Abstract
To study genome evolution in wheat, we have sequenced and compared two large physical contigs of 285 and 142 kb covering orthologous low molecular weight (LMW) glutenin loci on chromosome 1AS of a diploid wheat species (Triticum monococcum subsp monococcum) and a tetraploid wheat species (Triticum turgidum subsp durum). Sequence conservation between the two species was restricted to small regions containing the orthologous LMW glutenin genes, whereas >90% of the compared sequences were not conserved. Dramatic sequence rearrangements occurred in the regions rich in repetitive elements. Dating of long terminal repeat retrotransposon insertions revealed different insertion events occurring during the last 5.5 million years in both species. These insertions are partially responsible for the lack of homology between the intergenic regions. In addition, the gene space was conserved only partially, because different predicted genes were identified on both contigs. Duplications and deletions of large fragments that might be attributable to illegitimate recombination also have contributed to the differentiation of this region in both species. The striking differences in the intergenic landscape between the A and Am genomes that diverged 1 to 3 million years ago provide evidence for a dynamic and rapid genome evolution in wheat species.
INTRODUCTION
Within the Triticeae tribe, the grass genus Triticum includes diploid, tetraploid, and hexaploid species. Bread wheat (Triticum aestivum; 2n = 42, AABBDD) is allohexaploid and carries three different subgenomes, A, B, and D. There are two different A genomes in the wheat species: the Au genome in Triticum urartu and the closely related Am genome in Triticum monococcum (Dvorak et al., 1988). Modern hexaploid wheat resulted from two independent hybridization events. The first combined the Au genome of the wild diploid wheat T. urartu and the B genome of an unknown species (Feldman et al., 1995; Huang et al., 2002a). This resulted in the tetraploid ancestor of modern Triticum species, Triticum turgidum. The cultivated T. turgidum subsp durum is derived from this tetraploid ancestor. T. turgidum and the diploid donor of the D genome (Aegilops tauschii) hybridized ∼8000 years ago, resulting in hexaploid wheat (Feldman et al., 1995).
Wheat belongs to the grass family Poaceae, and members of this family display a high variability in genome size, for example, 450 Mb for rice, 2500 Mb for maize, 5000 Mb for barley, and 16,000 Mb for hexaploid wheat (Arumuganathan and Earle, 1991). This genome size variation is caused partly by differences in the ploidy level, but it is attributable mainly to differences in the amount of repetitive DNA (Flavell et al., 1974). In maize and wheat, genes were found to be organized in gene islands or as single genes separated by large regions of nested repetitive elements (reviewed by Feuillet and Keller, 2002). In addition to expressed genes, gene fragments have been found in several sequenced regions in grass genomes (Feuillet and Keller, 1999; Feuillet et al., 2001; Ramakrishna et al., 2002b). Illegitimate recombination and double-strand break repair mechanisms that introduce filler sequences into chromosomes might be responsible for genome restructuring and the formation of truncated genes (Ramakrishna et al., 2002b). The large genome size of wheat is attributable partially to the presence of a large amount of nested long terminal repeat (LTR) retrotransposons, very similar to the situation in maize (SanMiguel et al., 1996; Wicker et al., 2001; Ramakrishna et al., 2002a). The increase of genome size caused by retrotransposons as well as transposons (Wicker et al., 2003) is counteracted by mechanisms that reduce genome size. These include unequal inter- or intra-retroelement recombination events and deletions of apparently random fragments of repetitive elements (reviewed by Bennetzen, 2002). The conflicting mechanisms of genome expansion and reduction result in a dynamic process of genome evolution (Petrov, 2001; Bennetzen, 2002). The balance between these opposing forces might result in a stable genome size that can vary among species.
Molecular mechanisms of genome evolution can be identified by comparative analysis of the genomes of related species. Large-scale comparative studies have focused on sorghum and maize as well as barley and wheat, which are estimated to have diverged 16 and 11 million years ago, respectively. Analysis in rice, maize, and sorghum revealed a mosaic genome structure at orthologous loci, with conserved sequences interspersed with nonconserved sequences and gene amplification, gene movement, and retrotransposition accounting for most of the changes (Bennetzen and Ramakrishna, 2002; Song et al., 2002). A comparison between wheat and barley revealed that almost all of the intergenic regions are completely different, indicating that 11 million years is sufficient time to erase homology outside of the gene space (Ramakrishna et al., 2002a; SanMiguel et al., 2002).
The genus Triticum is ideally suited for comparative studies of genomes that diverged relatively recently. A number of closely related diploid and polyploid species are available, and the individual genomes as well as the evolutionary time scale of their divergence have been studied in detail (Dvorak and Zhang, 1992; Allaby et al., 1999; Huang et al., 2002a, 2002b). Based on the evolution of the Acc-1 gene, T. urartu (donor of the A genome of T. durum and T. aestivum) and T. monococcum (donor of the Am genome) were estimated to have diverged only 0.5 to 1 million years ago (Huang et al., 2002a). The genome of T. monococcum has been used successfully as a model for the A genome of hexaploid wheat, confirming the close relationship between the Au and the Am genomes (Stein et al., 2000).
To study genome evolution in wheat at the molecular level, we have identified orthologous loci from A genomes of T. monococcum and T. durum. Comparison of 285 kb from T. monococcum with 142 kb from T. durum showed very little sequence conservation between the two species. Only part of the gene space is conserved, and the intergenic regions show extensive rearrangements. In addition to retrotransposon and transposon insertions, comparisons of insertion and deletion (InDel) patterns in a large (54-kb) duplication revealed a high frequency of illegitimate recombination events that have shaped the wheat genomes.
RESULTS
Identification of Orthologous Loci on Chromosome 1AS of Two Wheat Species
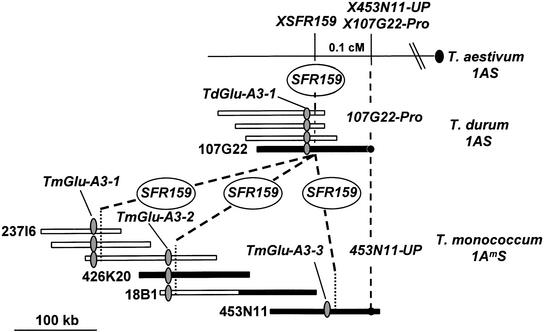
Orthologous genomic sequences of the Am and Au genomes were isolated from BAC libraries of T. monococcum cv DV92 (Lijavetzky et al., 1999) and the tetraploid wheat T. durum cv Langdon (A. Cenci, N. Chantret, Xy. Kong, Y. Gu, D.D. Anderson, T. Fahima, A. Distelfeld, and J. Dubcovsky, unpublished data). A BAC contig of 420 kb was established after screening of the T. monococcum BAC library with a probe corresponding to the coding region of a LMW glutenin gene, TaGlu-1D-1 (Colot et al., 1989) (Figure 1). The LMW glutenin genes encode wheat storage proteins and belong to a gene family located on the short arm of group 1 chromosomes (McIntosh et al., 1998). Hybridization experiments identified three LMW glutenin genes (TmGlu-A3-1, -2, and -3) (Figure 1) on the 420-kb contig. Low-pass shotgun sequencing of BAC 453N11 identified a 2-kb shotgun insert (453N11-159, hereafter called SFR159) that has a low copy number and shows no homology with any known sequence in the database. SFR159 identifies three loci in T. monococcum cv DV92 (Figures 1 and 2). Hybridization experiments showed that the three fragments are present on the 420-kb contig and are located close to each of the three LMW glutenin genes (Figure 1). Hybridization of SFR159 to genomic DNA of Langdon/Chinese Spring substitution lines showed the presence of a unique locus for SFR159 on chromosome 1A of T. durum cv Langdon (Figure 2). The screening of the T. durum BAC library using SFR159 as a probe yielded four BAC clones that form a single contig of ∼180 kb (Figure 1).
Figure 1.
Genetic and Physical Maps of Orthologous LMW Glutenin Loci in Diploid, Tetraploid, and Hexaploid Wheat Species.
BAC libraries from T. monococcum and T. durum were screened with a probe corresponding to the coding region of the TaGlu-1D-1 gene and with SFR159, a shotgun clone of T. monococcum BAC453N11, respectively. The positions of probes on the contigs are indicated. The black dot represents the centromere position. Shaded areas indicate BAC clones that were chosen for complete sequencing. cM, centimorgan.
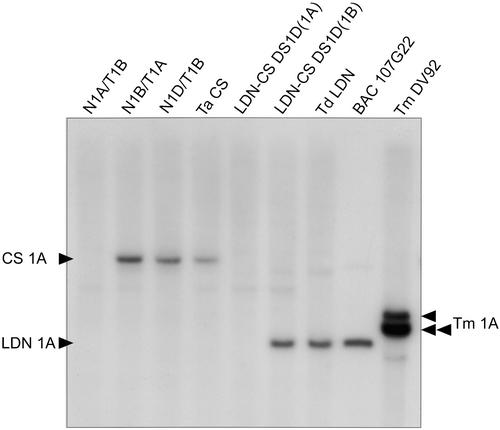
Figure 2.
Determination of the Copy Number and Chromosomal Location of SFR159 in Chinese Spring and T. durum cv Langdon by DNA Gel Blot Hybridization of HindIII-Digested Genomic DNA of Chinese Spring Nullitetrasomic Lines and of Langdon/Chinese Spring Substitution Lines.
TaCS represents the hexaploid wheat line Chinese Spring, TdLDN represents the tetraploid T. durum cv Langdon, and TmDV92 represents diploid T. monococcum line DV92. N1A/T1B, N1B/T1A, and N1D/T1B are nullitetrasomic lines of Chinese Spring. LDN-CS-1D(1A) and LDN-CS-1D(1B) are Langdon/Chinese Spring disomic substitution lines in which chromosome 1D of Chinese Spring replaces chromosome 1A or 1B of Langdon. Arrowheads at left indicate the SFR159 loci missing in the Chinese Spring nullitetrasomic line N1A/T1B (CS 1A) and in the substitution line LDN-CS-1D(1A) (LDN 1A). These loci are attributed to chromosome 1A of Chinese Spring (CS 1A) and to chromosome 1A of T. durum cv Langdon (LDN 1A), respectively. The T. durum fragment is present on Langdon BAC 107G22. Arrowheads at right indicate the three loci detected by SFR159 in T. monococcum line DV92 (Tm 1A).
The orthologous relationship between the T. monococcum and T. durum contigs was confirmed by genetic mapping in hexaploid wheat. High-resolution mapping was performed using three low-copy-number probes: SFR159, 453N11-UP from T. monococcum, and 107G22-Pro from T. durum (Figure 1). Interestingly, although SFR159 identified one polymorphic locus on chromosome 1A of both parents of the mapping population, Chul and Frisal, the 1A locus of 453N11-UP (a second locus was mapped on chromosome 1B) was present only in Chul and 107G22-Pro was present only on chromosome 1A in Frisal, indicating the existence of intervarietal differences at this locus in hexaploid wheat. 453N11-UP-1A and 107G22-Pro both mapped at the same position 0.1 centimorgan proximal to SFR159 on chromosome 1A (Figure 1). The orthologous contigs were characterized further by low-pass shotgun sequencing of BAC 237I6 (TmGlu-A3-1 locus) and by complete sequencing of the region of T. monococcum BAC clones 426K20, 18B1, and 453N11 (TmGlu-A3-2 and TmGlu-A3-3 loci) and of T. durum BAC 107G22.
Genes and Pseudogenes on the T. monococcum and T. durum Contigs
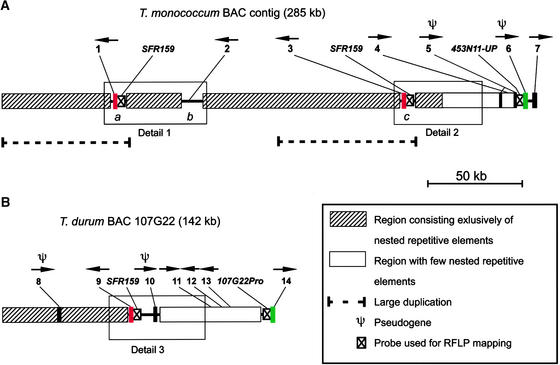
The sequenced T. monococcum contig covers a region of 285,444 bp (Figure 3A). The contig contains five putative genes, two pseudogenes (see supplemental data online), and >83% repetitive DNA. More than 54 kb are present in duplicate, with an overall sequence identity of 97.7%. The two duplicated units each contain one LMW glutenin gene (TmGlu-A3-2 and TmGlu-A3-3), separated from each other by >150 kb (Figure 3A). The coding sequences of these two genes are 99.4% identical. SFR159 is located ∼2 kb upstream of both glutenin genes. The relationship between these two genes and the third locus on the T. monococcum BAC contig, which also contains both a LMW glutenin gene (TmGlu-A3-1) and SFR159 (Figure 1), was investigated by partial sequencing of the TmGlu-A3-1 gene and SFR159 from BAC237I6. These sequences are 96 to 97% identical to the corresponding regions of the TmGlu-A3-2 and TmGlu3-A3-3 loci, confirming that the three LMW glutenin loci are paralogs.
Figure 3.
Sequence Organization of the T. monococcum and T. durum LMW Glutenin Contigs.
(A) T. monococcum sequence contig. a, b, and c indicate regions in which no repetitive elements were identified.
(B) T. durum sequence contig.
The left end of the contigs is distal on chromosome 1AS. Positions of genes, pseudogenes, and probes used for genetic mapping are indicated above the maps. Transcriptional orientations are indicated with horizontal arrows. The details shown in Figure 6 are indicated with boxes. Colored boxes and black boxes represent gene and pseudogene predictions supported by BLASTX homologies. Genes and pseudogenes are marked as follows: 1, TmGlu-A3-2 (red box); 2, TmHG-1; 3, TmGlu-A3-3 (red box); 4, TmHG-2; 5, TmPIK-1; 6, TmRGL-1 (green box); 7, TmSTF-1; 8, TdHbox-1; 9, TmGlu-A3-1 (red box); 10, TdLRR-1; 11, TdHG-1; 12, TdHG-2; 13, TdHG-3; and 14, TdRGL-1 (green box). RFLP, restriction fragment length polymorphism.
A third putative gene, the sulfotransferase-like gene TmSTF-1, is only partially covered, because only the 5′ region is present at the right end of the contig. Two hypothetical genes (TmHG-1 and TmHG-2) were predicted by the RiceGAAS annotation system (Sakata et al., 2002), and corresponding ESTs were found (see supplemental data online). Close to the right end of the contig and upstream of TmSTF-1, two pseudogenes were found: TmRGL-1 belongs to the NBS-LRR resistance gene analog type, and TmPIK-1 shows similarity to a phosphatidylinositol-3 kinase gene from soybean.
The sequence of T. durum BAC 107G22 has a size of 142,018 bp. The 75 kb on the right side of the T. durum BAC are poor in nested repetitive elements and contain six of seven predicted genes (Figure 3B). Two putative genes (TdGlu-A3-1 and TdRGL-1) and a pseudogene (TdLRR-1) were identified by BLASTX (Basic Local Alignment Search Tool) (Figure 3B; see also supplemental data online). The LMW glutenin gene (TdGlu-A3-1) lies in the center of the T. durum sequence. Very similar to the T. monococcum contig, the region corresponding to SFR159 is found 2 kb upstream of the gene. TdRGL-1 is similar to TmRGL-1 (see below), and only its 5′ region is covered by the contig. Approximately 12 kb upstream of TdGlu-A3-1, a truncated Leu-rich repeat (LRR) of 250 amino acids (TdLRR-1; Figure 3B) was identified that shows similarity to the LRR domains of NBS-LRR resistance gene analogs. Three hypothetical genes (TdHG-1, TdHG-2, and TdHG-3) were predicted in a 16-kb region between TdGlu-A3-1 and TdRGL-1 (Figure 3B). Finally, a pseudogene of 111 bp that has 70% homology with the protein sequence encoded by the last exon of a gene that encodes a Glabra2-like1 protein, TdHbox-1 (Figure 3B, gene 8), was found inside a gypsy retrotransposon, Fatima_107G22-2 (Wicker et al., 2001). It is likely that it has been acquired by Fatima_107G22-2. Acquisition of segments of cellular genes by LTR retrotransposons has been described previously (Jin and Bennetzen, 1994; Elrouby and Bureau, 2001).
Thus, only two genes (Glu-A3 and RGL-1) are conserved between T. monococcum and T. durum in the studied region, whereas the other genes are different, indicating rapid changes in overall gene organization in the two species.
Intergenic Regions
More than 230 kb (80% of the sequence) starting at the left end of the T. monococcum contig and the first 67 kb from the left end of the T. durum contig show a pattern of nested repetitive elements (Figure 3A). In T. monococcum, in addition to 25 LTR retrotransposons and four large foldback elements that we refer to as large inverted-repeat transposable elements (LITEs; Apollo_453N11-1, Rhea_426K20-1, Rhea_ 453N11-1, and Zeus_426K20-1), we also identified 8 class-II transposons and 10 non-LTR retrotransposons. Except for Emil_453N11-1, whose ends are truncated, all identified class-II transposons have terminal inverted repeats with a conserved CACTA motif. This makes them similar to the previously described TAT-1 element from T. aestivum (Feuillet et al., 2001). Five of the CACTA transposons encode proteins similar to known transposases, whereas the other three do not appear to have any coding capacity. Two of these deletion derivatives are small (892 and 1367 bp). We refer to these elements as small nonautonomous CACTA (SNAC) transposons. All identified CACTA transposons, except for Mandrake_426K20-1, were found nested into retrotransposons or other CACTA elements. On the T. durum contig, only one CACTA element was found in the region rich in nested repetitive elements, whereas six were found in the gene-rich 75-kb region at the right end, which contains only one non-LTR retrotransposon (Karin_107G22-1) and a highly degenerated fragment of a copia retrotransposon. Five of the class-II elements are of the CACTA type, one of which encodes a putative transposase (Isaac_ 107G22-1), whereas the other four are SNAC transposons (SNAC_107G22-1, -2, -3, and -4). The sixth transposon is similar to Mutator elements (Joseph_107G22-1). A detailed characterization of the identified class-II elements is provided by Wicker et al. (2003).
The last 50 kb at the right end of the T. monococcum contig have a sequence composition that differs from the rest of the contig. The TmSTF-1 gene, the TmPIK-1 and TmRGL-1 pseudogenes, and TmHG-2 form a “gene-enriched island” with only a few and highly degraded repetitive elements. In addition, this gene-rich region contains four miniature inverted-repeat transposable elements. For ∼18 kb of the 50-kb sequence, no obvious structures or similarities to known sequences could be identified. Similarly, on the T. durum contig, >25 kb of sequence in the 75-kb region with few nested elements have no obvious structures or similarities to known sequences.
Evolution of Paralogous Loci: Duplications and Deletions Have Shaped the TmGlu-A3 Loci
The T. monococcum sequence is characterized by the presence of a 54-kb tandem duplication containing the LMW glutenin genes. Interestingly, sequence conservation is not distributed equally over the entire duplicated sequence: the first 32 kb show an overall sequence identity of 96.9% and contain 79 InDels of 1 to 50 bp, whereas the next 22 kb (which contain the glutenin gene) are 98.8% identical and contain only 27 InDels of 1 to 33 bp.
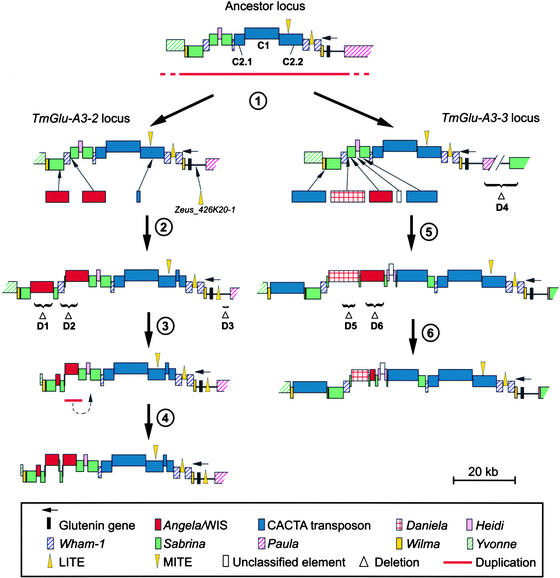
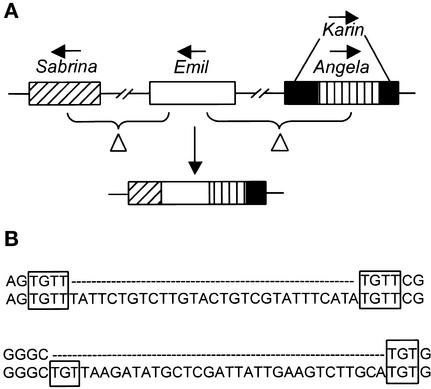
Comparison of the two duplicated sequences allowed the reconstruction of the evolutionary events that have shaped this region since the duplication. In addition to the glutenin gene, the ancestor locus must have contained numerous repetitive elements, including two copies of Sabrina (Shirasu et al., 2000), the retrotransposons Heidi, Paula, and Wham-1 (SanMiguel et al., 2002), two CACTA transposons, and two foldback elements, all of which are still present in the duplicated units. The duplicated sequences then were subjected independently to several insertions and deletions (Figure 4). The duplications and insertions postulated by the model in Figure 4 increased the size of the two loci by ∼65 kb (not including the initial 54-kb duplication). This increase in size was counteracted partially by six deletions (two intraelement recombination events and four random deletions) that resulted in the loss of at least 24 kb. Further indication of DNA loss through the deletion of random fragments was found close to the left end of the T. monococcum contig (positions 230,084 and 234,862; Figure 5A). Assuming a size of 8 kb for each of the four affected elements (Figure 5A), at least 16 kb must have been removed from this region in two deletion events. A similar pattern of repetitive elements truncated by two deletions was found close to the left end of the T. monococcum contig (data not shown).
Figure 4.
Model for the Evolution of the TmGlu-A3-2 and TmGlu-A3-3 Loci after the 54-kb Duplication in T. monococcum.
Nested repetitive elements are depicted raised above the elements into which they inserted (i.e., the CACTA element C1 has inserted into the CACTA element C2, producing the two fragments C2.1 and C2.2). Regions affected by deletions are indicated with braces. Step 1. A fragment of at least 54 kb is duplicated, creating the two paralogous loci TmGlu-A3-2 and TmGlu-A3-3. Step 2. A foldback element (Zeus_426K20-1) inserted ∼1 kb upstream of the coding region of the TmGlu-A3-2 gene and a CACTA transposon (SNAC_107G22-1) inserted 15 kb downstream of the gene. Two Angela elements inserted close to the left end of the contig. Step 3. The insertions were followed by three deletions (D1, D2, and D3). D1 was an unequal crossover event that resulted in a single LTR (Angela_426K20-1), whereas D2 and D3 were deletions of random fragments. Step 4. A small duplication of ∼4.5 kb resulted in a complex pattern of fragments of repetitive elements. Step 5. The insertions of two CACTA transposons, two LTR retrotransposons (WIS_453N11-1 and Daniela_453N11-1), and the unclassified XG_453N11-1 element increased the size of the TmGlu-A3-3 locus by >40 kb. At the same stage, a random deletion affected the flanking region of the originally duplicated fragment (D4), resulting in the truncated Sabrina_453N11-1 and a small 39-bp fragment of the Paula_453N11-1 element. Step 6. Two additional deletions (D5 and D6) affected two retrotransposons. D5 was a deletion of a random fragment within the internal domain of Daniela_453N11-1, and D6 was an unequal crossover between the two LTRs of WIS_453N11-1. MITE, miniature inverted-repeat transposable element.
Figure 5.
Patterns of DNA Deletions in Wheat.
(A) Truncated repetitive elements resulting from random fragment deletions. The deleted regions are indicated with braces. One deletion affected Sabrina_453N11-2 and the 3′ region of Emil_453N11-1 as well as the sequences located between them. A second, independent event affected the 5′ region of Emil_453N11-1 as well as a non-LTR retrotransposon (Karin_453N11-1) and Angela_453N11-1, which is nested inside Karin_453N11-1.
(B) Direct repeat structures in short InDels from the 54-kb duplication in T. monococcum. InDels were characterized by the alignment of the two duplicated units. Direct repeat structures that could serve as templates for illegitimate recombination are highlighted with boxes.
In total, 8 of the 10 large deletions in repetitive elements detected on the T. monococcum contig did not correspond to unequal crossover events between LTR sequences. Their breakpoints were searched for the presence of short direct repeats in their flanking sequences that can indicate illegitimate recombination events (Devos et al., 2002). Two breakpoints were flanked by a 2-bp direct repeat, and one was flanked by a 3-bp repeat. Two other breakpoints were flanked by a 6-bp direct repeat that carried one mismatch, and two (7 and 4 bp) carried a mismatch and were separated from one another by 1 and 2 bp, respectively. Additionally, among the 28 InDels of >3 bp that were revealed by comparison of the 54-kb tandem duplication units, 50% were associated with perfect short repeats (Figure 5B). The proportion increased to 82% (23 of 28) when repeats with one mismatch or 1 bp away from the InDel border were included. Four InDels consisted of different numbers of repeat units of simple sequence repeats and therefore might have originated from template slippage during DNA replication. Direct repeats could not be identified for only 1 of the 28 InDels. Four of the 28 analyzed InDels were duplications of 9 to 27 bp. In all four cases, the duplicated unit itself terminated in short direct repeats with sizes of 2, 3, 4, and 4 bp. Interestingly, both units of the 4.5-kb duplication (Figure 4, step 4) and of a 320-bp direct repeat upstream of TmRGL-1 also were flanked by 4-bp direct repeats. Therefore, illegitimate recombination also could be responsible for the duplication of large fragments.
Comparison of Orthologous Regions: Sequence Conservation between T. monococcum and T. durum Is Very Limited
Sequences conserved between the T. monococcum and T. durum contigs are restricted to the regions that contain the glutenin and RGL genes, whereas the rest of the two contigs (∼90% of the total sequence) shows no conservation at all. In the glutenin region, the T. durum sequence is 96% identical to the TmGlu-A3-2 and TmGlu-A3-3 loci. A region of ∼4.5 kb that includes the 1-kb coding sequence of the glutenin gene shows no major deletions or insertions in any of the three loci (Figure 6). All three genes appear to be functional.
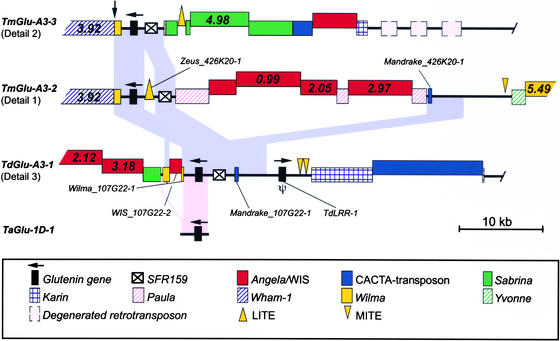
Figure 6.
Comparison between the Glu-A3 Loci of T. monococcum and T. durum and the TaGlu-1D-1 Locus from the D Genome of Hexaploid Wheat.
The depicted regions of the T. monococcum and T. durum sequence contigs correspond to details 1, 2, and 3 in Figure 3. Regions conserved between T. monococcum and T. durum are indicated by light blue areas. Regions conserved between T. monococcum, T. durum, and the TaGlu-1D-1 locus are indicated by pink areas. Repetitive elements nested into other elements are depicted raised above the elements into which they inserted. The estimated insertion times of the repetitive elements (in million years) are shown inside the elements. The vertical arrow indicates the breakpoint of a large deletion in T. monococcum. MITE, miniature inverted-repeat transposable element.
A solo-LTR of a Wilma retrotransposon was found at identical positions, 1.1 kb downstream of all three genes, indicating that this element inserted before the divergence of T. monococcum and T. urartu. Comparison of our sequences with a LMW glutenin gene isolated from chromosome 1D of T. aestivum (TaGlu-1D-1) showed 90% identity with each of the three loci over its entire length of 3165 bp. This sequence, however, is not interrupted by a Wilma element downstream of the glutenin gene (Figure 6), suggesting that the insertion of Wilma occurred only in the A genome lineage.
At greater distances from the glutenin gene, more differences can be found between T. monococcum and T. durum. The left border of the conserved region is marked in T. monococcum by a Wham-1 retrotransposon that has inserted into Wilma_ 426K20-1 and in T. durum by a WIS_107G22-2 single LTR interrupting Wilma_107G22-1 (Figure 6). Upstream of the glutenin gene, conservation of sequences between the TdGlu-A3-1 locus and the TmGlu-A3-2 locus is interrupted by the insertion of a foldback element (Zeus_426K20-1) followed by >35 kb of nested insertions of a non-LTR retrotransposon (Paula) and three LTR retrotransposons of the BARE-1 superfamily (Angela_ 426K20-4, Angela_426K20-5, and WIS_426K20-1) (Figure 6). After this interruption is a region of ∼4 kb that includes a small CACTA transposon (Mandrake_107G22-1 and Mandrake_ 426K20-1) that again shows 96% identity (Figure 6). In total, 12.4 kb surrounding the glutenin gene are conserved between the TdGlu-A3-1 and TmGlu-A3-2 loci. The size of the conserved region between the TdGlu-A3-1 and TmGlu-A3-3 loci is ∼8 kb, because the TmGlu-A3-3 locus is truncated by a deletion upstream of the glutenin gene (Figure 4, step 5).
Both contigs contain an NBS-LRR resistance-like gene close to their right ends. These genes are 85% identical in their coding regions, whereas their 5′ untranslated regions are completely different. Although the region between the LMW glutenin and the RGL genes shows no sequence conservation between the two species, the two genes are found at similar locations in both contigs (65 kb in T. monococcum and 75 kb in T. durum) (Figures 3A and 3B). In this region, both contigs contain different hypothetical genes and large regions in which no repetitive elements are found (Figures 3A and 3B). Therefore, the absence of sequence conservation here cannot be explained solely by insertions of repetitive elements.
The Glu-A3 Loci from T. monococcum and T. durum Diverged ∼1 to 3 Million Years Ago
Because of the mechanism of reverse transcription, the two LTRs of a retrotransposon are identical at the time of insertion into the genome. Therefore, the number of base substitutions between the two LTRs can be used to estimate the time of retrotransposon insertion (SanMiguel et al., 1998, 2002). Insertion times were estimated for 11 retrotransposons with intact LTRs using the average base substitution rate calculated from the grass adh1-adh2 region of 6.5 × 10−9 per site per year (Gaut et al., 1996). Our results show that all 11 elements inserted into the genome within the last 5.5 million years (Figure 6; see also supplemental data online). The estimated insertion times are consistent with the insertion order established from the positions of the individual elements in the nested structures (younger elements are inserted into older elements). The solo-LTR of Wilma (downstream of the glutenin gene; Figure 6) is the only retrotransposon conserved between the two species. Because the loci were identical at the time of their divergence, we also applied the dating method to the conserved Wilma LTR of the two species. According to this estimate, the T. monococcum and T. durum loci diverged ∼2.9 million years ago. Very similar values also were obtained when the entire region (8 kb) that is conserved in T. durum and in both T. monococcum Glu-A3 loci was used (3 to 3.13 million years ago) (Table 1). The same 8-kb region also was used to date the large 54-kb duplication in T. monococcum (1.23 million years ago; Table 1).
Table 1.
Estimated Divergence Time of the Two T. monococcum LMW Glutenin Loci Glu-A3-2 and Glu-A3-3 and the T. durum LMW Glutenin Glu-A3-1 Locus
| LMW Glutenin | Base Pairs Aligned |
Substitutions | Million Years Ago |
sd |
|---|---|---|---|---|
| TmGlu-A3-2/TmGlu-A3-3 | 7280 | 115 | 1.23 | 0.11 |
| TdGlu-A3-1/TmGlu-A3-2 | 7361 | 277 | 3.00 | 0.18 |
| TdGlu-A3-1/TmGlu-A3-3 | 7331 | 290 | 3.13 | 0.18 |
Our estimate for the A genome species divergence time differs from recently published data that wheat species carrying the A genome radiated during the last 0.5 to 1 million years (Huang et al., 2002a). A possible explanation for this discrepancy is that different haplotypes were present already in the ancestor population of the A genome species from which all other A genomes evolved. A second explanation is that different regions of the genome might evolve at different rates, resulting in differences in their estimated divergence times. LTRs in retrotransposons as well as other noncoding sequences show high mutation frequencies in CG/CNG sites because of the 5′ methylation of deoxycytidines (SanMiguel et al., 1998). In the LTR sequences used to date retrotransposon insertions, 65% of the transitions occurred in CG/CNG sites. This number does not differ significantly from the observed 60% of transitions in CG/CNG sites in the 8-kb region used to date the T. monococcum duplication as well as the T. monococcum/T. durum divergence (data not shown). Thus, it is likely that the relative dating of this sequence versus LTR sequences is accurate but that the absolute dates are overestimated.
Interestingly, 6 of the 11 dated retrotransposon insertions were predicted to have occurred before the divergence of the T. monococcum and T. durum loci (Figure 6; see also supplemental data online), but none of the 6 elements is conserved between the two species. It is likely that the absence of the oldest dated retroelements is caused by deletions that occurred in one of the two species. In T. monococcum, a breakpoint of such a putative deletion was identified (indicated by a vertical arrow in Figure 6). As a consequence of this large deletion, sequences that are not covered by the T. durum BAC were shifted into the window of observation provided by the T. monococcum contig. However, given the possible differences in evolution rates between the LTR sequences and other sequences (SanMiguel et al., 1998), in our case the 8-kb fragment, we cannot exclude the possibility that some of the nonconserved retroelements inserted only after the species divergence.
DISCUSSION
Mechanisms of Genome Divergence at the Glu-A3/SFR159 Loci in Wheat
The characterization of orthologous LMW Glu-A3/SFR159 loci from T. monococcum and T. durum and the subsequent analysis of large sequence stretches from both loci revealed a complex evolution of this region. Several mechanisms acted on the orthologous Glu-A3/SFR159 loci and resulted in a different genome landscape. The first differences were caused by a differential amplification of the glutenin loci. In T. monococcum, large duplication events resulted in the presence of three loci of the glutenin gene associated with the low-copy probe SFR159, whereas only one locus of a LMW glutenin in T. durum was associated with SFR159. Other LMW glutenin genes are present on chromosome 1A of T. durum (our unpublished results), but none of them is linked to SFR159. This finding indicates that different duplication events have occurred at the LMW glutenin locus in the T. urartu lineage compared with the T. monococcum lineage or that further deletion events may have eliminated the SFR159 region from the duplicated units. Our estimate of the TmGlu-A2 and TmGlu-A3 date of divergence favors the first hypothesis. To confirm this hypothesis, we performed restriction fragment length polymorphism (RFLP) analysis of SFR159 in different accessions of T. monococcum and T. urartu and in wild tetraploid wheat (Triticum dicoccoides; AABB). The findings of this analysis indicated that the triplicate pattern of SFR159 in cv DV92 is present in some, but not all, T. monococcum accessions. Moreover, we found that variation exists among T. monococcum and T. urartu genomes at this locus (our unpublished results).
In addition to differential gene amplification, transposable element insertions and subsequent deletions contributed to the differentiation of the regions studied. The duplicated fragments in T. monococcum include large stretches of transposable elements that were subsequently enlarged differentially by insertions of additional repetitive elements. Deletions and small duplication processes that act on these regions contributed further to the divergence of part of the duplicated sequence, whereas the glutenin gene regions were less affected by large-scale rearrangements. In T. durum, the distal side of the Wilma LTR/LMW-glutenin/SFR159 region is bordered by a region rich in retroelements with no similarity to those found at similar positions in T. monococcum. This finding indicates that similar mechanisms of transposable element insertions and deletions occurred independently in both species, resulting in large differences in the nongenic space. Previous studies, including recent work on small genomic fragments of high molecular weight glutenins in the Triticeae, have shown that retroelement insertions are a major mechanism of divergence of paralogous and orthologous intergenic regions (Anderson et al., 2002). This notion is demonstrated here on a much larger scale. In addition and in contrast to previous studies in the Triticeae, a large number of class-II elements also participated in the divergence of intergenic regions. They contribute ∼24 and 21% to the total sequence of our T. monococcum and T. durum contigs, respectively. These numbers do not necessarily reflect the global contribution of class-II elements to the entire wheat genome. However, they suggest that these elements may occur more frequently in the Triticeae genomes than was assumed previously. The observed high density of class-II elements may have contributed to an increased instability of the region, and some of the numerous small InDels identified on the T. monococcum contig could be footprints generated upon the excision of transposons from the genome.
In both species, a large region proximal to the glutenin genes with a low content of nested repetitive elements was identified. A pseudogene and a partial gene of the same RGL family were the only similar gene sequences identified between the two species in this region. The presence of different sets of predicted genes and the lack of similarity in the rest of the region indicate large rearrangements. The mosaic structure of conserved and nonconserved genes is similar to the observations in a comparative study of the zein locus of maize and orthologous regions in sorghum and rice (Song et al., 2002). Gene movement and translocations (Song et al., 2002), as well as large-scale deletions that occurred independently in T. monococcum and T. durum, likely are responsible for the actual structure of the Glu-A3 regions.
Genome Expansion and Contraction through Illegitimate Recombination
In addition to the insertions of repetitive elements, duplications can contribute to genome expansion. At the LMW glutenin locus on chromosome 1D, the detection of multiple loci by most RFLP probes suggested that gene duplication events have occurred throughout this chromosomal region (Spielmeyer et al., 2000). We found large duplication events leading to the three identified glutenin loci in T. monococcum and duplications of smaller fragments (from a few base pairs up to 4.5 kb). Our data indicate that, in addition to unequal recombination, illegitimate recombination (Devos et al., 2002) is a possible molecular mechanism that generates duplications. Illegitimate recombination requires only a few base pairs of sequence identity and therefore could explain the apparently random distribution, size, and sequence composition of the duplicated units.
The increase in genome size through duplications and insertions of repetitive elements is counteracted partially by the loss of DNA (Bennetzen, 2002). In addition to the loss of DNA through intraelement crossover that results in solo-LTRs, as reported previously in the Triticeae (reviewed by Bennetzen, 2002), a second type of deletion was observed. It affected large random fragments of repetitive DNA, and in one case, it probably resulted in the truncation of the TdLRR-1 pseudogene. In previous studies of Triticeae genomes, extensive deletions were reported that are not associated with intraelement crossover or intrastrand interelement recombination (Wicker et al., 2001; SanMiguel et al., 2002). In the T. durum and T. monococcum sequences studied here, the major deletion events that might be attributable to illegitimate recombination clearly outnumber the deletions caused by intraelement recombination. These data are consistent with findings in Arabidopsis, in which illegitimate recombination also is believed to be primarily responsible for the removal of nonessential DNA (Devos et al., 2002). Therefore, we suggest that illegitimate recombination is a major factor in the evolution of the wheat genome. We speculate that it is of importance in the creation of new genetic “raw material” through duplications of random fragments as well as in counteracting the increase in genome size through deletions.
The Intergenic Regions of the Wheat Genome Evolve Very Rapidly
Most of the studies on microcolinearity between genomes of grass species have been performed on species that diverged >10 million years ago (Bennetzen and Ramakrishna, 2002; Ramakrishna et al., 2002a; SanMiguel et al., 2002; Song et al., 2002). They have shown that insertions of new elements, deletions, and in some cases gene translocation and amplification may eliminate any similarity in intergenic regions. We found that similarities in the Triticum intergenic regions already were eliminated within 1 to 3 million years, which is at least three times more rapid than has been suggested previously (SanMiguel et al., 2002). This is likely the case in the entire wheat genome, although the ratio between conserved and nonconserved regions might show variations. In our case, only 8% of the analyzed region was conserved, including the gene space. In another study, comparison of a 340-kb colinear region of T. monococcum chromosome 5AmL with T. durum chromosome 5A in the Vrn-2 region revealed 100 kb of colinear sequence (97.3% identical). After excluding five conserved gene regions (16 kb), the conserved segments represented 26% of the intergenic regions of the T. monococcum sequence (J. Dubcovsky and P. SanMiguel, unpublished data), which indicates extensive and rapid changes in these regions as well. The various InDels of DNA that took place after the large 54-kb duplication in T. monococcum demonstrate that the wheat genome has undergone dramatic changes even in “recent” evolutionary times (<1.2 million years ago).
These data imply that significant differences can be expected even between different wheat varieties. Analysis of orthologous regions in different species usually relies on the assumption that the genotypes chosen for the analysis represent the entire species. However, exceptional haplotype variation was reported recently at orthologous bz loci in maize (Fu and Dooner, 2002), and in wheat, RFLP studies have detected the existence of different haplotypes in diploid wild wheat species that were transmitted to different genotypes of hexaploid wheat (Scherrer et al., 2002). We have found similar evidence for polymorphic haplotypes at the SFR159 loci in different T. monococcum, T. urartu, and T. dicoccoides accessions (our unpublished results), and we are currently investigating the presence of these haplotypes in hexaploid wheat.
We have shown that similarities in A genome intergenic regions are eliminated already within 1 to 3 million years. Given that the diploid Triticum/Aegilops species (A, B, and D genome species) radiated 5 to 6.9 million years ago (Allaby et al., 1999), we expect that the three genomes that form the polyploid wheat genome are highly differentiated in their intergenic regions by differential insertion and deletion events. This divergence probably is responsible for the size difference of the three genomes (Kellogg, 1998) and is increased by the elimination of low-copy sequences and retrotransposition events that occur during polyploidization (Ozkan et al., 2001). Additional studies of orthologous loci in diploid, tetraploid, and hexaploid wheat as well as in other Triticeae species are required to gain a better overview of their genomes and to determine whether our observations can be generalized.
METHODS
Plant Material
Genetic mapping was performed on an F2 population of 1340 plants from the cross between Triticum aestivum lines Chul and Frisal. For restriction fragment length polymorphism studies, leaf material was harvested from diploid, tetraploid, and hexaploid wheat lines. Diploid Triticum monococcum subsp monococcum line DV92 (Am genome) and tetraploid Triticum turgidum subsp durum cv Langdon (AB genome) were used. The copy numbers and chromosomal locations of restriction fragment length polymorphism probes were determined by DNA gel blot hybridization using aneuploid nullitetrasomic lines of hexaploid wheat, Chinese Spring (Sears, 1966), and Langdon/Chinese Spring substitution lines in which chromosomes of Langdon were substituted by the Chinese Spring D genome homologs (Joppa and Williams, 1988).
Restriction Fragment Length Polymorphism Analysis
Isolation of genomic DNA, DNA gel blot hybridization, labeling experiments, and linkage analysis were performed as described by Stein et al. (2000). Twenty micrograms of genomic DNA was used for hexaploid wheat lines, 13 μg was used for tetraploid lines, and 7 μg was used for diploid lines.
BAC Library Screening and BAC Analysis
Screening of the T. monococcum (Lijavetzky et al., 1999) and the T. durum BAC library (http://agronomy.ucdavis.edu/Dubcovsky/BAC-library/BAC_Langdon.htm) was performed by hybridization as described by Stein et al. (2000). BAC DNA preparation for fingerprint analysis and BAC end sequencing were performed as described previously (Stein et al., 2000). BAC clones were assembled into contigs based on their HindIII, NotI, and SalI restriction patterns and hybridization with the BAC ends.
Shotgun Sequencing
Preparation of shotgun clones from BAC clones 107G22 (T. durum) and 426K20, 453N11, and 237I6 (T. monococcum) was performed as described by Stein et al. (2000). A 95-kb NotI fragment from 18B1, which overlaps with BACs 453N11 and 426K20, was chosen for the construction of a partial shotgun library. BAC DNA (20 μg) was digested with NotI and run on a pulse field gel electrophoresis gel overnight. The 95-kb fragment was excised from the gel, and the DNA was eluted by electroelution and used directly for mechanical shearing. All subsequent steps of shotgun library production were performed as described for the other BAC clones. Plasmid DNA from shotgun clones was extracted with BioRobot 9600 (Qiagen, Valencia, CA) using the Wizard SV96 plasmid DNA purification system (Promega, Wallisellen, Switzerland) and used directly for cycle sequencing from both directions on an ABI PRISM 377 sequencer (Perkin-Elmer Applied Biosystems, Rotkreuz, Switzerland). Gaps between subcontigs were filled by PCR using 20-mer oligonucleotides with a minimal GC content of 60%. PCR products were cloned into the pGEM-T Easy Vector (Promega). Difficult sequences or large PCR products were sequenced using automatic DNA Sequencer 4200 (Li-Cor, Lincoln, NE).
Sequence Analysis
Base calling and quality of the shotgun sequences were processed using Phred (Ewing et al., 1998) and assembled using the Phrap assembly engine (version 0.990319; provided by P. Green and available at http://www.phrap.org). The BACs were sequenced up to nine times coverage. Subcontigs and singlet DNA sequences were analyzed using BLAST (Basic Local Alignment Search Tool) algorithms (Altschul et al., 1997) against public DNA and protein sequence databases. Detailed sequence analysis was performed with the GCG Sequence Analysis Software Package version 10.1 (Madison, WI) and by dot-plot analysis (DOTTER; Sonnhammer and Durbin, 1995). Analysis of repetitive sequences and transposable elements was performed by BLAST search against public databases, the database for Triticeae repetitive DNA (TREP; http://wheat.pw.usda.gov/ITMI/Repeats) (Wicker et al., 2002), and a local database for repetitive DNA. Direct and inverted repeat sequences were identified with the GCG program BESTFIT and dot-plot analysis of the query sequence against itself. For gene prediction in regions in which no putative genes or repetitive elements were identified by BLASTX or BLASTN, the RiceGAAS annotation system (Sakata et al., 2002) was used. Predicted genes were considered if they were predicted by at least two prediction programs. Genes and proteins are named with the first two letters representing the initial letters of the Latin binomial followed by the original symbol. For efficient processing of large sets of sequences, programs were written using the language PERL. Dating of retrotransposon insertions and divergence times between different loci was performed as described by SanMiguel et al. (1998).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The sequences presented in this study were deposited in GenBank under the accession numbers AY146587 (T. durum contig) and AY146588 (T. monococcum contig). GenBank accession numbers of other sequences mentioned in this article are P42347 (soybean phosphatidylinositol 3-kinase) and X13306 (low molecular weight glutenin gene TaGlu-1D-1). TrEMBL accession numbers for other sequences mentioned in this article are Q9M1V2 (Arabidopsis sulfotransferase-like protein) and Q9LFW5 (Arabidopsis homeodomain Glabra2-like1 protein).
Supplementary Material
Acknowledgments
We thank L.R. Joppa for providing seeds of the disomic substitution lines. We also are grateful to Catherine Feuillet and Clair Wicker for critical reading of the manuscript. We thank Stephanie Narain-Meier for her excellent technical assistance. This work was supported by Grant 31-65114.01 from the Swiss National Science Foundation.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.011023.
Footnotes
Online version contains Web-only data.
References
- Allaby, R.C., Banerjee, M., and Brown, T.A. (1999). Evolution of the high molecular weight glutenin loci of the A, B, D, and G genomes of wheat. Genome 42, 296–307. [PubMed] [Google Scholar]
- Altschul, S., Madden, T.L., Schaeffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, O.D., Larka, L., Christoffers, M.J., McCue, K.F., and Gustafson, J.P. (2002). Comparison of orthologous and paralogous DNA flanking the wheat high molecular weight glutenin genes: Sequence conservation and divergence, transposon distribution, and matrix-attachment regions. Genome 45, 367–380. [DOI] [PubMed] [Google Scholar]
- Arumuganathan, K., and Earle, E.D. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9, 208–218. [Google Scholar]
- Bennetzen, J., and Ramakrishna, W. (2002). Numerous small rearrangements of gene content, order and orientation differentiate grass genomes. Plant Mol. Biol. 48, 821–827. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L. (2002). Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica 115, 29–36. [DOI] [PubMed] [Google Scholar]
- Colot, V., Bartels, D., Thompson, R., and Flavell, R. (1989). Molecular characterization of an active wheat LMW glutenin gene and its relation to other wheat and barley prolamin genes. Mol. Gen. Genet. 216, 81–90. [DOI] [PubMed] [Google Scholar]
- Devos, K.M., Brown, J.K.M., and Bennetzen, J.L. (2002). Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12, 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak, J., McGuire, P.E., and Cassidy, B. (1988). Apparent sources of the A genomes of wheats inferred from polymorphism in abundance and restriction fragment length of repeated nucleotide sequences. Genome 30, 680–689. [Google Scholar]
- Dvorak, J., and Zhang, H.B. (1992). Reconstruction of the phylogeny of the genus Triticum from variation in repeated nucleotide sequences. Theor. Appl. Genet. 84, 419–429. [DOI] [PubMed] [Google Scholar]
- Elrouby, N., and Bureau, T.E. (2001). A novel hybrid open reading frame formed by multiple cellular gene transductions by a plant long terminal repeat retroelement. J. Biol. Chem. 276, 41963–41968. [DOI] [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. (1998). Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8, 175–185. [DOI] [PubMed] [Google Scholar]
- Feldman, M., Lupton, F.G.H., and Miller, T.E. (1995). Wheats. In Evolution of Crops, 2nd ed., J. Smartt and N.W. Simmonds, eds (London: Longman Scientific), pp. 184–192.
- Feuillet, C., and Keller, B. (1999). High gene density is conserved at syntenic loci of small and large grass genomes. Proc. Natl. Acad. Sci. USA 96, 8265–8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet, C., and Keller, B. (2002). Comparative genomics in the grass family: Molecular characterization of grass genome structure and evolution. Ann. Bot. 89, 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet, C., Penger, A., Gellner, K., Mast, A., and Keller, B. (2001). Molecular evolution of receptor-like kinase genes in hexaploid wheat: Independent evolution of orthologs after polyploidization and mechanisms of local rearrangements at paralogous loci. Plant Physiol. 125, 1304–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell, R.B., Bennett, M.D., Smith, J.B., and Smith, D.B. (1974). Genome size and proportion of repeated nucleotide sequence DNA in plants. Biochem. Genet. 12, 257–269. [DOI] [PubMed] [Google Scholar]
- Fu, H.H., and Dooner, H.K. (2002). Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99, 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S., Morton, B.R., McCaig, B.C., and Clegg, M.T. (1996). Substitution rate comparisons between grasses and palms: Synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. USA 93, 10274–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S., Sirikhachornkit, A., Su, X.J., Faris, J., Gill, B., Haselkorn, R., and Gornicki, P. (2002. a). Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA 99, 8133–8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S.X., Sirikhachornkit, A., Faris, J.D., Su, X.J., Gill, B.S., Haselkorn, R., and Gornicki, P. (2002. b). Phylogenetic analysis of the acetyl-CoA carboxylase and 3-phosphoglycerate kinase loci in wheat and other grasses. Plant Mol. Biol. 48, 805–820. [DOI] [PubMed] [Google Scholar]
- Jin, Y.K., and Bennetzen, J.L. (1994). Integration and non random mutation of a plasma-membrane proton ATPase gene fragment within the Bs1 retroelement of maize. Plant Cell 6, 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joppa, L.R., and Williams, N.D. (1988). Langdon durum disomic substitution lines and aneuploid analysis in tetraploid wheat. Genome 30, 222–228. [Google Scholar]
- Kellogg, E.A. (1998). Relationships of cereal crops and other grasses. Proc. Natl. Acad. Sci. USA 95, 2005–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lijavetzky, D., Muzzi, G., Wicker, T., Keller, B., Wing, R., and Dubcovsky, J. (1999). Construction and characterization of a bacterial artificial chromosome (BAC) library for the A genome of wheat. Genome 42, 1176–1182. [PubMed] [Google Scholar]
- McIntosh, R.A., Hart, G.E., Devos, K.M., Gale, M., and Rogers, W.J. (1998). Catalogue of gene symbols for wheat. In Proceedings of the 9th International Wheat Genetics Symposium. (Saskatoon, Saskatchewan, Canada: University Extension Press, University of Saskatchewan), pp. 99–108.
- Ozkan, H., Levy, A.A., and Feldman, M. (2001). Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13, 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov, D.A. (2001). Evolution of genome size: New approaches to an old problem. Trends Genet. 17, 23–28. [DOI] [PubMed] [Google Scholar]
- Ramakrishna, W., Dubcovsky, J., Park, Y.-J., Busso, C., Emberton, J., SanMiguel, P., and Bennetzen, J. (2002. a). Different types and rates of genome evolution detected by comparative sequence analysis of orthologous segments from four cereal genomes. Genetics 162, 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna, W., Emberton, J., Ogden, M., SanMiguel, P., and Bennetzen, J. (2002. b). Structural analysis of the maize Rp1 complex reveals numerous sites and unexpected mechanisms of local rearrangement. Plant Cell 14, 3213–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata, K., et al. (2002). RiceGAAS: An automated annotation system and database for rice genome sequence. Nucleic Acids Res. 30, 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, P., Gaut, B.S., Tikhonov, A., Nakajima, Y., and Bennetzen, J.L. (1998). The paleontology of intergene retrotransposons of maize. Nat. Genet. 20, 43–45. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., et al. (1996). Nested retrotransposons in the intergenic regions of the maize genome. Science 274, 765–768. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P.J., Ramakrishna, W., Bennetzen, J.L., Busso, C., and Dubcovsky, J. (2002). Transposable elements, genes and recombination in a 215-kb contig from wheat chromosome 5Am. Funct. Integr. Genomics 2, 70–80. [DOI] [PubMed] [Google Scholar]
- Scherrer, B., Keller, B., and Feuillet, C. (2002). Two haplotypes of resistance gene analogs have been conserved during evolution at the leaf rust resistance locus Lr10 in wild and cultivated wheat. Funct. Integr. Genomics 2, 40–50. [DOI] [PubMed] [Google Scholar]
- Sears, E.R. (1966). Nullisomic-tetrasomic combinations in hexaploid wheat. In Chromosome Manipulations and Plant Genetics, R. Riley and K.R. Lewis, eds (Edinburgh, UK: Oliver and Boyd), pp. 29–45.
- Shirasu, K., Schulman, A.H., Lahaye, T., and Schulze-Lefert, P. (2000). A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res. 10, 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., Llaca, L., and Messing, J. (2002). Mosaic organization of orthologous sequences in grass genomes. Genome Res. 12, 1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer, E.L.L., and Durbin, R. (1995). A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167, GC1–GC10. [DOI] [PubMed] [Google Scholar]
- Spielmeyer, W., Moullet, O., Laroche, A., and Lagudah, E.S. (2000). Highly recombinogenic regions at seed storage protein loci on chromosome 1DS of Aegilops tauschii, the D-genome donor of wheat. Genetics 155, 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, N., Feuillet, C., Wicker, T., Schlagenhauf, E., and Keller, B. (2000). Subgenome chromosome walking in wheat: A 450-kb physical contig in Triticum monococcum L. spans the Lr10 resistance locus in hexaploid wheat (Triticum aestivum L.). Proc. Natl. Acad. Sci. USA 97, 13436–13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker, T., Guyot, R., Yahiaoui, N., and Keller, B. (2003). CACTA transposons in Triticeae: A diverse family of high-copy repetitive elements. Plant Physiol., in press. [DOI] [PMC free article] [PubMed]
- Wicker, T., Matthews, D.E., and Keller, B. (2002). TREP: A database for Triticeae repetitive elements. Trends Plant Sci. 7, 561–562. [Google Scholar]
- Wicker, T., Stein, N., Albar, L., Feuillet, C., Schlagenhauf, E., and Keller, B. (2001). Analysis of a contiguous 211 kb sequence in diploid wheat (Triticum monococcum L.) reveals multiple mechanisms of genome evolution. Plant J. 26, 307–316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.