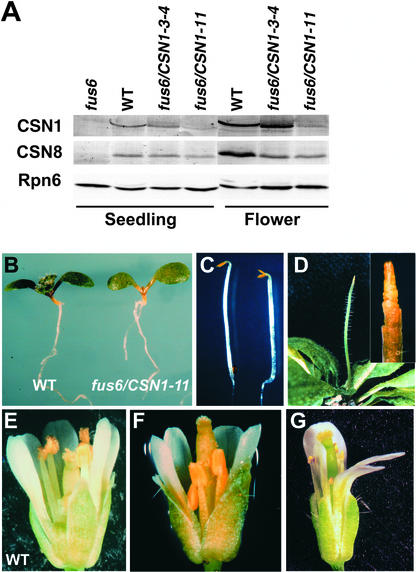
Most of the wild-type CSN1 transgenic lines (fus6/CSN1) completely rescued the mutant phenotype (Wang et al., 2002). However, the fus6/CSN1-11 line exhibited a low level of expression of the CSN1 transgene in both seedlings and flowers compared with other fus6/CSN1 lines, such as fus6/CSN1-3-4, and endogenous CSN1 in wild-type plants (Figure 2A). The level of CSN8, a subunit whose accumulation depends on CSN1 (Wang et al., 2002), displayed a slight reduction. This finding suggests that the amount of the complex as a whole is affected, but only to a limited extent. As in previous reports, we also found a higher abundance of CSN in flowers than in seedlings (Figure 2A).
Figure 2.
Characterization of fus6/CSN1-11 Phenotype.
(A) Anti-CSN1 and anti-CSN8 immunoblots showing the levels of CSN1 and CSN8 endogenous and transgene products in fus6/CSN1-11 compared with the wild type (WT), the original fus6-1 mutant, and the fus6 mutant containing a normal CSN1 transgenic allele, fus6/CSN1-3-4. Extracts were made from seedlings and flowers as indicated. Equal amounts of total protein were loaded in each lane and was verified by immunoblotting against proteasome subunit Rpn6 using a membrane containing identical samples.
(B) and (C) Seedling morphology of fus6/CSN1-11 (right) compared with the wild type (left) grown in the light (B) or in the dark (C).
(D) A fus6/CSN1-11 plant showing the meristem phenotype resembling the pin1 mutant. An enlarged image is shown in the inset.
(E) to (G) An early-arising (F) and a later-arising (G) fus6/CSN1-11 flower are shown in comparison with a wild-type flower (E).
Consistent with the lower expression levels of the CSN1 transgene product, fus6/CSN1-11 displayed abnormal phenotypes that were not found in other fus6/CSN1 lines. The light-grown fus6/CSN1-11 seedlings grew at a slower rate, judging by the initial appearance of the true leaves (Figure 2B), an event indicating the transition from seedling to the vegetative/juvenile stage. In the dark, fus6/CSN1-11 developed opened cotyledons (Figure 2C), indicating a slight defect in its ability to undergo etiolation in the dark. Under normal growth conditions, the fus6/CSN1-11 plants frequently were found to exhibit a shoot apex phenotype similar to that of the pin-formed1 (pin1) mutant (Gälweiler et al., 1998), in which the shoot apex aborts its normal developmental program and terminates (Figure 2D). PIN1 is a putative auxin carrier required for organ outgrowth, separation, and positioning (Vernoux et al., 2000). This phenotype accounted for ∼10% of the fus6/CSN1-11 shoots. Lateral shoots eventually grew and enabled the plant to enter the reproductive stage. The fus6/CSN1-11 plants were able to flower, but with poor seed yield. Upon closer inspection of the flowers, we found that stamens usually were shorter or reduced in number and contained considerably fewer pollen grains (Figures 2F and 2G). In addition, the number of petals was reduced in the later-arising flowers (Figure 2G).
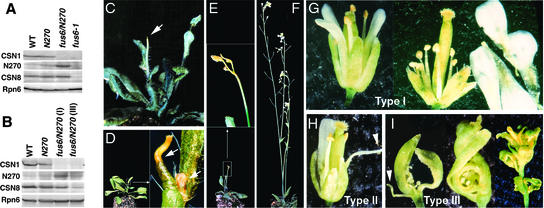
Both the shoot apex and flower phenotypes also were evident in the fus6/N270 line, which carries a transgene expressing a CSN1 N-terminal 270–amino acid fragment in the absence of endogenous CSN1 (Wang et al., 2002). The N270 fragment contains a partial proteasome, COP9, initiation factor 3 domain critical for complex assembly but lacks the C-terminal domain that also plays a structural role. The fus6/N270 seedlings contained normal levels of the CSN complex, as determined by assessment of the CSN8 level (Figure 3A), and they displayed a seedling morphology indistinguishable from that of the wild type (Wang et al., 2002). However, after the seedling stage, fus6/N270 starts to exhibit a range of phenotypes that is more severe than that of fus6/CSN1-11 (Figures 3C to 3I). Approximately 30% of fus6/N270 plants were unable to flower and arrested at the vegetative stage, largely as a result of senescence of the inflorescence apex at various stages of bolting (indicated by arrows in Figures 3C to 3E). The rosette or cauline leaves appeared normal.
Figure 3.
Characterization of fus6/N270 Phenotype.
(A) and (B) Immunoblot analyses to determine the levels of N270 transgene expression and endogenous CSN1 and CSN8 in seedlings (A) and flowers (B). The genotypes of the plant samples are labeled above each lane. The anti-CSN1 antibody can react with both endogenous CSN1 and the N270 transgene product. Equal amounts of total proteins were loaded and verified by anti-Rpn6 blotting. WT, wild type.
(C) to (I) Adult phenotypes of fus6/N270. The fus6/N270 plants in (C) to (E) are 5 to 7 weeks old and display premature senescence of the apical region of the inflorescence apices before (C) or at the onset of (E) flower initiation. An enlarged image is shown in the inset in (E). The plant shown in (D) exhibits senescence of both the primary and secondary inflorescence apices (indicated by arrows), as shown in the enlarged image in the inset. The plant shown in (F) displays normal vegetative growth but develops a nearly sterile flower phenotype, as shown in (G) to (I). Type-I flowers (G) contain a complete set of floral organs, but their stamens are shorter and produce fewer mature pollen grains. The petals of the flower at right were pulled out manually and were placed on the side to expose shortened stamens. Type-II flowers (H) have reduced numbers of petals and stamens. Type-III flowers (I) exhibit defects in all floral organs. Arrowheads mark the filamentous structure.
Only ∼20% of fus6/N270 flowers were able to set seed, but they yielded fewer than 10 seeds per silique. According to the severity of the phenotype, we classified these flowers into three groups. Type-I flowers contained all floral organs but produced very few mature pollen grains and often had shorter stamens (Figure 3G), similar to fus6/CSN1-11 flowers. Weak and shorter stamens also have been observed in the axr1-12;ECR1-C1 double mutants for the Rub1-activating enzymes in Arabidopsis (del Pozo et al., 2002). Type-II flowers (Figure 3H) had drastically reduced numbers of stamens and petals. Sometimes these organs were replaced by filamentous structures similar to those found in ufo mutant flowers (Levin and Meyerowitz, 1995; Wilkinson and Haughn, 1995). The most severe fus6/N270 flowers (type III) exhibited abnormalities in all floral organs (Figure 3I). These completely sterile flowers were devoid of petals and stamens. Carpels were not fused and were deformed. Sepals were missing or reduced in number. In many cases, the entire inflorescence contained multiple deformations, so that it was difficult to discern organ identities or whorls (Figure 3I). To facilitate characterization, we counted organ numbers from inflorescences that contained primarily type-II flowers, because these flowers displayed clear defects, but their organ identities were easily distinguishable (Table 1). These data indicate that petal and stamen formation are most sensitive to the mutation, although all flower organs were affected in type-III flowers.
Table 1.
Floral Organ Numbers in fus6/N270 Type-II Flowers
| Flower Position | Wild Type (Standard) | fus6/N270 (n = 50) |
|---|---|---|
| First whorl | ||
| Sepal/sepal-like | 4 | 3.58 |
| Second whorl | ||
| Petal/petal-like | 4 | 1.10 |
| Filament | 0 | 0.52 |
| Total | 4 | 1.62 |
| Third whorl | ||
| Stamen/stamen-like | 6 | 2.66 |
| Filament | 0 | 0.14 |
| Total | 6 | 2.80 |
| Fourth whorl | ||
| Carpel/carpel-like | 2 | 1.86 |
Although CSN8 accumulated to wild-type levels in the fus6/N270 seedlings (Figure 3A), both CSN8 and N270 were reduced drastically in type-III severe flowers but were reduced only slightly in type-I flowers (Figure 3B). The CSN complex in the fus6/N270 flowers did not differ from that in wild-type flowers in terms of gel-filtration profiles (data not shown). Note that the N270 level is significantly higher in the mutant background (fus6/N270) than in the wild-type background (N270). This is because, in wild-type plants, N270 is unable to compete with the endogenous CSN1 for integration into the complex and therefore is unstable (Wang et al., 2002).
Together, both fus6/CSN1-11 and fus6/N270 plants contained lower amounts of CSN1 throughout or at the adult stage of plant development, respectively; therefore, they represent weak csn1 mutants. No abnormalities were found when the transgenes were in the wild-type background (data not shown). Two major developmental defects of both mutants were in apical meristems and in flowers. In this study, we focus on flower development to further investigate the mechanism of regulation by CSN.
AP3 Expression Is Impaired in fus6/CSN1-11 Flowers
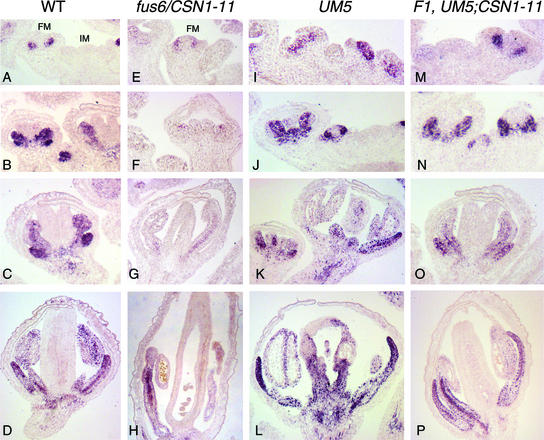
The development of petals and stamens requires the B-class organ identity genes AP3 and PI (Jack et al., 1992; Goto and Meyerowitz, 1994). Because these two organ types are among the most affected in the weak csn1 mutants, we examined the expression of AP3 in fus6/CSN1-11 flowers by immunostaining with anti-AP3 antibody (Jenik and Irish, 2001). In wild-type flowers, AP3 expression was detected starting from stage 3 in the presumptive petal and stamen primordia (Figure 4A) (Jack et al., 1992; Krizek and Meyerowitz, 1996; Lee et al., 1997; Jenik and Irish, 2001). This expression persisted in whorls 2 and 3 throughout flower development (Figures 4B to 4D). In fus6/CSN1-11 flowers, the level of AP3 was reduced throughout the developmental stages (Figures 4E to 4H), whereas the whorl-2 and whorl-3 domain-specific pattern of weak AP3 immunostaining appeared normal. These results indicate that CSN is required for the full expression of AP3.
Figure 4.
AP3 Expression in Wild-Type, fus6/CSN1-11, UM5, and UM5;CSN1-11 F1 Flowers.
AP3 expression was determined by immunostaining.
(A), (E), (I), and (M) Inflorescence meristem and early-stage flowers (stages 2 to 4).
(B), (F), (J), and (N) Stage-5 and -6 flowers.
(C), (G), (K), and (O) Stage-7 to -10 flowers.
(D), (H), (L), and (P) Late-stage flowers that are about to mature.
In fus6/CSN1-11 flowers ([E] to [H]), AP3 level was significantly lower than in the wild type ([A] to [D]). Strong AP3 staining was observed in UM5 flowers ([I] to [L]), which clearly extended into the fourth whorl. However, this ectopic expression of AP3 was abolished by the CSN1-11 allele, and the wild-type pattern was restored in the F1 flowers of UM5;CSN1-11 ([M] to [P]). FM, floral meristem; IM, inflorescence meristem; WT, wild type.
UFO is necessary for AP3 expression in whorls 2 and 3 (Levin and Meyerowitz, 1995; Wilkinson and Haughn, 1995), and it encodes an F-box protein that has been postulated to be a component of SCF ubiquitin ligases (Samach et al., 1999; Zhao et al., 2001). In light of the findings that CSN interacts with and regulates SCF complexes, we set out to determine whether CSN affects AP3 expression by modulating UFO function in the context of SCFUFO.
UFO Interacts with Arabidopsis CUL1, ASK1, and CSN in Vivo
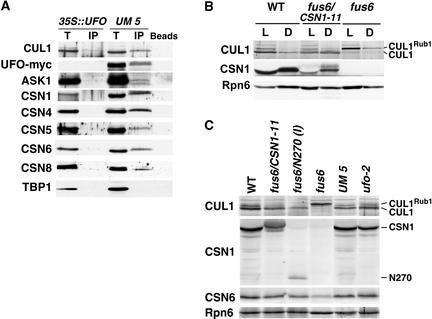
We generated a transgenic line (UM5) that overexpresses a C-terminal myc epitope–tagged UFO protein (UFO-myc). UM5 exhibited a dominant UFO overexpression (UFO-OE) phenotype similar to that described for 35S::UFO plants, in which the flowers are composed primarily of petals and stamens and the leaves are lobed (Lee et al., 1997). To examine the physical association of CSN with UFO in vivo, we immunoprecipitated the myc-tagged UFO protein from the flower extracts. As shown in Figure 5A, UFO-myc readily pulled down endogenous CUL1 as well as ASK1, a SKP1-like molecule in Arabidopsis, strongly supporting the existence of an SCFUFO ubiquitin ligase complex in vivo. Furthermore, all of the CSN subunits tested, but not the TATA Binding Protein (TBP), were coprecipitated with UFO-myc (Figure 5A). These proteins were not pulled down in the 35S:: UFO extract, demonstrating the specificity of UFO-myc in the immunoprecipitation (Figure 5A). Thus, CSN is associated with UFO-myc or SCFUFO in vivo.
Figure 5.
Interaction of UFO-myc with Arabidopsis CUL1 and CSN, and CUL1 Neddylation Level in the csn1 Mutants.
(A) UFO-myc was immunoprecipitated from UM5 and 35S::UFO plant extracts using the anti-myc antibody. The immunocomplexes were examined by immunoblot analysis using the antibodies indicated at left, including Arabidopsis CUL1 (CUL1), myc (UFO-myc), the CSN subunits (CSN1, CSN4, CSN5, CSN6, and CSN8), and TATA Binding Protein1 (TBP1). “Beads” indicates protein A beads alone without charging with antibodies. IP, immunoprecipitation; T, total extract.
(B) Examination of Nedd8/Rub1 modification of Arabidopsis CUL1 in the seedlings of the wild type (WT), fus6/CSN1-11, and fus6-1 by anti-CUL1 immunoblotting. D, dark grown; L, light grown.
(C) Examination of Nedd8/Rub1 modification of Arabidopsis CUL1 in flower extracts from the wild type (WT), fus6/CSN1-11, fus6/N270 type-I flowers (I), the UM-5 transgenic line, and the ufo-2 mutant by anti-CUL1 immunoblotting. Seedling extract of fus6-1 was used to mark the Nedd8/Rub1-modified form of CUL1. Equal amounts of the total protein were loaded. The Nedd8/Rub1-modified and unmodified forms of CUL1 are labeled. The anti-CSN1 blot shows the expression of endogenous CSN1 in the wild type, the transgenic product in fus6/CSN1-11 samples, and the N270 fragment. The anti-Rpn6 blot is shown at bottom.
CUL1 accumulates primarily in the neddylated form in the fus6-1 mutant as a result of a lack of CSN-mediated cullin deneddylation. fus6/CSN1-11 seedlings, like fus6/N270 seedlings (Wang et al., 2002), displayed a CUL1 Nedd8/Rub1 modification pattern similar to that in the wild type (Figure 5B), indicating that the deneddylation activity has been rescued in these mutants seedlings. Moreover, CUL1 deneddylation was not defective in the flowers of these weak csn1 mutants (Figure 5C). These results indicate that the amount of the CSN complex, although lower as a result of the partial deficiency of CSN1, nonetheless is sufficient to effectively deconjugate Nedd8/Rub1 from CUL1. To the contrary, the flowers of fus6/CSN1-11 and fus6/N270 showed a notable decline in the level of CUL1, including the Rub1-modified form (CUL1Rub1), compared with wild type, UM5, and ufo-2 mutant flowers (Figure 5C). However, this effect seems to be flower specific, because no detectable decrease of CUL1 levels was found in the seedlings of these mutants (Figure 5B) (Wang et al., 2002). A decrease in CUL1 abundance likely affects the activity of CUL1-containing SCF complexes, including SCFUFO, and may contribute to the ufo-like phenotype of fus6/CSN1-11 and fus6/N270.
Suppression of the UFO Overexpression Phenotype by CSN1 Weak Transgenic Alleles
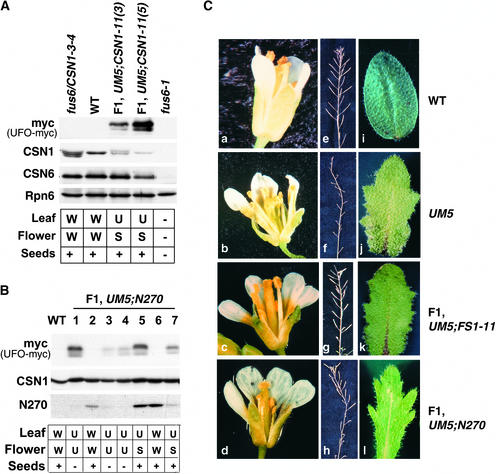
If CSN1 partial deficiency can decrease the activity of SCFUFO, it may abolish the gain-of-function effect caused by UFO-OE. To test this hypothesis, we generated double transgenic plants of UM5 and fus6/CSN1-11 or fus6/N270. Segregation of the transgenes in the F1 plants was followed individually by protein gel blot analysis (Figure 6). Similar to 35S::UFO (Lee et al., 1997), the UM5 plants exhibited a dominant UFO-OE phenotype characterized by missing carpels, supernumerary stamens and petals, lobed leaves, and infertility (Figures 6Cb, 6Cf, and 6Cj). Remarkably, in the F1 plants harboring UM5 plus the CSN1-11 or N270 transgene, carpel formation was rescued completely (Figures 6Cc and 6Cd, Table 2). As a result, fertility was improved significantly, as indicated by the considerable seed yield from these F1 plants (Figures 6Cg and 6Ch). It is remarkable that those F1 plants have a wild-type copy of the FUS6/CSN1 gene, and CSN1-11 and N270 exhibited no phenotype in heterozygous fus6 plants. However, the lobed-leaf phenotype typical of UFO-OE remained unchanged (Figures 6Ck and 6Cl). As shown in Figures 6A and 6B, such a “suppressed UFO-OE phenotype” (S) segregated strictly with the presence of both UM5 and N270 (Figure 6B, lanes 5 and 7) or UM5 and CSN1-11 (Figure 6A, F1 plants). We also made a similar cross between UM5 and a normal CSN1 expression line, fus6/CSN1-3-4 (Figure 6A). The resulting F1 plants exhibited a typical UFO-OE phenotype with characteristic lobed leaves, missing carpels, and infertility (data not shown). Therefore, the UFO-OE phenotype was suppressed only by the CSN1-11 and N270 transgene alleles that cause partial deficiency of CSN1.
Figure 6.
Characterization of the F1 Flower Phenotype between UM5 and fus6/CSN1-11 or fus6/N270 Genetic Crosses.
(A) Phenotype summary of F1 plants from the cross between UM5 and fus6/CSN1-11. Expression of UFO-myc, CSN1, and CSN6 is shown at top. The genotypes of the plants are labeled above the lanes, and their phenotypes are summarized at bottom.
(B) Phenotype summary of F1 plants from the cross between UM5 and fus6/N270. Seven individual F1 plants were genotyped by immunoblot analysis of transgene expression: the myc blot detects UFO-myc; the CSN1 blot detects N270 and the endogenous CSN1. The corresponding phenotype for each F1 plant is summarized at bottom.
W denotes the wild-type phenotype (as shown in [Ca], [Ce], and [Ci]); U denotes the UFO-OE phenotype (as shown in [Cb], [Cf], and [Cj]); and S denotes the suppressed UFO-OE phenotype (as shown in [Cc], [Cd], [Cg], [Ch], [Ck], and [Cl]. + and − indicate the fertility or infertility, respectively, for seed production.
(C) Representative images of flowers ([a] to [d]), siliques showing the seed yield ([e] to [h]), and cauline leaves ([i] to [l]) from the four different genotypes (labeled at right).
Table 2.
Summary of Mutant Transgenic Lines and Their Corresponding Phenotypes
| Genotype
|
||||
|---|---|---|---|---|
|
FUS6 Locus (or CSN1 Locus) |
Transgene UM5 |
Transgene CSN1-11 or N270 |
Transgene CSN1-3-4 |
Flower Phenotype |
| FUS6/fus6 | +/− | −/− | −/− | High and ectopic AP3 expression, UFO-overexpression phenotype |
| FUS6/fus6 | −/− | +/− | −/− | Wild-type AP3 expression pattern, wild-type–like phenotype |
| FUS6/fus6 | +/− (F1) | +/− (F1) | −/− | Wild-type AP3 expression pattern, wild-type–like phenotype |
| FUS6/fus6 | −/− | −/− | +/− | Wild-type–like phenotype |
| FUS6/fus6 | +/− (F1) | −/− | +/− (F1) | UFO-overexpression phenotype |
| fus6/fus6 | +/− | −/− | −/− | Plant not viable |
| fus6/fus6 | −/− | +/− | −/− | Low AP3 expression, defective flowers, ufo mutant–like phenotype |
| fus6/fus6 | +/− (F2) | +/− (F2) | −/− | Defective flowers, ufo mutant–like floral phenotype |
| fus6/fus6 | −/− | −/− | +/− | Wild-type–like phenotype |
AP3 expression requires UFO; therefore, it was used as a downstream marker to determine whether UFO activity is suppressed by CSN1-11. Similar to the 35S::UFO flowers (Lee et al., 1997), UM5 exhibited a broader expression pattern of AP3 (Figures 4I to 4L) that extended beyond the presumptive petal and stamen primordia, compared with the wild type (Figures 4A to 4D). At later stages, the ectopic expression of AP3 in the center of the flower was more evident (Figures 4J to 4L), which correlated with the transformation of carpels into stamens in the UM5 flowers. However, in the presence of the CSN1-11 allele, the AP3 expression domain was restricted to whorls 2 and 3 throughout floral development (Figures 4M to 4P), as it was in wild-type flowers (Figures 4A to 4D), correlating with the rescue of carpels and fertility. The observation that the CSN1-deficient alleles dominantly suppressed the UFO OE phenotype in the F1 plants suggests that UFO activity may be sensitive to the level of CSN1 or CSN. The level and pattern of UFO-myc expression were similar among all of the genotypes, based on our anti-myc antibody immunostaining (data not shown), indicating that it is the activity, and not the expression, of the UM5 transgene that requires full function of CSN1.
The CSN1 Deficiency Phenotype Overrides That Caused by UFO Overexpression
We followed the genetic crosses between UM5 and the weak csn1 mutants to the F2 generation, in which we obtained plants that contained the UM5 transgene in the fus6/N270 background (fus6/fus6; N270; UM5) (Figure 7, lane 8-18). These F2 plants exhibited a characteristic premature arrest, as seen in fus6/N270 plants, resulting in very few plants able to reach the flowering stage. As summarized in Table 2, the flowers from these plants were essentially identical to those from fus6/N270, as described above (Figure 3), and differed drastically from those of UM5 (Figure 6Cb), despite the expression of the UM5 transgene. Nonetheless, all of the F2 plants carrying UM5 developed lobed leaves (Figure 7, bottom). We conclude from these data that the floral phenotype caused by UFO OE is suppressed completely by CSN1 deficiency. These genetic data strongly support the idea that CSN1 level is critical for the function of UFO or SCFUFO in flowers.
Figure 7.
Characterization of Individual F2 Plants from the Cross between UM5 and fus6/N270.
The segregation of phenotypes and the transgenes among F2 plants was analyzed. The segregation of UFO-myc, the N270 transgene product, and the endogenous CSN1 (indicating fus6/fus6 homozygosity) in each of the nine F2 plants was determined by immunoblot analysis. The corresponding phenotype of each F2 plant is summarized at bottom. Note that a fus6/N270 plant harboring the UM5 transgene was obtained (lane 8-18). This plant displayed a typical fus6/N270 phenotype except in the leaves (see text). WT, wild type.
DISCUSSION
In this report, we examined two reduction-of-function lines for CSN1, fus6/CSN1-11 and fus6/N270, both of which contained lower amounts of CSN1 throughout or at the adult stage of plant development, respectively. Analysis of these two lines revealed the essential roles of CSN in apical meristem and flower development. Although those phenotypes are pleiotropic in nature, they share many defects with the ufo mutants. A detailed analysis of the relationship of CSN with UFO-mediated flower development suggests that CSN plays a role in flower development through interaction with and possibly regulation of SCFUFO.
Evidence That UFO Forms an SCFUFO Complex with Arabidopsis CUL1 and ASK1
The SCF ubiquitin ligase complexes represent a major type of E3 enzyme that regulates multiple signaling pathways, mostly by targeting important regulators for degradation in a cell-signaling-dependent manner. In Arabidopsis, there are 11 cullin-related genes, 19 SKP1-like genes, including ASK1 and ASK2, and ∼700 F-box–containing proteins (Callis and Vierstra, 2000; del Pozo and Estelle, 2000; Gagne et al., 2002; Shen et al., 2002). At present, SCFTIR1 and SCFCOI1, which contain CUL1 and ASK1 with the F-box proteins TIR1 or COI1, respectively, have been demonstrated experimentally to exist in vivo (Gray et al., 1999; Xu et al., 2002). UFO has been shown to interact with ASK1 and ASK2 in two-hybrid assays and in vitro (Samach et al., 1999). Additionally, UFO and ASK1 interact genetically to regulate AP3 and PI gene expression (Zhao et al., 2001). In this study, we show that UFO-myc is associated with Arabidopsis CUL1 and ASK1 in flowers (Figure 5A). These lines of evidence collectively lead us to propose that UFO forms an SCFUFO complex in vivo to regulate flower development. Thus, flower development represents another physiological process, in addition to embryogenesis (Shen et al., 2002) and the auxin (Gray et al., 1999) and jasmonate (Xu et al., 2002) responses, that is regulated by Arabidopsis CUL1.
CSN Interacts Physically with SCFUFO and Affects Flower Development
CSN is highly abundant in Arabidopsis floral tissues, and its role in flower development has been suggested by the phenotypes of antisense/cosuppression mutants of csn3 and csn6 (Peng et al., 2001a, 2001b). Using the csn1 weak mutants, we revealed in this study that one of the mechanisms by which CSN regulates flower development is the modulation of SCFUFO activity. Several lines of evidence support this conclusion. First, the CSN expression pattern in flowers overlaps that of UFO, although it is more ubiquitous (Samach et al., 1999) (Figure 1). Second, both ufo and fus6/CSN1-11 mutants show decreased AP3 expression, which indicates that AP3 activation requires UFO (or SCFUFO) as well as normal levels of CSN1. Correspondingly, both CSN1 and UFO are necessary for the proper formation of second- and third-whorl floral organs, as indicated by their respective mutant phenotypes (Levin and Meyerowitz, 1995; Wilkinson and Haughn, 1995) (Figures 1 and 2). Third, there is a strong biochemical interaction between the CSN complex and UFO-myc, likely through an SCFUFO complex (Figure 5). We showed that UFO-myc can coimmunoprecipitate the entire CSN complex from flower extracts (Figure 5A). Fourth, genetic interaction data presented in this study (Figures 4, 6, and 7) indicate that the overexpression of UFO requires wild-type levels of CSN1 to manifest its gain-of-function effect with respect to AP3 ectopic expression and the homeotic transformation of fourth-whorl organs. Fifth, fus6/CSN1-11 and fus6/N270 flowers contain decreased levels of total and Rub1-conjugated CUL1, which may compromise the overall activity of CUL1-containing SCF ubiquitin ligases such as SCFUFO. Thus, the level of CSN1 is important for the proper functioning of SCF complexes in flowers. The UFO OE–mediated leaf phenotype is unaffected by CSN1 partial deficiency, possibly because leaves may have a relatively lower demand for CSN, because they normally contain the lowest level of CSN1 (Staub et al., 1996). Thus, our data provide another case in which CSN regulates a development process by interacting with and modulating a specific SCF E3 ligase, similar to the other two cases involving SCFTIR1 (Schwechheimer et al., 2001) and SCFCOI1 (Feng et al., 2003).
It should be emphasized that SCFUFO is unlikely to be the only effector of CSN in flowers. Additional F-box proteins and their SCF E3 ligases may be affected by the CSN1 deficiency and may contribute to the observed flower phenotype. Moreover, CSN is highly expressed in the inner carpel wall, pollen mother cells, and tapetal cells, which suggests a role in gametogenesis and in the development of reproductive structure. The phenotypes of the two csn1 mutants described here as well as that of fus6/C231 (Wang et al., 2002) also indicate an important function(s) of CSN1 in organ initiation and differentiation from the apical meristems. Characterization of the molecular mechanism of CSN action in these developmental events awaits further investigation.
Distinct Role of CSN1 in Regulating SCF Function
Two CSN subunits, CSN5 and CSN2 (Cope et al., 2002; Yang et al., 2002), have been implicated directly in CSN-mediated cullin deneddylation. Deletion of Rri1, the ortholog of CSN5 in budding yeast, abolishes Rub1/Nedd8 deconjugation activity on Cdc53 (Lyapina et al., 2001). Partial deficiency in Arabidopsis csn5 also impairs the deneddylation activity and results in the preferential accumulation of CUL1Rub1 (Schwechheimer et al., 2001). By contrast, the csn1 weak mutants fus6/CSN1-11 and fus6/N270 showed no defect in CUL1 deneddylation, indicating that the level of the CSN complex was restored to sufficient levels with regard to CUL1 deneddylation (Figures 5B and 5C) (Wang et al., 2002). Previous studies of CSN1 showed that most of the activities involving CSN1 are contained in its N-terminal region, which is dispensable for cullin deneddylation, and that CSN1 is critical for complex assembly (Tsuge et al., 2001; Wang et al., 2002). Thus, we conclude that the deneddylation defect of the csn1 null mutant (fus6) can be attributed largely to the inability of CSN5 to associate with the CSN complex, whereas the role of CSN1 in CUL1 deneddylation appears to be more complex. The apparent decrease in CUL1Rub1 in the csn1 mutants is not understood. It is possible that the CSN1 subunit somehow negatively regulates the complex's own deneddylation activity. Alternatively, it may influence CUL1 neddylation level through direct binding to Hrt1/Rbx1/Roc1, a core SCF component required for CUL1 neddylation (Furukawa et al., 2000).
Nevertheless, the level of CSN1 is critical for the activity of SCFUFO, as indicated by our genetic data. Similarly, the level of CSN5 has been shown to be important for the degradation of PSIAA6, a substrate of SCFTIR1 (Schwechheimer et al., 2001). However, the deficiencies in CSN1 and CSN5 lead to strikingly different effects on CUL1, suggesting that these subunits modulate SCF by distinct mechanisms. In fus6/CSN1-11 and fus6/N270 flowers, we unexpectedly observed a reduction in total CUL1 level, indicating a role of CSN1 in the regulation of CUL1 abundance. Regulation of CUL1 gene expression in mammalian cells has been linked to the cell-proliferating activity of c-myc. Forced overexpression of CUL1 in c-myc–deficient mouse embryo fibroblasts, in which CUL1 expression is undetectable, enhanced SCFSKP2-mediated proteolysis of p27kip1 and rescued the slow-growth phenotype associated with the c-myc null cells (O'Hagan et al., 2000). We favor the hypothesis that a partial deficiency in CSN1 causes a decrease in the level of CUL1 and CUL1Rub1, which contributes to the phenotype of fus6/CSN1-11 and fus6/N270 with respect to AP3 expression and petal/stamen formation as well as to the suppression of UFO-OE phenotypes in the UM5; CSN1-11 or N270 (F1) and fus6/N270; UM5 (F2) flowers.
Comparison of the Weak Mutants of csn1 and Other csn Subunits
The two csn1 mutants described here, fus6/CSN1-11 and fus6/N270, were generated by rescuing fus6-1 (csn1 null mutant) with stable transgenes of full length or the deletion mutant of CSN1, respectively (Wang et al., 2002). Like other wild-type or truncated CSN1 proteins we have examined, the CSN1-11 and N270 transgene products are stabilized by complex assembly and accumulate only as part of the CSN complex in vivo. In addition, CSN1-11 and N270 alleles do not cause any obvious phenotype in the wild-type background, suggesting that the phenotypes of fus6/CSN1-11 and fus6/N270 are not caused by free CSN1 and N270 proteins. However, these alleles are able to suppress the UFO-OE phenotype in F1 plants, which are genotypically heterozygous at the FUS6 locus (FUS6/fus6) (Table 2). It is possible that these two transgenic alleles may have a slight cosuppression effect on the endogenous CSN1 gene, which can lead to suppression of the UFO-OE phenotype but not enough to cause an obvious CSN1-deficient phenotype or to be detectable by our immunoblot analysis.
We noted that the csn1 weak mutants are more similar to the csn3 (Peng et al., 2001a) and csn6 (Peng et al., 2001b) antisense/cosuppression lines in having abnormal flowers and meristems but differ from the csn5 antisense/cosuppression lines that produce healthy flowers and are fertile (Schwechheimer et al., 2001). The csn5 low-expression lines exhibit loss of apical dominance, a leaf phenotype, and a defect in CUL1 deneddylation that are not found in csn1 weak mutants. Although the levels of the CSN complex are lower than wild-type levels to various extents in all of these weak mutants, considering the differences in their phenotypes, it seems that the decrease of the specific subunit, rather than the decrease of the entire complex, may account for many aspects of their phenotypes. It should be mentioned that CSN5, unlike CSN1, accumulates in both the CSN complex–associated form and the free form (Kwok et al., 1998).
METHODS
Plant Materials and Antibody Reagents
The fus6-1 mutation of Arabidopsis thaliana (ecotype Wassilewskija) was caused by a T-DNA insert that carries a kanamycin resistance marker (Castle and Meinke, 1994). This mutant allele is a null fus6 (or csn1) mutant and has been used in combination with CSN1-11 and N270 transgenes. The wild-type plants used in this study were of the Wassilewskija ecotype. Arabidopsis seeds were surface-sterilized, washed, and plated on a dish containing growth medium (Sigma) with 1% sucrose. The seeds were incubated at 4°C for 3 to 8 days and then placed in a standard long-day growth chamber at 22°C. At 5 to 10 days after germination, seedlings were transferred to soil in a standard long-day growth room.
The antibodies used include anti-CSN1 (Staub et al., 1996), anti-CSN4 (Serino et al., 1999), anti-CSN5 (Kwok et al., 1998), anti-CSN6 (Peng et al., 2001a), anti-CSN8 (Wei et al., 1994), anti-CUL1 (Wang et al., 2002), anti-TBP (Schwechheimer et al., 2001), anti-AP3 (Jenik and Irish, 2001), anti-Rpn6 (Kwok et al., 1999), and anti-myc (Convance, Berkeley, CA).
Constructs and Transgenic Plants
To make the UFO-myc construct, the UFO coding region was amplified by PCR from p35SUFO (Lee et al., 1997) using primers UFOFW1 (5′-TCTAGACCATGGATTCAACTGTGTTCATC-3′) and UFOMYCRV1 (5′-GCTTTTGTTCGTGACCATAACAGACTCCAGGAAATG-3′). The 5× myc tag was amplified from pRbx-myc9 (Schwechheimer et al., 2002) using primers UFOMYCFW2 (5′-CATTTCCTGGAGTCTGTTATGGTCACGAAC-AAAAGC-3′) and MYCRV2 (5′-GATATCGAGCTCATTCAAGTCTTCTTC-TGAGAT-3′). The PCR fragments obtained from these reactions were combined and used as the template in a new PCR with primers UFOFW1 and MYCRV2. The resulting PCR fragment was cloned into pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA), sequenced, and then subcloned into the pPZPY122 plant transformation vector, which contains a gentamycin selection marker and a 35S promoter to drive the expression of the transgene. Wild-type Arabidopsis ecotype Columbia was used in the transformation of the UFO-myc transgene, as described previously (Kang et al., 2000). The construction and generation of full-length subunit 1 (CSN1) and its N-terminal mutant (N270) transgenic lines have been described (Wang et al., 2002).
Immunostaining of Flower Sections
The procedure for immunostaining was essentially as described (Jenik and Irish, 2000). Briefly, the inflorescence was cut and fixed overnight at 4°C in 4% paraformaldehyde in PBS. The samples were dehydrated and embedded in wax molds. Sections (8 μm thick) were cut and placed onto ProbeOn Plus slides (Fisher Biotechnology). After dewaxing and rehydration, the sections were treated with 10 μg/mL proteinase K in PBS for 10 min at room temperature and then washed three times in PBSS (PBS + 0.02% saponin). After blocking in 0.5% BSA and 1% normal goat serum in PBSS for 1 h at room temperature, the slides were incubated with a 1:500 dilution of anti-AP3 antibody (Jenik and Irish, 2001) in PBSS + 0.5% BSA for 5 h at 4°C. The slides then were washed three times in PBSS and incubated overnight with a 1:2000 dilution of the secondary antibody (alkaline phosphatase–conjugated goat anti-rabbit antibody; Sigma). Subsequently, the slides were washed three times in PBSS and twice in TNM (10 mM Tris-HCl, pH 9.5, 100 mM NaCl, and 5 mM MgCl2) and developed with 0.33 mg/mL nitroblue tetrazolium and 0.16 mg/mL 5-bromo-4-chloro-3-indolyl phosphate (Boehringer Mannheim) in TNM. Sections were examined under bright-field optics (Carl Zeiss, Jena, Germany). Images were made with a digital camera (Carl Zeiss) and assembled using Photoshop (Adobe Systems, Mountain View, CA).
Immunoblot Analysis and Immunoprecipitation
Flowers or seedlings were homogenized in extraction buffer containing 50 mM Tris-HCl, pH 7.4, 10 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and complete protease inhibitor cocktail (Roche Molecular Biochemicals, Indianapolis, IN). The extract was centrifuged at 4°C for 10 min, and the protein concentration in the supernatant was measured by Bradford assay (Bio-Rad). In all cases in which equal loading of the samples was required, the same samples or the blot also were probed with anti-Rpn6 to confirm equal loading.
Immunoprecipitation was performed by adding 20 μL of anti-myc bead slurry (Convance) to the protein extract. The sample was incubated at 4°C for 3 h with gentle mixing in the immunoprecipitation buffer containing 150 mM NaCl, 20 mM Tris, pH 7.5, 10 mM MgCl2, 10% glycerol, complete protease inhibitor cocktail, and phenylmethylsulfonyl fluoride (1 mM). The beads then were washed three times with the immunoprecipitation buffer and once with PBS containing 0.1% Tween 20 before immunoblot analysis.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank Claus Schwechheimer, Giovanna Serino, and Rebecca Lamb for discussion and technical advice and Giuliana Gusmaroli, Tomohiko Tsuge, and Elizabeth Strickland for critically reading the manuscript. This work was supported by grants from the National Institutes of Health to N.W. (GM-61812-01) and from the U.S. Department of Agriculture (2001-35304-10852) and the National Science Foundation (MCB-0077217 and MCB-0115870) to X.W.D.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.009936.
References
- Bech-Otschir, D., Seeger, M., and Dubiel, W. (2002). The COP9 signalosome: At the interface between signal transduction and ubiquitin-dependent proteolysis. J. Cell Sci. 115, 467–473. [DOI] [PubMed] [Google Scholar]
- Callis, J., and Vierstra, R.D. (2000). Protein degradation in signaling. Curr. Opin. Plant Biol. 3, 381–386. [DOI] [PubMed] [Google Scholar]
- Castle, L.A., and Meinke, D.W. (1994). A FUSCA gene of Arabidopsis encodes a novel protein essential for plant development. Plant Cell 6, 25–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamovitz, D.A., Wei, N., Osterlund, M.T., von Arnim, A.G., Staub, J.M., Matsui, M., and Deng, X.W. (1996). The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86, 115–121. [DOI] [PubMed] [Google Scholar]
- Coen, E.S., and Meyerowitz, E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353, 31–37. [DOI] [PubMed] [Google Scholar]
- Cope, G.A., Suh, G.S., Aravind, L., Schwarz, S.E., Zipursky, S.L., Koonin, E.V., and Deshaies, R.J. (2002). Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of NEDD8 from CUL1. Science 298, 608–611. [DOI] [PubMed] [Google Scholar]
- del Pozo, J.C., Dharmasiri, S., Hellmann, H., Walker, L., Gray, W.M., and Estelle, M. (2002). AXR1-ECR1–dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell 14, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, J.C., and Estelle, M. (2000). F-box proteins and protein degradation: An emerging theme in cellular regulation. Plant Mol. Biol. 44, 123–128. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., et al. (2000). Unified nomenclature for the COP9 signalosome: An essential regulator of development. Trends Genet. 16, 289. [DOI] [PubMed] [Google Scholar]
- Deshaies, R.J. (1999). SCF and cullin/ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Feng, S., Ma, L., Wang, X., Xie, D., Dinesh-Kumar, S.P., Wei, N., and Deng, X.W. (2003). The COP9 signalosome interacts physically with SCFCOI1 and modulates jasmonate responses. Plant Cell 15, 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa, M., Zhang, Y., McCarville, J., Ohta, T., and Xiong, Y. (2000). The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol. Cell. Biol. 20, 8185–8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne, J.M., Downes, B.P., Shiu, S., Durski, A.M., and Vierstra, R.D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gälweiler, L., Guan, C., Muller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, K., and Meyerowitz, E.M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8, 1548–1560. [DOI] [PubMed] [Google Scholar]
- Ingram, G.C., Goodrich, J., Wilkinson, M.D., Simon, R., Haughn, G.W., and Coen, E.S. (1995). Parallels between UNUSUAL FLORAL ORGANS and FIMBRIATA, genes controlling flower development in Antirrhinum. Plant Cell 7, 1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish, V.F. (1999). Patterning the flower. Dev. Biol. 209, 211–220. [DOI] [PubMed] [Google Scholar]
- Jack, T., Brockman, L.L., and Meyerowitz, E.M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. [DOI] [PubMed] [Google Scholar]
- Jenik, P.D., and Irish, V.F. (2000). Regulation of cell proliferation patterns by homeotic genes during Arabidopsis floral development. Development 127, 1267–1276. [DOI] [PubMed] [Google Scholar]
- Jenik, P.D., and Irish, V.F. (2001). The Arabidopsis floral homeotic gene APETALA3 differentially regulates intercellular signaling required for petal and stamen development. Development 128, 13–23. [DOI] [PubMed] [Google Scholar]
- Kang, D., Wang, X., Cao, K., Sun, C., Deng, X.W., and Wei, N. (2000). Functional conservation of COP9 signalosome among diverse organisms revealed by expressing heterologous subunit genes in Arabidopsis. Plant J. 23, 597–608. [DOI] [PubMed] [Google Scholar]
- Krizek, B.A., and Meyerowitz, E.M. (1996). The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122, 11–22. [DOI] [PubMed] [Google Scholar]
- Kwok, S.F., Solano, R., Tsuge, T., Chamovitz, D., Ecker, J.R., Matsui, M., and Deng, X.W. (1998). Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell 10, 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok, S.F., Staub, J.M., and Deng, X.W. (1999). Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J. Mol. Biol. 285, 85–95. [DOI] [PubMed] [Google Scholar]
- Lamb, R.S., Hill, T.A., Tan, Q.K., and Irish, V.F. (2002). Regulation of APETALA3 floral homeotic gene expression by meristem identity genes. Development 129, 2079–2086. [DOI] [PubMed] [Google Scholar]
- Lee, I., Wolfe, D.S., Nilsson, O., and Weigel, D. (1997). A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr. Biol. 7, 95–104. [DOI] [PubMed] [Google Scholar]
- Levin, J.Z., and Meyerowitz, E.M. (1995). UFO: An Arabidopsis gene involved in both floral meristem and floral organ development. Plant Cell 7, 529–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina, S., Cope, G., Shevchenko, A., Serino, G., Tsuge, T., Zhou, C., Wolf, D.A., Wei, N., Shevchenko, A., and Deshaies, R.J. (2001). Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292, 1382–1385. [DOI] [PubMed] [Google Scholar]
- Mundt, K.E., Liu, C., and Carr, A.M. (2002). Deletion mutants in COP9/signalosome subunits in fission yeast Schizosaccharomyces pombe display distinct phenotypes. Mol. Biol. Cell 13, 493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, M., and Yanofsky, M.F. (2001). Activation of the Arabidopsis B class homeotic genes by APETALA1. Plant Cell 13, 739–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan, R.C., Ohh, M., David, G., de Alboran, I.M., Alt, F.W., Kaelin, W.G., Jr., and DePinho, R.A. (2000). Myc-enhanced expression of Cul1 promotes ubiquitin-dependent proteolysis and cell cycle progression. Genes Dev. 14, 2185–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Z., Serino, G., and Deng, X.W. (2001. a). Molecular characterization of subunit 6 of the COP9 signalosome and its role in multifaceted developmental processes in Arabidopsis. Plant Cell 13, 2393–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Z., Serino, G., and Deng, X.W. (2001. b). A role of Arabidopsis COP9 signalosome in multifaceted developmental processes revealed by the characterization of its subunit 3. Development 128, 4277–4288. [DOI] [PubMed] [Google Scholar]
- Samach, A., Klenz, J.E., Kohalmi, S.E., Risseeuw, E., Haughn, G.W., and Crosby, W.L. (1999). The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 20, 433–445. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., and Deng, X.W. (2001). COP9 signalosome revisited: A novel mediator of protein degradation. Trends Cell Biol. 11, 420–426. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., Callis, J., Crosby, W.L., Lyapina, S., Deshaies, R.J., Gray, W.M., Estelle, M., and Deng, X.W. (2001). Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 292, 1379–1382. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., and Deng, X.W. (2002). Multiple ubiquitin ligase–mediated processes require COP9 signalosome and AXR1 function. Plant Cell 14, 2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serino, G., Su, H., Peng, Z., Tsuge, T., Wei, N., Gu, H., and Deng, X.W. (2003). Characterization of the last subunit of the Arabidopsis COP9 signalosome: Implications for the overall structure and origin of the complex. Plant Cell 15, 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serino, G., Tsuge, T., Kwok, S.F., Matsui, M., Wei, W., and Deng, X.W. (1999). Arabidopsis cop8 and fus4 mutations define the same locus that encodes subunit 4 of the COP9 signalosome. Plant Cell 11, 1967–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W., Parmentier, Y., Hellmann, H., Lechner, E., Dong, A., Masson, J., Granier, F., Lepiniec, L., Estelle, M., and Genschik, P. (2002). Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol. Biol. Cell 13, 1916–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub, J.M., Wei, N., and Deng, X.W. (1996). Evidence for FUS6 as a component of the nuclear-localized COP9 complex in Arabidopsis. Plant Cell 8, 2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh, G.S., Poeck, B., Chouard, T., Oron, E., Segal, D., Chamovitz, D.A., and Zipursky, S.L. (2002). Drosophila JAB1/CSN5 acts in photoreceptor cells to induce glial cells. Neuron 33, 35–46. [DOI] [PubMed] [Google Scholar]
- Tsuge, T., Matsui, M., and Wei, N. (2001). The subunit 1 of the COP9 signalosome suppresses gene expression through its N-terminal domain and incorporates into the complex through the PCI domain. J. Mol. Biol. 305, 1–9. [DOI] [PubMed] [Google Scholar]
- Vernoux, T., Kronenberger, J., Grandjean, O., Laufs, P., and Traas, J. (2000). PIN-FORMED 1 regulates cell fate at the periphery of the shoot apical meristem. Development 127, 5157–5165. [DOI] [PubMed] [Google Scholar]
- Wang, X., Kang, D., Feng, S., Serino, G., Schwechheimer, C., and Wei, N. (2002). CSN1 N-terminal-dependent activity is required for Arabidopsis development but not for Rub1/Nedd8 deconjugation of cullins: A structure-function study of CSN1 subunit of COP9 signalosome. Mol. Biol. Cell 13, 646–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., Chamovitz, D.A., and Deng, X.W. (1994). Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell 78, 117–124. [DOI] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.W. (1996). The role of the COP/DET/FUS genes in light control of Arabidopsis seedling development. Plant Physiol. 112, 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.W. (1999). Making sense of the COP9 signalosome, a regulatory protein complex conserved from Arabidopsis to human. Trends Genet. 3, 98–103. [DOI] [PubMed] [Google Scholar]
- Wilkinson, M.D., and Haughn, G.W. (1995). UNUSUAL FLORAL ORGANS controls meristem identity and organ primordia fate in Arabidopsis. Plant Cell 7, 1485–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L., Liu, F., Ma, H., Peng, W., Huang, D.F., and Xie, D.X. (2002). The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14, 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., Menon, S., Lykke-Andersen, K., Tsuge, T., Xiao, D., Wang, X., Rodriguez-Suarez, R.J., Zhang, H., and Wei, N. (2002). The COP9 signalosome inhibits p27kip1 degradation and impedes G1-S phase progression via deneddylation of SCF Cul1. Curr. Biol. 12, 667–672. [DOI] [PubMed] [Google Scholar]
- Zhao, D., Yu, Q., Chen, M., and Ma, H. (2001). The ASK1 gene regulates B function gene expression in cooperation with UFO and LEAFY in Arabidopsis. Development 128, 2735–2746. [DOI] [PubMed] [Google Scholar]
- Zhou, C., Seibert, V., Geyer, R., Rhee, E., Lyapina, S., Cope, G., Deshaies, R.J., and Wolf, D.A. (2001). The fission yeast COP9/signalosome is involved in cullin modification by ubiquitin-related Ned8p. BMC Biochem. 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]