Abstract
The Arabidopsis FLOWERING LOCUS C (FLC) gene is a key floral repressor in the maintenance of a vernalization response. In vernalization-sensitive genetic backgrounds, FLC levels are high, and they decline after exposure to long cold periods. Four FLC paralogs (MAF2 [MADS AFFECTING FLOWERING2] to MAF5) are arranged in a tandem array on the bottom of Arabidopsis chromosome V. We used a reverse genetics approach to analyze their functions. Loss-of-function and gain-of-function studies indicate that MAF2 acts as a floral repressor. In particular, maf2 mutant plants display a pronounced vernalization response when subjected to relatively short cold periods, which are insufficient to elicit a strong flowering response in the wild type, despite producing a large reduction in FLC levels. MAF2 expression is less sensitive to vernalization than that of FLC, and its repressor activity is exerted independently or downstream of FLC transcription. Thus, MAF2 can prevent premature vernalization in response to brief cold spells. Overexpression of MAF3 or MAF4 produces alterations in flowering time that suggest that these genes also act as floral repressors and might contribute to the maintenance of a vernalization requirement. However, the final gene in the cluster, MAF5, is upregulated by vernalization. Therefore, MAF5 could play an opposite role to FLC in the vernalization response.
INTRODUCTION
Genetic and molecular studies of Arabidopsis have revealed that the timing of flowering is influenced by a large number of different genes (Martinez-Zapater and Somerville, 1990; Koornneef et al., 1991, 1998a, 1998b; Martinez-Zapater et al., 1994; Levy and Dean, 1998; Simpson et al., 1999; Ratcliffe and Riechmann 2002; Simpson and Dean, 2002). Such loci ensure that the switch from vegetative to reproductive growth takes place at the most appropriate time with respect to a variety of abiotic and biotic variables. Among the most intensively studied effects are the responses to daylength and prolonged exposure to low temperatures (vernalization).
Arabidopsis flowers rapidly in long-day photoperiodic conditions of 16 h or continuous light. However, under short-day conditions of 8 to 10 h of light, the plants display a much more extensive period of vegetative growth before flowering. Genes that control this daylength response were identified originally via mutations that cause late flowering under long days but that do not alter flowering time in short-day conditions. Examples of photoperiod pathway genes include CONSTANS, GIGANTEA, FE, FD, and FHA. A second group of genes, which includes LUMINIDEPENDENS, FCA, FVE, FY, and FPA, form an autonomous pathway that monitors the developmental state of the plant and is active under all photoperiodic conditions. Mutants for this second class of genes flower later than wild-type controls irrespective of daylength (Koornneef et al., 1991, 1998a, 1998b; Martinez-Zapater et al., 1994).
Mutants from the photoperiod and autonomous pathways show a differential response to vernalization (Vince-Prue, 1975). Via a vernalization response, Arabidopsis accessions from Northern latitudes, such as Stockholm, are adapted to flower in the spring after exposure to cold winter conditions (Napp-Zinn, 1957). This avoids flowering in late summer, when seed maturation might be curtailed by the onset of winter (Reeves and Coupland, 2000). When such accessions are grown in the laboratory, they flower late, but they flower much earlier if subjected to a cold period of 4 to 8 weeks while the seed is germinating. In a comparable manner, mutants from the autonomous pathway exhibit a very marked reduction in flowering time when subjected to vernalization. By contrast, mutants from the photoperiod pathway show only a minor response to cold treatments (Martinez-Zapater and Somerville, 1990; Koornneef et al., 1991, 1998a; Bagnall, 1992; Burn et al., 1993; Lee et al., 1993; Clarke and Dean, 1994; Chandler et al., 1996). Thus, vernalization can overcome the requirement for the autonomous pathway (Martinez-Zapater and Somerville, 1990; Reeves and Coupland, 2000).
Genetic and molecular analyses have revealed that a MADS box protein, FLOWERING LOCUS C (FLC), is a major determinant of the vernalization response (Koornneef et al., 1994; Lee et al., 1994; Sanda and Amasino, 1996; Michaels and Amasino, 1999, 2001, Sheldon et al., 1999, 2000, 2002; Rouse et al., 2002). High levels of FLC transcript and protein are present in mutants for the autonomous pathway and also in naturally late-flowering Northern accessions, which contain active alleles of a second locus, FRIGIDA (FRI) (Clarke and Dean, 1994; Johanson et al., 2000). By contrast, mutants from the photoperiod pathway, and backgrounds lacking an active FRI allele, show relatively low levels of FLC. Furthermore, null alleles of flc completely suppress the late flowering caused by autonomous pathway mutations and active FRI alleles but have no effect on the delayed flowering in photoperiod pathway mutants (Michaels and Amasino, 2001). Recently, it was shown that two other floral repressors, EARLY IN SHORT DAYS4 (ESD4) and VERNALIZATION INDEPENDENCE4 (VIP4), also act to maintain FLC levels (Reeves et al., 2002; Zhang and van Nocker, 2002). Therefore, FLC expression appears to be supported by genes such as FRI, VIP4, and ESD4 and is repressed strongly by floral activators within the autonomous pathway.
During vernalization, FLC transcript and protein levels decrease, and the plants become competent to flower (Michaels and Amasino, 1999, 2001; Sheldon et al., 1999, 2000; Johanson et al., 2000; Rouse et al., 2002). Additionally, overexpression of FLC from a 35S promoter of Cauliflower mosaic virus in the Landsberg accession (which lacks an active FRI allele) is sufficient to severely delay or prevent flowering and renders the plants insensitive to vernalization (Michaels and Amasino, 1999; Sheldon et al., 1999). These findings indicate that FLC is a potent floral repressor. It has now been shown that such repression is achieved by FLC inhibiting downstream genes that promote flowering, including SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1) and FLOWERING LOCUST (Borner et al., 2000; Lee et al., 2000; Onouchi et al., 2000; Samach et al., 2000; Michaels and Amasino, 2001). Thus, promotion of flowering by either the autonomous pathway or vernalization involves the repression of FLC and the subsequent derepression of FLC targets. Recently, regions within the FLC gene and its promoter that are required for its vernalization-induced repression were defined (Sheldon et al., 2002). However, the molecular signaling events that lead to a decrease in FLC levels during vernalization remain unclear. The products of VERNALIZATION2 and VERNALIZATION1 maintain the repression of FLC once the levels of FLC transcript have declined (Gendall et al., 2001; Levy et al., 2002), but it is not known how this decline is achieved initially.
A number of additional questions regarding the molecular basis of vernalization remain unanswered. First, it has been observed that null flc mutants are responsive to vernalization (Michaels and Amasino, 2001). Therefore, vernalization can promote flowering by other mechanisms as well as via the repression of FLC. In addition, vernalization is a quantitative response to prolonged periods of cold (Sheldon et al., 2000); therefore, a mechanism must exist to ensure that vernalization does not occur in response to short periods of cold that last for only a few days.
Examination of the Arabidopsis genome sequence has revealed the existence of five MADS box genes that encode proteins that are highly related to FLC (Alvarez-Buylla et al., 2000b; Ratcliffe et al., 2001). Based on their similarity to FLC, we theorized that these loci might play a related role in the regulation of flowering time. The first of the genes to be analyzed, MADS AFFECTING FLOWERING1 (MAF1), which also has been referred to as FLOWERING LOCUS M (FLM; Scortecci et al., 2001) and AGAMOUS-LIKE 27 (AGL27; Alvarez-Buylla et al., 2000b), like FLC, was shown to be a floral repressor (Ratcliffe et al., 2001; Scortecci et al., 2001). MAF1 expression is not influenced as strongly by vernalization as that of FLC, and the gene potentially acts downstream or independently of FLC transcription (Ratcliffe et al., 2001). In this study, we analyze the function of the remaining four FLC-related genes and demonstrate that they too influence the timing of flowering. In particular, our results reveal a mechanism that prevents Arabidopsis plants from becoming vernalized by short cold spells.
RESULTS
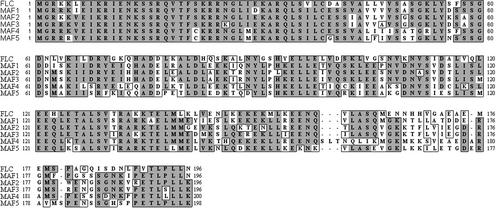
The Arabidopsis genome contains four genes that are highly related to FLC and MAF1; they are arranged in a tight cluster at the bottom of chromosome V (Ratcliffe et al., 2001). The gene cluster occupies ∼22 kb and comprises At5g65050 (which corresponds to AGL31 [Alvarez-Buylla et al., 2000b]), At5g65060, At5g65070, and At5g65080. An alignment of the putative full-length protein sequences encoded by cDNAs for MAF1, FLC, and these four genes (see Methods for details of the isolation of cDNA clones) shows between 53 and 87% identity across the entire sequences, depending on the pair-wise combination (Figure 1). This strong similarity suggested that the additional genes might play related roles to those of MAF1 and FLC in the regulation of flowering time. Based on the results described here, we have renamed the four loci MAF2 to MAF5, respectively.
Figure 1.
Sequence Comparison of the FLC and MAF1 to MAF5 Proteins.
Sequences for MAF1 to MAF5 are derived from the corresponding putatively full-length cDNA clones. MAF2 to MAF5 all exhibit alternative splicing (see Methods). Dark shading indicates identical amino acids.
Identification of a T-DNA Insertion Mutant for MAF2
A single plant hemizygous for a T-DNA insertion within At5g65050 was identified by PCR screening of an in-house collection of random insertion lines. Sequencing from a primer matching the left T-DNA border revealed that the T-DNA resided within the predicted final intron of the gene (a position corresponding to nucleotide 3443 of At5g65050). Selfed seeds were collected from the individual containing the insertion, and these progeny were examined. The progeny plants were genotyped by PCR, and 4 of 20 individuals were identified as being homozygous for the T-DNA insertion. These four individuals all showed visible flower buds at 13 days (continuous light conditions), at least 2 days earlier than any of the wild-type Columbia control plants or any of their hemizygous or wild-type siblings growing side by side. Thus, it appeared that At5g65050 might function as a repressor of the floral transition. Therefore, we renamed At5g65050 MAF2. Homozygous seeds were collected from the four early-flowering plants. To examine the effects of the T-DNA insertion on MAF2 expression, RNA was extracted from pooled 10-day-old seedlings in the next generation. Semiquantitative reverse transcriptase–mediated (RT) PCR was performed using primers specific to MAF2; we were unable to detect transcript in the maf2 seedlings but detected strong MAF2 expression in the wild-type controls, demonstrating that MAF2 activity had been reduced substantially or eliminated in the mutant (Figure 2). Furthermore, RT-PCR expression profiles of the other three genes in the cluster appeared unaffected by the insertion in MAF2 (data not shown).
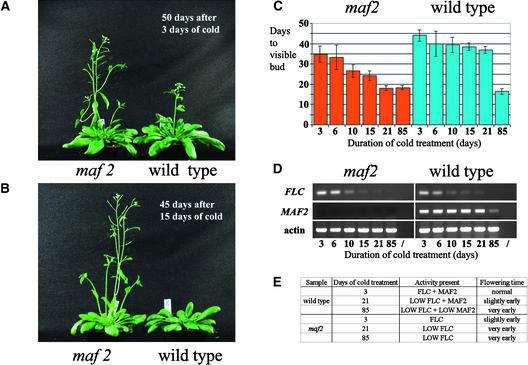
Figure 2.
Effect of Vernalization on the maf2 Mutant.
(A) The maf2 mutant is marginally earlier flowering than wild-type Columbia in the absence of vernalization. Plants are shown after 50 days of growth under a 12-h photoperiod. Water-imbibed seeds were stratified for 3 days at 4°C before transfer to the growth room.
(B) The maf2 mutant is considerably earlier flowering than wild-type Columbia after a short vernalization treatment. Plants are shown after 45 days of growth under a 12-h photoperiod. Imbibed seeds were cold treated for 15 days at 4°C before transfer to the growth room.
(C) The maf2 mutant responds prematurely to vernalization. Data depicted by the graph are available in the supplemental data online. Bars represent days to the appearance of a visible flower bud under a 12-h photoperiod after 4°C cold treatments of 3, 6, 10, 15, 21, and 85 days on imbibed seeds. Error bars indicate standard errors to which 95% confidence limits have been attached. A 10-day cold treatment significantly reduced the time to flowering in the maf2 mutant but not in wild-type Columbia.
(D) Expression of FLC and MAF2 in maf2 and wild-type Columbia seedlings after cold treatments of 3, 6, 10, 15, 21, and 85 days on imbibed seeds. RNA was extracted from pools of 10 whole seedlings after 10 days of growth under 12 h of light, and expression was monitored by RT-PCR over 20, 25, 30, and 35 cycles (products are shown after the following numbers of cycles: FLC, 30; MAF2, 35; and actin, 25). Slashes indicate water controls. The premature vernalization response of maf2 seen in (C) does not seem to be correlated with a premature decline in FLC levels. MAF2 transcript is absent from the maf2 seedlings but is present at a constant level in the 3-, 6-, 10-, 15-, and 21-day time points in the wild type, then declines by the 85-day sample. The MAF2 RT-PCR product is a doublet corresponding to splice variants I and II (see Methods). Equivalent results were obtained when this experiment was repeated for a second time on independent sets of seedlings (data not shown). We also performed RT-PCR with SOC1 primers (data not shown). However, given the fact that SOC1 levels increase very rapidly after germination (Borner et al., 2000; Lee et al., 2000; Michaels and Amasino, 2001), we were unable to detect any differences in SOC1 mRNA levels between the different samples, which were already 13 or more days after germination at the time of harvest.
(E) Summary of the relationship between the duration of cold treatment (days), MAF2/FLC activity, and flowering time for wild-type versus maf2 plants. Twenty-one-day cold-treated maf2 plants flowered equally early as 85-day cold-treated wild-type plants, indicating that the very low levels of MAF2 present in the wild type after the 85-day treatment did not provide significant repression of flowering.
To show that the early-flowering phenotype segregated consistently with homozygosity for the T-DNA insertion within MAF2, a maf2 homozygote was crossed to a wild-type background. F1 plants flowered at the same time as controls. Twenty-two individuals then were examined in the F2 population; five of these plants flowered 2 to 3 days earlier than the others (under continuous light conditions) and were verified by PCR genotyping to be the only individuals that were homozygous for the MAF2 insertion. The F3 population from an F2 homozygote then was compared with the F3 population from an F2 plant lacking the insertion: the homozygous maf2 plants flowered significantly earlier than the wild-type F3 population, confirming that the T-DNA insertion cosegregated with the phenotype.
MAF2 Functions as a Floral Repressor That Prevents Vernalization In Response to Short Cold Periods
Populations of homozygous maf2 and wild-type plants were grown under continuous light, a 12-h photoperiod, and an 8-h photoperiod. In each case, the maf2 plants on average flowered slightly earlier than the controls in terms of both days to visible flower buds and total leaf number (see supplemental data online). Thus, MAF2 acts as a floral repressor but appeared to play a relatively minor role in determining flowering time under the conditions of these experiments.
A null mutant for flc shows a much weaker response to vernalization than the wild type (Michaels and Amasino, 2001). However, the fact that the vernalization response is not abolished completely in this mutant demonstrates that other factors, in addition to FLC, must contribute to the maintenance of a vernalization requirement. To determine whether MAF2 could be one of those factors, batches of germinating wild-type and maf2 seedlings were subjected to an extensive cold treatment of 52 days. The seedlings then were grown alongside nonvernalized controls under a 12-h photoperiod (see supplemental data online). In this experiment, nonvernalized maf2 plants produced flower buds ∼6 days sooner than nonvernalized wild-type plants (28.7 ± 0.9 days versus 34.7 ± 1.6 days), confirming our earlier observations. However, maf2 seedlings showed a similar vernalization response to the wild type. Vernalization of maf2 seedlings reduced the time to bud emergence by 31% and the total leaf number by 47% (with respect to nonvernalized maf2 plants). In the wild-type seedlings, vernalization produced 34% (time to bud emergence) and 53% (total leaf number) reductions. Thus, although MAF2 acts as a floral repressor, either it does not directly maintain a vernalization requirement in the same manner as FLC or it plays a more minor role in maintaining that requirement.
An additional experiment was performed in which batches of maf2 and wild-type seedlings were subjected to a cold treatment for a period of only 10 days (see supplemental data online). After this treatment, the maf2 population flowered proportionally much earlier than in any of our previous experiments, with a mean total leaf number of 11.1 ± 0.7 versus 19.0 ± 0.8 in the wild type (i.e., the mutant showed an ∼42% decrease in leaf number relative to the wild type compared with decreases of ∼16 and ∼25% in experiments 1 and 2). To confirm this result, batches of maf2 and wild-type plants were given a range of different cold treatments: 3, 6, 10, 15, 21, or 85 days (Figures 2A to 2C; see also supplemental data online). maf2 plants given intermediate cold periods of 10, 15, or 21 days (which are not sufficiently long to elicit full vernalization in the wild type) showed a strong response and flowered disproportionately early. Thus, a specific role for MAF2 appears to be the repression of premature vernalization in response to brief cold spells.
To determine whether the observed acceleration of flowering in the maf2 mutant was accompanied by a decline in FLC mRNA levels, we extracted RNA from whole seedlings at 10 days after the cold treatments. RT-PCR experiments showed that FLC transcript levels declined progressively in direct relation to the duration of the cold treatment (Figure 2D), confirming the results of Sheldon et al. (2000). However, for each of the time points, there were no clear discernible differences in FLC levels between maf2 and the wild-type controls (Figure 2D). Thus, the premature vernalization response in the maf2 seedlings apparently was induced independently of changes in FLC transcription.
We observed that by the 10-day cold time point, FLC levels already had decreased very substantially in both the maf2 and wild-type samples compared with levels at the 3- and 6-day time points (Figure 2D). However, despite this decline in FLC levels, there was very little difference in the flowering time of wild-type plants that had received 10 or 15 days of cold compared with those that had been subjected to 6 days of cold. Although a slight reduction in flowering time was seen after 21 days of cold, a very marked reduction in flowering time was seen only in the wild-type plants that had been given an extensive 85-day cold treatment. After this treatment, wild-type plants flowered as early as 85-day cold-treated maf2 plants. In the maf2 mutant, a 10-day treatment accelerated flowering substantially, and a 21-day treatment produced an equivalent effect to that caused by 85 days of cold. Analysis of MAF2 expression in the wild-type samples by RT-PCR showed that MAF2 mRNA levels were similar between the 3-, 6-, 10-, 15-, and 21-day time points but that they declined in the 85-day sample, for which a pronounced reduction in flowering time was observed. These data suggest that in wild-type plants, MAF2 expression might have compensated partly for the decline in FLC levels caused by the 10-, 15-, or 21-day cold treatments, thus maintaining flowering at a time similar to that seen for the 3- and 6-day treatments (Figure 2E).
Overexpression of MAF2, MAF3, MAF4, or MAF5 Modifies Flowering Time
To further investigate the role of MAF2 as a floral repressor and determine whether the other three genes in the cluster (At5g65060, At5g65070, and At5g65080) also could affect flowering time, we analyzed transgenic Arabidopsis lines each containing full-length cDNA expressed from the 35S promoter of Cauliflower mosaic virus.
Overexpression lines for each of the FLC/MAF1 paralogs displayed alterations in the time of flowering compared with wild-type control plants (see supplemental data online). Accordingly, we renamed At5g65060, At5g65070, and At5g65080 MAF3, MAF4, and MAF5, respectively. Flowering time was monitored in primary transformants and/or in a number of independent lines in the second generation (see below and supplemental data online).
Overexpression of MAF2 Produces Similar Phenotypes to Overexpression of FLC or MAF1
A total of 39 35S:MAF2 primary transformants in the Columbia accession were obtained in separate experiments (transgene expression was verified by RT-PCR on leaf tissue). Approximately half (21 of 39 lines) of the plants flowered early, displaying visible flower buds ∼1 week earlier and producing significantly fewer leaves than controls that lacked the transgene. Thirteen of the lines flowered within the wild-type range, whereas the remainder (5 of 39 lines) flowered distinctly later than controls (see supplemental data online).
The progeny of two late-flowering 35S:MAF2 lines and two early-flowering 35S:MAF2 Columbia lines were examined in the T2 generation (see supplemental data online). All individuals from the two late lines (T2-16 and T2-24) also flowered markedly later than wild-type controls in the T2 generation under conditions of either 24 or 12 h of light. By contrast, the phenotype of the early-flowering lines was less consistent between generations. Under continuous light conditions, no significant difference in flowering time was observed in terms of either days to visible flower bud or total leaf number. Under the less inductive condition of 12 h of light, however, a very marginal acceleration of flowering was noted. Thus, the most consistent effect of MAF2 overexpression between generations was a delay in flowering, even though the majority of lines were early flowering as primary transformants.
These MAF2 overexpression data parallel those that have been described for FLC and MAF1 (Sheldon et al., 1999; Ratcliffe et al., 2001). Although the established role of FLC and MAF1 is to repress flowering, when overexpressed in accessions other than Landsberg, they induced flowering in a high proportion of lines (Sheldon et al., 1999; Ratcliffe et al., 2001; Scortecci et al., 2001). In the Landsberg accession, FLC or MAF1 overexpression produced late flowering, but no early-flowering lines were noted (Michaels and Amasino, 1999; Ratcliffe et al., 2001; Scortecci et al., 2001). Transformation of Landsberg with the 35S:MAF2 construct produced a similar result: only late-flowering lines were obtained (see supplemental data online). Therefore, the MAF2 overexpression data, combined with the acceleration of flowering observed in the maf2 mutant, indicate that the native function of MAF2 is to act as a repressor of flowering.
MAF2 Can Prevent Vernalization, Acts Independently of FLC Transcription, but Represses SOC1 Expression
To determine whether MAF2 could delay or prevent the vernalization response, we tested whether the 35S:MAF2 Columbia seedlings were responsive to extensive cold treatments (Figure 3; see also supplemental data online). Two separate studies were performed. In the first instance, batches of 35S:MAF2 (T2 from the late-flowering line 16) and wild-type germinating seedlings were placed in a cold room at 4°C for a period of 76 days and then transferred to a growth room (12-h photoperiod at 20 to 25°C) alongside a freshly sown nontreated batch. In the repeat experiment, seedlings were cold treated for 56 days and then transferred to a second growth room (24-h photoperiod at 20 to 25°C). In both experiments, the 35S:MAF2 plants were completely unresponsive to the cold treatment and flowered as late as the nonvernalized specimens grown alongside them. The wild-type control plants (and wild-type segregants from the 35S:MAF2 population) verified the effectiveness of the vernalization treatment; in both experiments, cold-treated wild-type plants flowered significantly earlier than nontreated individuals.
Figure 3.
Effects of MAF2 Overexpression in the Columbia Accession.
(A) 35S:MAF2 plants are late flowering and do not respond to vernalization. Plants are shown after ∼50 days of growth under a 12-h photoperiod after a 76-day 4°C cold treatment of the imbibed seeds.
(B) Expression of FLC, MAF2, and SOC1 in wild-type Columbia, 35S:MAF2, and 35S:FLC seedlings. RNA was extracted from pools of 10 whole seedlings after 10 days of growth under 12 h of light after either a 3-day cold stratification (−; nonvernalized) or a 76-day vernalization treatment (+; vernalized). Expression was monitored by RT-PCR over 20, 25, 30, and 35 cycles. Products are shown after 25 cycles. No significant changes in FLC mRNA levels are apparent in the 35S:MAF2 samples relative to the wild-type control, whereas SOC1 levels are reduced in the 35S:MAF2 plants in a manner comparable to that observed in the 35S:FLC samples.
To determine if this absence of a vernalization response was caused by MAF2 overexpression influencing FLC transcript levels, we performed RT-PCR experiments on cold-treated and nontreated 35S:MAF2 Columbia seedlings that had been harvested at 10 days after transfer to the growth room (Figure 3). Levels of FLC transcript in the nonvernalized 35S:MAF2 plants were much lower than those in 35S:FLC controls. Furthermore, although the cold-treated 35S:MAF2 seedlings were as late flowering as their nontreated siblings, FLC transcript abundance was reduced to levels similar to those found in vernalized wild-type Columbia plants. Thus, although MAF2 is capable of preventing a premature vernalization response, it does not appear to act through FLC transcription and cannot inhibit the depletion of FLC transcript by long cold treatments. The fact that cold-treated 35S:MAF2 plants were late flowering, despite containing levels of FLC comparable to those of vernalized Columbia plants, indicates that MAF2 can repress flowering via pathways independent or downstream of FLC.
A major target of repression by FLC is the MADS box gene SOC1 (Michaels and Amasino, 2001; Hepworth et al., 2002). To determine whether the mechanism by which MAF2 acts also influences downstream targets of the FLC repression pathway, we examined levels of SOC1 expression in 35S:MAF2 seedlings (Figure 3). As observed in 35S:FLC lines, SOC1 levels were low in both vernalized and nonvernalized 35S:MAF2 plants compared with wild-type Columbia controls. Thus, MAF2 overexpression was sufficient to maintain the repression of SOC1, even when repression by FLC had been reduced via extensive cold treatments.
Effects of MAF3, MAF4, or MAF5 Overexpression
In a comparable manner to the overexpression of MAF1, MAF2, or FLC, overexpression of MAF3, MAF4, or MAF5 produced a consistent delay in flowering in the Landsberg accession, demonstrating that each of these genes can act as a floral repressor (see supplemental data online). When overexpressed in Columbia, however, the effect of each of these genes was less consistent. In particular, significantly late-flowering 35S:MAF3 or 35S:MAF5 Columbia lines were not observed, and only lines that flowered early or at the same time as the wild type were established (see supplemental data online). Therefore, it is possible that MAF3 and MAF5 represent weaker repressors than the other FLC-like genes, such that overexpression levels in the Columbia lines were not sufficiently high to delay flowering. Alternatively, MAF3 or MAF5 might require a partner to act as a repressor in Columbia, but alone it could be sufficient to act as a repressor in Landsberg.
Vernalization Influences MAF3, MAF4, or MAF5 Expression
Mutant analysis of maf2 along with overexpression studies of MAF2 to MAF5 demonstrated that each of these genes could influence flowering time and that MAF2 prevents premature vernalization. In RT-PCR experiments, we observed that all of these genes are expressed across a wide range of tissue types (data not shown), similar to what has been described for FLC and MAF1 (Michaels and Amasino, 1999; Sheldon et al., 1999; Alvarez-Buylla et al., 2000a; Ratcliffe et al., 2001; Scortecci et al., 2001). A key feature of the mechanism by which FLC acts is that FLC transcript and protein levels decrease in response to long cold treatments of 4 to 6 weeks, thereby allowing the floral transition to occur (Michaels and Amasino, 1999; Sheldon et al., 1999, 2000; Rouse et al., 2002). The expression levels of MAF1 also are affected by vernalization in certain genetic backgrounds (Ratcliffe et al., 2001), and in the Columbia background, MAF2 levels decreased after a very prolonged cold treatment (Figures 2 and 3).
To determine whether and how the expression levels of MAF2 to MAF5 are influenced by vernalization, we compared by semiquantitative RT-PCR the expression of each of the genes in vernalized and nonvernalized seedlings from a number of different genetic backgrounds (Figure 4). Germinating seeds were vernalized in a cold room for 6 weeks and then transferred to a growth chamber along with freshly sown nonvernalized controls. After 8 days in continuous light conditions, whole seedlings were harvested and RNA was extracted. FLC transcript levels were substantially higher in nonvernalized versus vernalized seedlings in all of the backgrounds, confirming the efficacy of the treatment. MAF2 displayed no consistent differences in expression level between nonvernalized plants and those given a 6-week cold treatment (Figure 4), whereas after excessively long cold treatments of 10 to 12 weeks, MAF2 transcript levels eventually were reduced (Figures 2 and 3). MAF3, and to some extent MAF4, appeared to be responsive to a 6-week vernalization; in each of these backgrounds, transcript levels generally were lower in the vernalized than in the nonvernalized samples. However, the expression patterns of MAF3 and MAF4 also showed some differences from FLC, because transcript levels were not consistently higher in nonvernalized samples from each of the three late-flowering backgrounds relative to nonvernalized Columbia. Finally, expression of MAF5 showed an opposite response to vernalization than that of FLC. In each of the genetic backgrounds, MAF5 transcript levels were lower in nonvernalized samples than in seedlings that had been vernalized. Thus, although MAF2 to MAF5 are arranged in a very tight cluster, their expression appears to be under distinct modes of regulation, which suggests functional differences among them.
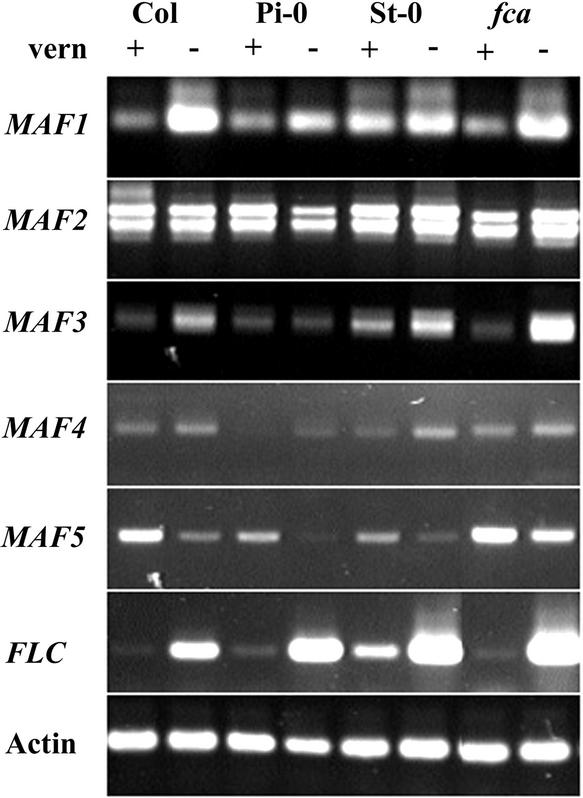
Figure 4.
Effects of Vernalization on the Expression of MAF2 to MAF5 in Different Genetic Backgrounds.
RNA was extracted from pools of 10 to 20 8-day-old seedlings grown under continuous light conditions. Expression was monitored by RT-PCR over 20, 25, 30, and 35 cycles for each of the genes. Products are shown after 30 cycles (FLC and MAF1 to MAF5) and 25 cycles (actin). Vernalized (+) samples were cold treated for 6 weeks at 4°C, whereas nonvernalized (−) samples were stratified for only 3 days at 4°C as imbibed seeds. Note that FLC levels are lower in vernalized than in nonvernalized samples, confirming the efficacy of the treatment. MAF2 levels are similar between vernalized and nonvernalized seedlings from all backgrounds. MAF3 and MAF4 levels are lower in vernalized than in nonvernalized samples for each of the different backgrounds. MAF5 levels, however, in contrast to FLC levels, are higher in vernalized compared with nonvernalized samples for each of the different backgrounds. Equivalent expression patterns were obtained for each of the genes when the experiment was repeated for a second time on different batches of seedlings that were vernalized and grown independently of those represented here. The upregulation of MAF5 by vernalization also was confirmed in two additional independent experiments performed on wild-type Columbia seedlings (data not shown). Col, Columbia; Pi-0, Pitztal; St-0, Stockholm; fca, fca-9 mutant.
DISCUSSION
The level of FLC transcript and protein has been proposed as the molecular basis for whether an Arabidopsis plant will show a vernalization response (Sheldon et al., 2000). However, the fact that the flc null mutant still responds to vernalization indicates that other factors are involved in maintaining the response (Michaels and Amasino, 2001). Given the close similarity of FLC to MAF1/FLM and to MAF2 to MAF5, it is possible that these related factors also participate in the control of vernalization (Ratcliffe et al., 2001). We used a reverse genetics approach to analyze the function of MAF2 to MAF5. Our data implicate each of these genes in the control of flowering time, probably through regulation of the vernalization response.
MAF2 encodes a floral repressor, which appears to participate in a previously unrecognized mechanism that prevents the plants from being vernalized by short cold periods. The early-flowering phenotype of the maf2 mutant, along with the result that MAF2 can delay flowering when overexpressed, indicates that the gene functions to repress the floral transition. Although the acceleration of flowering observed in the maf2 mutant was marginal after a 3-day cold stratification, the phenotype was much more pronounced after moderately short cold periods of 10, 15, or 21 days. Such treatments elicited only a weak response in wild-type plants, in which a substantial reduction in flowering time required a longer period of cold. Thus, MAF2 plays a particular role in the prevention of a premature vernalization response.
The premature response to cold in the maf2 mutant, however, was not correlated with a premature decline in FLC transcript levels. This finding, combined with the observation that 35S:MAF2 lines were unresponsive to extremely long cold treatments and remained late flowering, despite the concomitant reduction in their FLC transcript levels, indicates that MAF2 blocks the vernalization pathway(s) of floral promotion independently or downstream of FLC transcription. The existence of an FLC-independent response has been inferred from the finding that flc null mutants are partly responsive to vernalization (Michaels and Amasino, 2001). MAF2 might play such a repressive role within a pathway parallel to that containing FLC or within a pathway that converges on the same targets as FLC. Examination of an flc maf2 double mutant would allow this possibility to be assessed. However, given the fact that SOC1 expression was reduced in 35S:MAF2 plants, it is likely that MAF2 directly or indirectly inhibits at least some of the same downstream floral activator genes as FLC.
MAF2 expression levels were not markedly different in the Stockholm (carrying an active FRI allele) or fca-9 (deficient in the autonomous pathway) background compared with those of wild-type Columbia. Thus, unlike FLC, it appears that MAF2 expression is neither activated by functional FRI alleles (or by high FLC levels) nor inhibited by the autonomous promotion pathway. In addition, MAF2 transcript levels were not altered noticeably by moderate vernalization treatments of up to 6 weeks. Therefore, pathways different from those that control FLC expression could regulate MAF2, at least in part. The blocking of a response to short periods of cold, which MAF2 appeared to mediate, might be achieved by MAF2 compensating for the decline in FLC levels produced by such cold spells. After very long periods of low temperature, however, MAF2 levels decreased and a marked acceleration of flowering occurred (Figure 2E).
It should be noted that the effects of MAF2 overexpression resembled those seen when MAF1 was overexpressed. MAF1 also behaves as a floral repressor, and late-flowering overexpression lines for this gene do not respond to vernalization (Ratcliffe et al., 2001; Scortecci et al., 2001). Additionally, overexpression of MAF1, like that of MAF2, does not influence FLC transcript levels notably, suggesting that MAF1 can act independently or downstream of FLC (Ratcliffe et al., 2001). Similarly, MAF1 levels are not increased measurably by the presence of active FRI alleles or autonomous pathway mutations (Ratcliffe et al., 2001; Scortecci et al., 2001). On the other hand, MAF1 levels decline in response to moderate vernalization treatments in some genetic backgrounds (Ratcliffe et al., 2001), whereas MAF2 did not show such a pronounced response. Furthermore, the early-flowering phenotype of the maf1/flm null mutant is exacerbated substantially by short-day conditions, as is the phenotype of an flc null mutant (Michaels and Amasino, 2001; Scortecci et al., 2001), but we observed no such effect in maf2 mutants. Thus, it appears that there are parallels and differences between the activities of MAF1, MAF2, and FLC. All three genes appear to have similar repression activity, but divergence has occurred in the way in which their expression is regulated. Their activities likely overlap in part, but the extent to which this occurs remains unknown. However, based on the current data, it seems that MAF1 plays a role more similar to that of FLC than of MAF2.
Although FLC, MAF1/FLM, and MAF2 function as floral repressors, the overexpression of any of these genes in backgrounds other than Landsberg (which cycles very rapidly) results in a substantial portion of the transgenic lines flowering early (Sheldon et al., 1999; Ratcliffe et al., 2001; this study). In our experiments, overexpression of either MAF3 or MAF4 in Columbia produced early flowering in a significant number of lines, but overexpression in Landsberg caused only late flowering, similar to what has been observed upon overexpression of FLC, MAF1, or MAF2. Together with the observation that MAF3 and MAF4 transcript levels declined in response to cold treatments, such results suggest that both genes most likely function as floral repressors and contribute to the maintenance of a vernalization response in a similar manner to FLC.
In contrast to FLC and MAF1 to MAF4, MAF5 might act as a floral activator. The main support for this argument comes from the finding that MAF5 transcript levels are higher in vernalized versus nonvernalized seedlings. The results of overexpression experiments, however, did not indicate such an activator function; rather, in certain conditions (genetic backgrounds), MAF5 can act as a repressor, albeit with a potentially weaker activity than FLC, MAF1, or MAF2. Given the fact that MAF5 is highly related to FLC and the other MAF proteins, it could contend for the same target promoters or perhaps heterodimerize with them. Thus, in certain genetic backgrounds, MAF5 could act as a floral activator simply by competing with other more potent FLC-like proteins. It is possible that MAF5 becomes effective at blocking the activity of such repressors only when their levels have decreased below a certain threshold, thereby “locking” the vernalization response in place. If MAF5 does act by competing with FLC or MAF floral repressors in this manner, the phenotype of a maf5 null mutant might be relatively subtle and show no clear difference from the wild type in particular accessions or in the absence of vernalization. In fact, we have obtained a putative T-DNA insertion mutant for MAF5 in the Columbia accession, but to date, we have been unable to draw firm conclusions regarding its phenotype (data not shown).
Overall, the work described in this and our previous study (Ratcliffe et al., 2001) implicates the MAF1 to MAF5 genes in the regulation of flowering time and the vernalization response. This conclusion has been reached based on the phenotypes of MAF1 to MAF5 overexpression lines, the maf2 mutant, and the changes in MAF1 to MAF5 transcript levels that occur upon vernalization. Thus, it seems likely that, at least in certain genetic backgrounds, the response to vernalization is not achieved solely through the downregulation of FLC but that MAF1 to MAF5 also contribute to it. However, the molecular mechanism of MAF1 to MAF5 action remains to be elucidated. In addition, numerous different splice variants exist for each of the MAF genes (see Methods) (Ratcliffe et al., 2001; Scortecci et al., 2001). Analysis of the relative roles of different non-full-length variants, if any, was beyond the scope of this study. Nonetheless, given the fact that at least two of the autonomous pathway genes, FCA and FPA, encode RNA binding proteins (Macknight et al., 1997, 2002; Schomburg et al., 2001), the regulation of RNA splicing might represent a mechanism by which the autonomous pathway can influence the MAF genes.
In conclusion, the most significant outcome of the current study is that MAF2 prevents a flowering response from being triggered by short cold spells, perhaps by compensating for the decrease in FLC levels that occurs after such conditions occur. In some environments, the promotion of flowering in response to a few days of cold weather might be advantageous. However, winter annual strains of Arabidopsis from northern latitudes have evolved to overwinter vegetatively and commence flowering in the spring only after a sustained period of low temperature (Michaels and Amasino, 2000; Reeves and Coupland, 2000). Individuals without MAF2-like activity would be more susceptible to transient cold spells in the autumn, when conditions for seed set are unfavorable. Thus, there likely would be a selective advantage for the plant to evolve MAF2 function.
METHODS
Plant Materials and Manipulations
Experiments were performed using Arabidopsis thaliana of accession Columbia except where indicated otherwise. Stockholm (CS6863) and Pitztal (CS6832) lines were supplied by the ABRC (Ohio State University, Columbus). The fca-9 allele was in a Columbia background (Page et al., 1999) (kindly provided as a gift to O.J.R. by Caroline Dean). 35S:FLC lines were generated as described by Ratcliffe et al. (2001).
Arabidopsis plants were transformed by the floral-dip method (Bechtold and Pelletier, 1998; Clough and Bent, 1998) using Agrobacterium tumefaciens carrying a standard transformation vector. The vector contained a kanamycin resistance selectable marker system driven by a nopaline synthase promoter and either the MAF1 to MAF5 or the FLC cDNA downstream from the 35S promoter of Cauliflower mosaic virus. Transgene expression was verified by reverse transcriptase–mediated (RT) PCR experiments on RNA extracted from rosette leaves of primary transformants.
In all experiments, seeds were sterilized by a 2-min ethanol treatment followed by 20 min in 30% bleach and 0.01% Tween and five washes in distilled water. Seeds were sown on Murashige and Skoog (1962) (MS) agar in 0.1% agarose and stratified for 3 days at 4°C before transfer to growth rooms with a temperature of 20 to 25°C. MS medium was supplemented with 50 mg/L kanamycin for the selection of transformed plants. Plants were transplanted to soil after 8 days of growth on plates when grown under continuous light and after 10 days when grown under 8- or 12-h photoperiods. For vernalization treatments, seeds were sown on MS agar plates, sealed with micropore tape, and placed in a 4°C cold room with low light levels. The plates then were transferred to the growth rooms alongside plates containing freshly sown nonvernalized control seeds, which had received a short cold stratification of 3 days (to synchronize germination). Time to flowering was measured as days to a flower bud becoming visible and/or in terms of the total number of leaf nodes formed by the primary shoot meristem. Rosette leaves were counted when a visible inflorescence of ∼3 cm was apparent.
Isolation and Characterization of MAF2 to MAF5 cDNA Clones
cDNAs for each of the FLC/MAF1-like genes were identified either among clones in a library derived from leaf RNA or by a combination of rapid amplification of cDNA ends (RACE) and RT-PCR performed on RNA derived from mixed tissue samples of the Columbia accession. Alternative transcripts were detected for each of the four genes. At least four variants of At5g65050 (MAF2) were identified. Variant I, which corresponds to the full-length transcript, encodes a 196–amino acid protein. Variants II and III differ in their 3′ regions but both generate a protein of 145 amino acids, the last 20 residues of which are different from those of the 196–amino acid full-length version. Variant IV comprises a truncated form of variant I and gives rise to a small polypeptide of 80 amino acids. At5g65060 (MAF3) and At5g65070 (MAF4) both displayed at least five variants. The longest MAF3 product, encoded by variant I, is 196 amino acids in length. MAF3 variants II and III encode 185– and 118–amino acid proteins, respectively, whereas variants IV and V both generate products of 77 amino acids but differ in their 3′ regions. The longest MAF4 clone identified, variant I, encodes a protein of 200 amino acids. MAF4 variant II encodes a 136–amino acid product, whereas MAF4 variants III, IV, and V encode very short polypeptides of 63, 66, and 69 amino acids, respectively. Finally, two alternative variants of At5g65080 (MAF5), variants I and II, were identified that generate 198– and 184–amino acid proteins, respectively. The significance of these alternative transcripts is unclear; alternative splicing for MAF1 has been described previously (Ratcliffe et al., 2001; Scortecci et al., 2001), although it has not been reported for FLC.
MAF2 variants II and III were isolated randomly from an in-house library derived from Arabidopsis leaf mRNA. All other MAF clones were isolated by RT-PCR on RNA extracted from whole vegetative Columbia seedlings. RNA was extracted from plant tissue using a cetyl-trimethyl-ammonium bromide–based protocol (Jones et al., 1995), poly(A+) RNA was purified using oligo(dT) cellulose (Gibco BRL), and first-stand cDNA synthesis was performed using a SuperScript kit (Gibco BRL).
To confirm the gene boundaries, 3′ RACE was first performed using a SMART RACE cDNA amplification kit (Clontech, Palo Alto, CA), and two rounds of PCR (30 and 25 cycles) were performed using the following first- and second-round gene-specific primers: MAF2, 5′-AAGAAGCAA-AAAACATTGTGGGTCTCCG-3′ and 5′-GTCTCCGGCTCCGGAAAA-CTCTACAAG-3′; MAF3, 5′-CTGTTGTCGCCGTCTCCGGTTCCGGAAA-3′ and 5′-ACTCTACGACTCTGCCTCCGGTGACAA-3′; MAF4, 5′-ATC-AAACGAATTGAGAACAAAAGCTCTC-3′ and 5′-CTTATCATCATCTCTGCC-ACCGGAAGAC-3′; and MAF5, 5′-GGGGATTAGATGTGTCGGAAG-AGTGAAG-3′ and 5′-AACTCTACAACTCCTCCTCCGGCGACAG-3′.
RACE products then were cloned to the pGEM-T Easy vector (Promega) and sequenced. After RACE analysis, MAF cDNA clones were isolated by PCR from cDNA using the primers listed below and then ligated into the overexpression vector after digestion with the restriction enzymes indicated: MAF2 (KpnI-NotI), 5′-GAGGGGTACCACATT-GTGGGTCTCCGGTGATTAGGATC-3′ and 5′-GGGAAAGCGGCCGCA-ATCAGGCTGTAAGTTTAAGGTGAAAGC-3′; MAF3 (KpnI-NotI), 5′-GAG-GGGTACCAGAAAAAAAGCAAACACATTTTGGGTCC-3′ and 5′-GGG-AAAGCGGCCGCACAAGAACTCTGATATTTGTCTACTAAG-3′; MAF4 (SalI-NotI), 5′-GCACGCGTCGACCAAATTAGGTCAGAAGAATTAGTC-GGAG-3′ and 5′-GGGAAAGCGGCCGCTCTCCTTGGATGACTTTTC-CGTAGCAGG-3′; and MAF5 (SalI-NotI), 5′-GCACGCGTCGACGGGGATTAGATGTGTCGGAAGAGTGAAG-3′ and 5′-GGGAAAGCGGCC-GCGATCCTGTCTTCCAAGGTAACACAAAGG-3′.
RT-PCR Analyses
For semiquantitative RT-PCR expression studies, the following primers were used: FLC, 5′-TTAGTATCTCCGGCGACTTGAACCCAAACC-3′ and 5′-AGATTCTCAACAAGCTTCAACATGAGTTCG-3′; MAF2, 5′-ACA-TTGTGGGTCTCCGGTGATTAGGATC-3′ and 5′-AATCAGGCTGTA-AGTTTAAGGTGAAAGC-3′; MAF3, 5′-GAAGAAAAAAAGCAAACACAT-TTTGGGTCC-3′ and 5′-AAGAACTCTGATATTTGTCTACTAAGGTAC-3′; MAF4, 5′-ATTAGGTCAGAAGAATTAGTCGGAGAAAAC-3′ and 5′-CTT-GGATGACTTTTCCGTAGCAGGGGGAAG-3′; MAF5, 5′-GGGGATTAG-ATGTGTCGGAAGAGTGAAG-3′ and 5′-GATCCTGTCTTCCAAGGTAAC-ACAAAGG-3′; Actin, 5′-AGAGATTCAGATGCCCAGAAGTCTTGTTCC-3′ and 5′-AACGATTCCTGGACCTGCCTCATCATACTC-3′; and SOC1, 5′-GGCATACTAAGGATCGAGTCAGCACCAAAC-3′ and 5′-ACCCAATGA-ACAATTGCGTCTCTACTTCAG-3′. General RT-PCR procedures were as described previously (Ratcliffe et al., 2001).
Identification and Isolation of the maf2 T-DNA Mutant
The T-DNA insertion event within MAF2 was detected initially in a pooled collection of ∼3000 lines and then dereplicated to a single plant by multiple rounds of PCR using the following pairs of T-DNA left-border (LB) and gene-specific (GS) primers: first round (40 cycles), 5′-CTC-ATCTAAGCCCCCATTTGGACGTGAATG-3′ (LB) and 5′-CAGGCTGTA-AGTTTAAGGTGAAAGCTCA-3′ (GS); second round (40 cycles), 5′-TTG-CTTTCGCCTATAAATACGACGGATCG-3′ (LB nested) and 5′-TGATGA-TGGTGATTACTTGAGCAGCGGA-3′ (GS nested). The insertion positions were confirmed by sequencing of the PCR products. Homozygous plants for the MAF2 insertion then were identified by the absence of a band after 40 cycles of PCR with the following pair of gene-specific primers, 5′-AAGACAGAACTAATGATGGGGGAAGTGAAGTCC-3′ and 5′-TACGAAGGTACAATAAAGATCTACTATAGC-3′, which resided on either side of the insertion locus.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The GenBank accession numbers for the sequences mentioned in this article are as follows: AY231441 (MAF2 variant I), AY231442 (MAF2 variant II), AY231443 (MAF2 variant III), AY231444 (MAF2 variant IV), AY231445 (MAF3 variant I), AY231446 (MAF3 variant II), AY231447 (MAF3 variant III), AY231448 (MAF3 variant IV), AY231449 (MAF3 variant V), AY231450 (MAF4 variant I), AY231451 (MAF4 variant II), AY231452 (MAF4 variant III), AY231453 (MAF4 variant IV), AY231454 (MAF4 variant V), AY231455 (MAF5 variant I), and AY231456 (MAF5 variant II).
Supplementary Material
Acknowledgments
We thank Luc Adam and Marsha Pilgrim for provision of the T-DNA insertion collection in which the maf2 mutant was identified. Valuable experimental assistance was provided by Natalie Irwin, Elsa Eysteinsdottir, Tony Reyes, and Sky Eales. We also are grateful to Elliot Meyerowitz, Frederick Hempel, Lynne Reuber, Neal Gutterson, Jacqueline Heard, Bill Goure, and Matthew Kaser for discussions and comments on the manuscript. We thank the Monsanto Company for financial support of this project.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.009506.
Footnotes
Online version contains Web-only data.
References
- Alvarez-Buylla, E.R., Liljegren, S.J., Pelaz, S., Gold, S.E., Burgeff, C., Ditta, G.S., Vergara-Silva, F., and Yanofsky, M.F. (2000. a). MADS-box gene evolution beyond flowers: Expression in pollen, endosperm, guard cells, roots and trichomes. Plant J. 24, 457–466. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla, E.R., Pelaz, S., Liljegren, S.J., Gold, S.E., Burgeff, C., Ditta, G.S., Ribas de Pouplana, L., Martinez-Castilla, L., and Yanofsky, M.F. (2000. b). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 97, 5328–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnall, D.J. (1992). Control of flowering in Arabidopsis thaliana by light, vernalization and gibberellins. Aust. J. Plant Physiol. 19, 401–409. [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Borner, R., Kampmann, G., Chandler, J., Gleissner, R., Wisman, E., Apel, K., and Melzer, S. (2000). A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 24, 591–599. [DOI] [PubMed] [Google Scholar]
- Burn, J.E., Bagnall, D.J., Metzger, J.D., Dennis, E.S., and Peacock, W.J. (1993). DNA methylation, vernalization, and the initiation of flowering. Proc. Natl. Acad. Sci. USA 90, 287–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, J., Wilson, A., and Dean, C. (1996). Arabidopsis mutants showing an altered response to vernalization. Plant J. 10, 637–644. [DOI] [PubMed] [Google Scholar]
- Clarke, J.H., and Dean, C. (1994). Mapping FRI, a locus controlling flowering time and vernalization response in Arabidopsis thaliana. Mol. Gen. Genet. 242, 81–89. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Gendall, A.R., Levy, Y.Y., Wilson, A., and Dean, C. (2001). The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107, 525–535. [DOI] [PubMed] [Google Scholar]
- Hepworth, S.R., Valverde, F., Ravenscroft, D., Mouradov, A., and Coupland, G. (2002). Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 21, 4327–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R., and Dean, C. (2000). Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347. [DOI] [PubMed] [Google Scholar]
- Jones, A., Davies, H.M., and Voelker, T.A. (1995). Palmitoyl-acyl carrier protein (ACP) thioesterase and the evolutionary origin of plant acyl-ACP thioesterases. Plant Cell 7, 359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Alonso-Blanco, C., Blankestijn-de Vries, H., Hanhart, C.J., and Peeters, A.J. (1998. a). Genetic interactions among late-flowering mutants of Arabidopsis. Genetics 148, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Alonso-Blanco, C., Peeters, A.J.M., and Soppe, W. (1998. b). Genetic control of flowering time in Arabidopsis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 345–370. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Blankestijn-de Vries, H., Hanhart, C., Soppe, W., and Peeters, A.J. (1994). The phenotype of some late-flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild-type. Plant J. 6, 911–919. [Google Scholar]
- Koornneef, M., Hanhart, C.J., and van der Veen, J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66. [DOI] [PubMed] [Google Scholar]
- Lee, H., Suh, S.-S., Park, E., Cho, E., Ahn, J.H., Kim, S.-G., Lee, J.S., Kwon, Y.M., and Lee, I. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I., Bleecker, A., and Amasino, R. (1993). Analysis of naturally occurring late flowering in Arabidopsis thaliana. Mol. Gen. Genet. 237, 171–176. [DOI] [PubMed] [Google Scholar]
- Lee, I., Michaels, S.D., Masshardt, A.S., and Amasino, R.M. (1994). The late-flowering phenotype of FRIGIDA and mutations in LUMINI-DEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J. 6, 903–909. [Google Scholar]
- Levy, Y.Y., and Dean, C. (1998). The transition to flowering. Plant Cell 10, 1973–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, Y.Y., Mesnage, S., Mylne, J.S., Gendall, A.R., and Dean, C. (2002). Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 297, 243–246. [DOI] [PubMed] [Google Scholar]
- Macknight, R., Bancroft, I., Page, T., Lister, C., Schmidt, R., Love, K., Westphal, L., Murphy, G., Sherson, S., Cobbett, C., and Dean, C. (1997). FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89, 737–745. [DOI] [PubMed] [Google Scholar]
- Macknight, R., Duroux, M., Laurie, R., Dijkwel, P., Simpson, G., and Dean, C. (2002). Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell 14, 877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Zapater, J.M., Coupland, G., Dean, C., and Koornneef, M. (1994). The transition to flowering in Arabidopsis. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 403–433.
- Martinez-Zapater, J.M., and Somerville, C.R. (1990). Effect of light quality and vernalization on late flowering mutants of Arabidopsis thaliana. Plant Physiol. 92, 770–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (2000). Memories of winter: Vernalization and the competence to flower. Plant Cell Environ. 23, 1145–1153. [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13, 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Napp-Zinn, K. (1957). Untersuchungen zur genetik des kaltebedurfnisses bei Arabidopsis thaliana (L.) Heynh. Z. Indukt. Abstammungs Vererbungsl. 88, 253–285. [Google Scholar]
- Onouchi, H., Igeño, M.I., Perilleux, C., Graves, K., and Coupland, G. (2000). Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12, 885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, T., Macknight, R., Yang, C.H., and Dean, C. (1999). Genetic interactions of the Arabidopsis flowering time gene FCA with genes regulating floral initiation. Plant J. 17, 231–239. [DOI] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Nadzan, G.C., Reuber, T.L., and Riechmann, J.L. (2001). Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 126, 122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe, O.J., and Riechmann, J.L. (2002). Arabidopsis transcription factors and the regulation of flowering time: A genomic perspective. Curr. Issues Mol. Biol. 4, 77–91. [PubMed] [Google Scholar]
- Reeves, P.H., and Coupland, G. (2000). Response of plant development to environment: Control of flowering by daylength and temperature. Curr. Opin. Plant Biol. 3, 37–42. [DOI] [PubMed] [Google Scholar]
- Reeves, P.H., Murtas, G., Dash, S., and Coupland, G. (2002). Early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development 129, 5349–5361. [DOI] [PubMed] [Google Scholar]
- Rouse, D.T., Sheldon, C.C., Bagnall, D.J., Peacock, W.J., and Dennis, E.S. (2002). FLC, a repressor of flowering, is regulated by genes in different inductive pathways. Plant J. 29, 183–191. [DOI] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. [DOI] [PubMed] [Google Scholar]
- Sanda, S.L., and Amasino, R.M. (1996). Interaction of FLC and late-flowering mutations in Arabidopsis thaliana. Mol. Gen. Genet. 251, 69–74. [DOI] [PubMed] [Google Scholar]
- Schomburg, F.M., Patton, D.A., Meinke, D.W., and Amasino, R.M. (2001). FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell 13, 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scortecci, K.C., Michaels, S.D., and Amasino, R.M. (2001). Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 26, 229–236. [DOI] [PubMed] [Google Scholar]
- Sheldon, C.C., Burn, J.E., Perez, P.P., Metzger, J., Edwards, J.A., Peacock, W.J., and Dennis, E.S. (1999). The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, C.C., Conn, A.B., Dennis, E.S., and Peacock, W.J. (2002). Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14, 2527–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, C.C., Rouse, D.T., Finnegan, E.J., Peacock, W.J., and Dennis, E.S. (2000). The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. USA 97, 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, G.G., and Dean, C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289. [DOI] [PubMed] [Google Scholar]
- Simpson, G.G., Gendall, A.R., and Dean, C. (1999). When to switch to flowering. Annu. Rev. Cell Dev. Biol. 15, 519–550. [DOI] [PubMed] [Google Scholar]
- Vince-Prue, D. (1975). Vernalization. In Photoperiodism in Plants. (London: McGraw-Hill), pp. 263–291.
- Zhang, H., and van Nocker, S. (2002). The VERNALIZATION INDEPENDENCE 4 gene encodes a novel regulator of FLOWERING LOCUS C. Plant J. 31, 663–673. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.