Abstract
The CLAVATA1 (CLV1) receptor kinase controls stem cell number and differentiation at the Arabidopsis shoot and flower meristems. Other components of the CLV1 signaling pathway include the secreted putative ligand CLV3 and the receptor-like protein CLV2. We report evidence indicating that all intermediate and strong clv1 alleles are dominant negative and likely interfere with the activity of unknown receptor kinase(s) that have functional overlap with CLV1. clv1 dominant-negative alleles show major differences from dominant-negative alleles characterized to date in animal receptor kinase signaling systems, including the lack of a dominant-negative effect of kinase domain truncation and the ability of missense mutations in the extracellular domain to act in a dominant-negative manner. We analyzed chimeric receptor kinases by fusing CLV1 and BRASSINOSTEROID INSENSITIVE1 (BRI1) coding sequences and expressing these in clv1 null backgrounds. Constructs containing the CLV1 extracellular domain and the BRI1 kinase domain were strongly dominant negative in the regulation of meristem development. Furthermore, we show that CLV1 expressed within the pedicel can partially replace the function of the ERECTA receptor kinase. We propose the presence of multiple receptors that regulate meristem development in a functionally related manner whose interactions are driven by the extracellular domains and whose activation requires the kinase domain.
INTRODUCTION
The generation of aerial organs in plants is dependent on the activity of shoot and flower meristems. Shoot meristems maintain organogenesis by balancing the proliferation of a population of undifferentiated stem cells at their centers and directing appropriately positioned descendant cells toward organ formation and eventual differentiation. Some mutants have been found to disrupt this balance, altering the structure and function of shoot and flower meristems. Mutations in the Arabidopsis CLAVATA loci (CLV1, CLV2, and CLV3) lead to ectopic accumulation of stem cells at shoot and flower meristems (Clark et al., 1993, 1995; Kayes and Clark, 1998). Enlarged clv meristems result in extra flower organs, club-shaped siliques, and enlarged inflorescence stems. Molecular genetic studies indicate that the primary function of the CLV loci is to restrict the expression domain of the stem cell–promoting factor WUSCHEL (WUS). WUS encodes a putative homeodomain transcription factor that has been shown to be both necessary and sufficient for stem cell identity (Laux et al., 1996; Mayer et al., 1998). The WUS expression domain expands in clv mutants, and ectopic expression of WUS within the meristem is sufficient to promote stem cell identity (Brand et al., 2000, 2002; Schoof et al., 2000; Lenhard et al., 2002).
clv1 alleles vary greatly in phenotypic severity, ranging from weak alleles with only modest increases in stem cell number to strong alleles with >1000-fold more undifferentiated cells compared with the wild type (Clark et al., 1993). Mutant alleles of clv1, clv2, or clv3 show strong correlations between shoot and flower meristem defects. Increases in flower meristem size are reflected in an increase in the number of organs per flower, especially the number of carpels, as well as in the extent of continued proliferation and organogenesis of the flower meristem beyond the initiation of the whorl-four carpels (Clark et al., 1993). Carpel number typically has been used to measure the severity of clv mutant defects.
The receptor-like kinases (RLKs), of which CLV1 is a member, represent one of the largest gene families in the Arabidopsis genome. One-third of the ∼610 RLKs contain Leu-rich repeats (LRRs) in their extracellular domains (Shiu and Bleecker, 2001). LRRs are found in tandem repeats that each contain ∼24 amino acids with conserved leucines. LRRs are present in a variety of proteins with diverse functions, from yeast to animals and plants, and many are involved in protein–protein interactions. Among the ∼220 transmembrane LRR receptors in Arabidopsis, only a few have known functions. The challenge now is to identify their signaling components, ligands, and mechanisms of activation. Substantial efforts have been devoted to a few LRR-RLKs that play critical roles in development. These include CLV1, BRASSINOSTEROID INSENSITIVE1 (BRI1), which is involved in the perception and signaling of the steroid hormone brassinolide (BL), and ERECTA (ER), which regulates organ shape (Torii et al., 1996; Clark et al., 1997; Li and Chory, 1997; He et al., 2000). The LRR-RLKs can be subdivided into subfamilies on the basis of numbers and locations of LRRs in their extracellular domains (Shiu and Bleecker, 2001). This classification puts CLV1 and ER in the same group of LRR-RLKs with 21 LRRs in their extracellular domains. On the other hand, BRI1 belongs to a separate subfamily because it possesses 25 LRRs and an island of 70 amino acids between the 20th and 21st LRRs. This island is proposed to mediate BRI1 binding to BR (Li and Chory, 1997; He et al., 2000). The ligand for the ER receptor remains unknown.
The CLV1 pathway, although still poorly understood, is one of the best characterized receptor kinase pathways in plants. The other two CLV genes, CLV2 and CLV3, have been cloned and encode components of the same signal transduction pathway that regulates stem cell behavior. CLV2 belongs to the receptor-like protein family and contains LRR motifs in its predicted extracellular domain but no intracellular kinase domain (Jeong et al., 1999). CLV3 encodes a small polypeptide that has been shown to be secreted (Fletcher et al., 1999; Rojo et al., 2002). The CLV1, CLV2, and CLV3 genes function in the same pathway, the formation of active CLV1 complexes requires CLV3, CLV3 overexpression phenotypes require CLV1 and CLV2, and CLV3 is secreted from cells adjacent to those expressing CLV1—all suggesting that CLV3 acts as the ligand for CLV1 (Clark et al., 1995, 1997; Fletcher et al., 1999; Trotochaud et al., 1999; Brand et al., 2000; Rojo et al., 2002).
Since the first description of the clv mutant phenotype more than a decade ago, 10 different alleles of clv1 have been discovered and described (Leyser and Furner, 1992; Medford et al., 1992; Clark et al., 1993, 1997; Pogany et al., 1998). They exhibit phenotypes ranging from weak (clv1-6 and clv1-7) to intermediate (clv1-1, clv1-2, and clv1-9) to strong (clv1-4, clv1-8, and clv1-10). All of the alleles that exhibit strong or intermediate phenotypes contain missense mutations within the LRR or the kinase coding sequences (Clark et al., 1997). Interestingly, the weakest alleles, clv1-6 and clv1-7, contain a frameshift and a nonsense mutation, respectively, near the beginning of the kinase domain that could eliminate most of the kinase domain. Although the clv1-6 and clv1-7 alleles suggested possible dominant-negative properties of the intermediate and strong alleles, it has not been possible to exclude a low level of read-through or the notion that the truncated proteins performed significant signaling activity.
Here, we provide evidence that all strong and intermediate clv1 alleles are dominant negative. We have isolated three insertional alleles that all exhibit weak phenotypes. We show that cosuppression of clv1-1 mRNA expression partially suppresses Clv1− phenotypes and that chimeric CLV1/BRI1 receptors can act as dominant-negative proteins in meristem development. We also show that CLV1 expressed within pedicels can regulate their development. Finally, we propose that multiple receptor kinases with functional overlap act within the meristem and perhaps in other organs as well.
RESULTS
Characterization of Three T-DNA Insertions in the CLV1 Locus
Despite the sequencing of over 10 clv1 alleles, no clear null alleles have been identified (Clark et al., 1997). All of the clv1 alleles that exhibit a strong or intermediate phenotype contain missense mutations within the coding sequence (Figures 1A and 1C). Even clv1-10, which was identified as an intragenic enhancer of clv1-1, contains a second missense mutation in the LRR domain (see Methods). The only alleles with nonsense or frameshift mutations are clv1-6 and clv1-7, which were isolated in pollen mutagenic screens for additional clv1 alleles (Clark et al., 1993). However, the lesions in clv1-6 and clv1-7 both occur after the transmembrane domain, leaving the possibility that the proteins these alleles encode are expressed, located to the plasma membrane, and capable of signaling (Figures 1A and 1C). Precedence for this idea comes from the disease resistance receptor kinase Xa-21, which is capable of functioning without a kinase domain (Wang et al., 1998).
Figure 1.
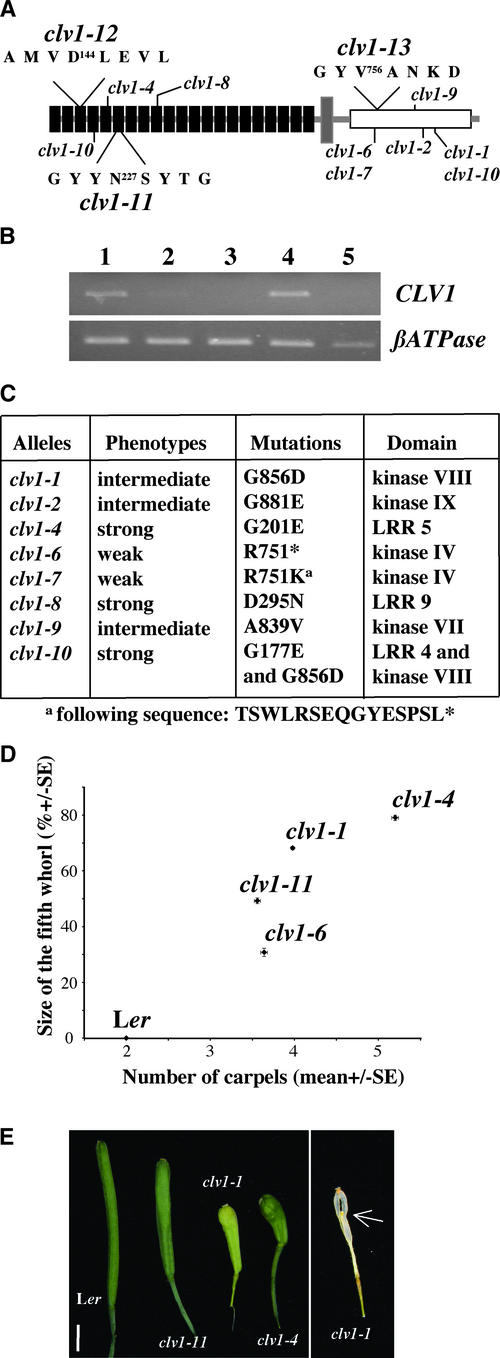
Alleles of clv1.
(A) Scheme of known alleles of clv1 and locations of the DsE insertion (clv1-11) and the T-DNA insertions (clv1-12 and clv1-13) of three novel null alleles of clv1. The amino acid sequences surrounding the integration sites are indicated. The LRRs (black boxes), transmembrane domain (gray box), and kinase domains (white box) are represented.
(B) Ethidium bromide agarose gels from the RT-PCR performed on Col (lane 1), clv1-12 (lane 2), clv1-13 (lane 3), Ler (lane 4), and clv1-11 (lane 5) samples. mRNA isolated from inflorescences was analyzed by RT-PCR to monitor the transcript accumulation of CLV1. Each PCR amplification of the cDNA was generated in parallel with specific primers for CLV1 (top gel) and control (βATPase; bottom gel). RT-PCR products from Col and Ler plants were examined as controls.
(C) For each allele, the phenotypic severity, the amino acid mutated, and the domain affected are indicated.
(D) Relative size of the fifth whorl compared with the number of carpels in clv1-1, clv1-6, clv1-11, clv1-4, and Ler. Ten flowers from at least 20 primary inflorescences of 30-day-old plants were scored for the number of carpels (means ± se). The size of the silique represents the length between the attachment site for the sepals, petals, and stamens and the top of the gynoecium. The same references were used for the measurement of fifth-whorl length. The measured size of the fifth whorl is represented as a percentage of total silique length. The vertical and horizontal bars represent standard errors for the relative fifth-whorl size and the number of carpels, respectively. Note that there is no fifth whorl in Ler and that the number of carpels does not vary from two, so the standard error is 0.
(E) Comparison of siliques from Ler plants. The shape of the siliques from Ler, clv1-11, clv1-1, and clv1-4 varies with the severity of the Clv1− phenotype. Note the club shape of the intermediate clv1-1 and strong clv1-4 alleles compared with the weak clv1-11 silique shape. At right is shown a dry clv1-1 silique revealing the fifth whorl of organ growing inside the gynoecium and representing ∼70% of the size of the silique. Bar = 2 mm.
To determine the null phenotype for CLV1, we identified one Ds transposable element (DsE) and two T-DNA insertion clv1 alleles. We named these three new alleles clv1-11, clv1-12, and clv1-13 (see Methods). clv1-12 was isolated in the Wassilewskija-2 (Ws-2) background and contains a T-DNA insertion in the third LRR. clv1-13 was isolated in the Ws-2 background, and a T-DNA insertion was identified between kinase conserved motifs IV and V. clv1-11 was isolated in the Landsberg erecta (Ler) background and contains a DsE insertion in the sixth LRR (Figure 1A). The location of each insertion was determined by PCR amplification of flanking regions followed by sequencing of the PCR products (data not shown). Reverse transcriptase (RT)–PCR was performed on wild-type, clv1-11, clv1-12, and clv1-13 RNA samples to verify that these were null alleles. We detected no accumulation of transcripts corresponding to sequences downstream of the clv1-11 and clv1-12 insertions (Figure 1B).
clv1 mutant plants accumulate stem cells at their shoot and flower meristems, leading to enlarged meristems and additional floral organs. Although wild-type plants develop exclusively two carpels in whorl four of the flower, flowers of the strongest clv1 mutants develop up to eight carpels, and a fifth whorl of ectopic organs develops inside the gynoecium generated by the whorl four of carpels (Clark et al., 1993). The number of carpels formed per flower and the extent of growth of the ectopic whorls reflect an early increase in flower meristem size and are sensitive indicators of clv1 mutant severity (Clark et al., 1993, 1995). We compared clv1-11 in the Ler background with clv1-6, clv1-1, and clv1-4, our reference weak, intermediate, and strong alleles, respectively. Both the number of carpels per flower and the size of the fifth whorl were analyzed. The phenotype of clv1-11 was less severe than that of clv1-1 and was comparable to those of the weak alleles clv1-6 and clv1-7 (Figures 1D and 1E) (Clark et al., 1993). Note the club shape of the siliques of the intermediate clv1-1 and strong clv1-4 alleles compared with those of the weak allele clv1-11 (Figure 1E). clv1-12 and clv1-13 displayed similarly weak phenotypes (see below). Thus, the clv1 null phenotype is weak, suggesting that strong and intermediate clv1 alleles may be dominant negative.
Generation of CLV1-BRI1 Chimeric Receptors
To determine the requirement for individual CLV1 domains in regulating meristem development, we generated various chimeric receptors (Figure 2). The coding sequences for portions of CLV1 were fused to those for the steroid receptor kinase BRI1 under the control of the ER promoter. ER encodes a receptor kinase that regulates organ shape and that is expressed broadly within the meristems and developing organ primordia (Yokoyama et al., 1998). We chose the ER regulatory sequences to drive expression for several reasons. First, ER expression appears to entirely encompass the CLV1 expression domain (Yokoyama et al., 1998). Second, despite using extended upstream and downstream sequences, we failed to isolate CLV1 regulatory sequences that can rescue clv1 mutants or mimic CLV1 expression (data not shown). Third, ER encodes a receptor kinase, so we would not be driving expression of the chimeric receptors at unusually high levels. The ER:CLV1 construct provided close to complete rescue of the Clv1− phenotype when transformed into clv1-1 plants (Figure 3A). The mean number of carpels per flower in clv1-1 ER:CLV1 plants is only slightly higher than that of clv1-1/+ heterozygous plants (Clark et al., 1996), suggesting that the ER promoter likely drives expression within the meristem close to the level of the endogenous CLV1 gene.
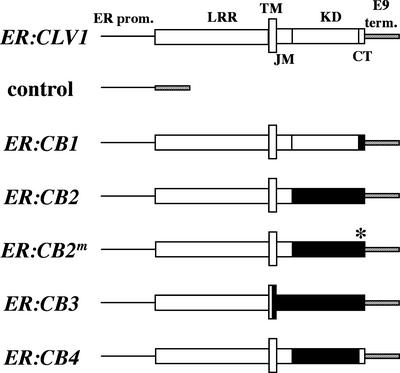
Figure 2.
Schemes (Not to Scale) of the CLV1 cDNA (ER:CLV1), ER:CB1, ER:CB2, ER:CB2m, ER:CB3, and ER:CB4 Chimeric Receptor Kinase, and Control Transgenes.
The CLV1 and BRI1 protein structures are shown as white and black boxes, respectively. The junction between CLV1 and BRI1 in each chimeric construct did not alter the amino acid sequence of CLV1 or BRI1. The mutation introduced in the BRI1 kinase domain to produce the ER:CB2m transgene (asterisk) is located in domain IX and is the same as the bri1-101 mutation, which results in a loss of kinase activity (Friedrichsen et al., 2000). These constructs are driven by the ERECTA promoter (black line; 1678 bp of sequence upstream of ER), contain the E9 terminator (hatched box), and are cloned into pCB302 (Xiang et al., 1999) (see Methods). CT, C-terminal tail; ER prom., ER promoter; JM, juxtamembrane domain; KD, kinase domain; E9 term., E9 terminator; TM, transmembrane domain.
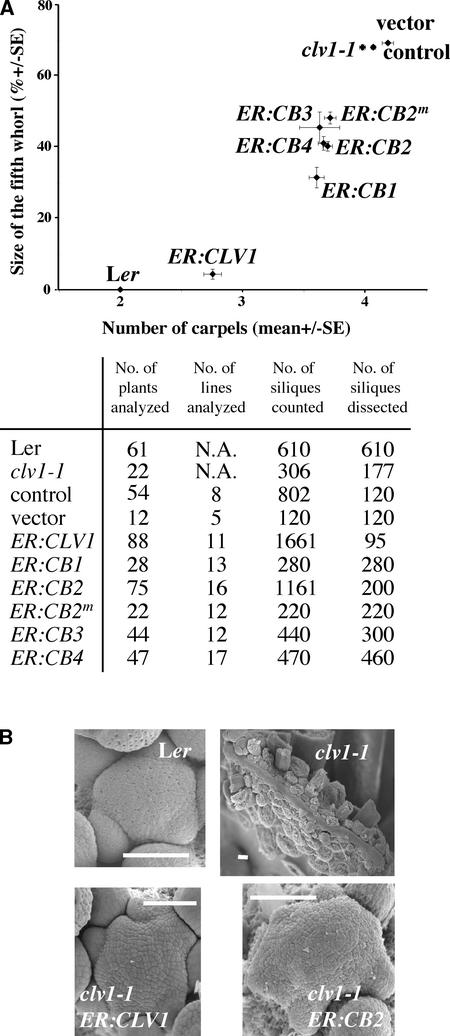
Figure 3.
CLV1/BRI1 (CB) Chimeric Receptors Rescue the clv1-1 Mutant Phenotype.
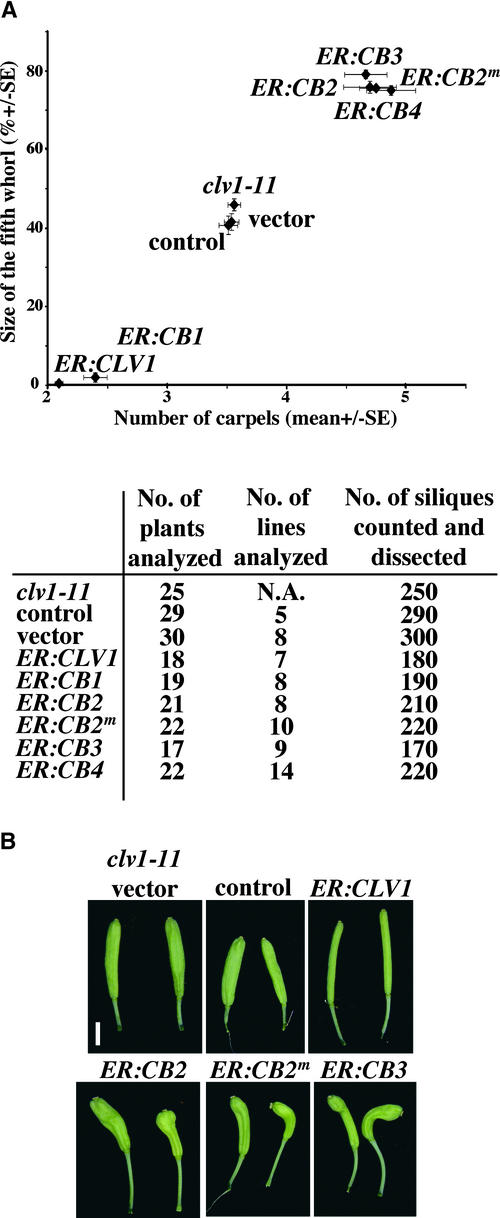
(A) Relative size of the fifth whorl compared with the number of carpels in Ler, clv1-1, and clv1-1 transformed with the ER:CLV1, ER:CB1, ER:CB2, ER:CB2m, ER:CB3, ER:CB4, empty vector, and control transgenes. The number of independent transgenic lines and the number of T3 plants analyzed are shown in the table at bottom. Between 10 and 20 fully expanded siliques of 30-day-old plants were counted for the number of carpels and dissected for the size of the fifth whorl before desiccation. The size of the fifth whorl growing inside the gynoecium was measured and compared with the size of the silique to give its relative size (% ± se) (cf. Figure 1D). The vertical and horizontal bars represent standard errors for the fifth whorl size and the number of carpels, respectively.
(B) Scanning electron micrographs of the shoot apical meristems of 30-day-old plants. Note that the shape of the meristems of clv1-1 ER:CLV1 and clv1-1 ER:CB2 transgenic plants is closer to that of Ler than to the fasciated clv1-1 meristem. Bars = 500 μm.
The coding sequences for CLV1 and BRI1 were divided into the extracellular LRR domain (Figure 2, LRR), the transmembrane domain (Figure 2, TM), the juxtamembrane domain (between the transmembrane domain and the first conserved kinase domain sequences; Figure 2, JM), the conserved kinase domain (Figure 2, KD), and the C-terminal tail lacking specific kinase domain motifs (Figure 2, CT). The ER:CB2m construct included the Glu-to-Lys mutation in domain IX of the kinase domain, which is found in the loss-of-function bri1-101 mutation (Li and Chory, 1997) (Figure 2).
clv1-1 Cosuppression Rescues the Clv1− Phenotype
Direct evidence that clv1-1 is dominant negative came unexpectedly from experiments with these CLV1/BRI1 chimeric receptors. The ER:CLV1, ER:CB1, ER:CB2, ER:CB2m, ER:CB3, and ER:CB4 constructs were transformed into clv1-1 plants and tested for the ability to rescue the Clv1− phenotype. As described above, the number of carpels per flower and the extent of whorl-five growth were used as measures of meristem size and CLV1 function.
When ER:CB2, ER:CB2m, ER:CB3, and ER:CB4 were transformed into clv1-1, we observed that 15 to 50% of the T1 plants for each construct exhibited partial suppression of the Clv1− phenotype (Table 1). When analyzed in detail in subsequent generations, these lines exhibited a clear reduction in mean carpel number and fifth-whorl growth (Figure 3A). The sizes of the shoot apical meristems of the affected transgenic lines also were suppressed compared with the fasciated clv1-1 shoot meristems (Figure 3B).
Table 1.
clv1-1 and BRI1 Cosuppression in Transgenic clv1-1 Plants
| Plant Lines | No. of T1 Plants | No. of T1 Transgenic Plants with Bri1− Phenotypea |
No. of Lines with Clv1− Suppressionb |
No. of Lines with Only Bri1− Suppressiona |
No. of Lines with Only Clv1− Suppressionb |
|---|---|---|---|---|---|
| clv1-1 | NAc | 0/22 | 0/22 | 0/22 | 0/22 |
| Control | 145 | 0/145 | 0/145 | 0/145 | 0/145 |
| Vector | 30 | 0/30 | 0/30 | 0/30 | 0/30 |
| ER:CLV1 | 47 | 0/47 | 24/47 (51%) | 0/47 | 24/47 (51%) |
| ER:CB1 | 36 | 2/36 (5.5%) | 16/36 (44%) | 0/36 | 14/36 (39%) |
| ER:CB2 | 150 | 33/150 (22%) | 36/150 (24%) | 4/150 (2.66%) | 7/150 (4.66%) |
| ER:CB2m | 73 | 21/73 (28.7%) | 20/73 (27.4%) | 4/73 (5.48%) | 3/73 (4.1%) |
| ER:CB3 | 67 | 10/67 (14.9%) | 10/67 (14.9%) | 0/67 | 0/67 |
| ER:CB4 | 27 | 4/27 (14.8%) | 14/27 (51.8%) | 1/27 (3.7%) | 11/27 (40.7%) |
Number of dwarf-looking plants.
Number of plants with fewer than four carpels.
Not applicable.
We suspected that the Clv1− phenotypic suppression was attributable to cosuppression of the endogenous clv1-1 transcript for two reasons. First, all of the lines gave rise to the same phenotype, regardless of the specific construct transformed (e.g., the kinase-active ER:CB2 and the kinase-inactive ER:CB2m chimeric constructs provided similar clv1-1 suppression). Second, the phenotypically suppressed transgenic clv1-1 plants were very similar in phenotype to the clv1 null mutants.
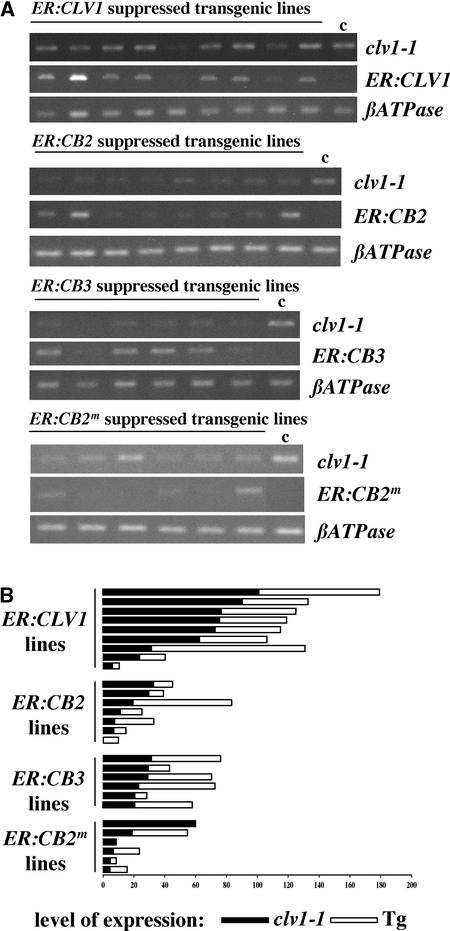
To determine the role of clv1-1 cosuppression in Clv1− phenotype suppression, the expression of clv1-1 and each transgene was analyzed by RT-PCR in phenotypically suppressed transgenic lines (Figures 4A and 4B). In every phenotypically suppressed line analyzed, we observed one of two expression patterns. (1) For most suppressed clv1-1 ER:CLV1 lines, we detected significant ER:CLV1 transgene expression. (2) For two suppressed clv1-1 ER:CLV1 lines and all suppressed clv1-1 ER:CB2, ER:CB2m, and ER:CB3 lines tested, the expression of the endogenous clv1-1 gene was reduced compared with that in controls (Figures 4A and 4B).
Figure 4.
clv1-1 Is Dominant Negative.
(A) Ethidium bromide agarose gels from RT-PCR performed on individual suppressed transgenic lines (see Methods). T1 inflorescence RNA samples for clv1-1 ER:CLV1, clv1-1 ER:CB2, clv1-1 ER:CB3, and clv1-1 ER:CB2m transgenic plants were analyzed by RT-PCR to monitor transcript accumulation of the endogenous clv1-1 and transgene mRNA. Each lane corresponds to one transgenic line. RT-PCR products from clv1-1 nontransgenic plants, clv1-1 control transgenic plants, and no RNA were examined as controls (c) in all cases. Each PCR amplification of the cDNA was generated in parallel with specific primers for clv1-1 (top gel), the appropriate transgene (middle gel), and the control (βATPase; bottom gel). Top panel, clv1-1 ER:CLV1 transgenic lines; second panel, clv1-1 ER:CB2; third panel, clv1-1 ER:CB3; fourth panel, clv1-1 ER:CB2m.
(B) Quantification and normalization of the RT-PCR agarose gel data (see Methods). Black boxes indicate clv1-1 expression, and white boxes indicate transgene (Tg) expression.
These data suggested that cosuppression of endogenous clv1-1 gene expression could lead to Clv1− phenotypic suppression. Consistent with this finding, we observed a correlation between Clv1− phenotype suppression and a weak/intermediate Bri1− phenotype, suggesting cosuppression of both the clv1-1 and BRI1 genes in these lines (Table 1). In subsequent generations, some plants lost BRI1 cosuppression with no effect on Clv1− phenotypic suppression (data not shown). Moreover, a portion of the T1 transgenic plants exhibited only clv1-1 or BRI1 cosuppression. Together, these data suggest that the Clv1− phenotype suppression was not attributable to a reduction in BRI1 activity.
Dominant-Negative Chimeric Receptors
Because the dominant-negative nature of the endogenous clv1-1 allele complicated the analysis of chimeric receptor function, we chose to transform all chimeric constructs into the clv1-11 null background. At least 30 primary transformants were obtained for each construct, and 5 to 14 lines were examined in detail in the T2 generation. The goal was to assess chimeric receptor function by monitoring suppression of the Clv1− phenotype; however, we observed that many of the constructs resulted in an enhancement of the Clv1− phenotype.
The two constructs that did provide Clv1− rescue were ER:CLV1 and ER:CB1. clv1-11 ER:CLV1 plants were nearly wild type in meristem phenotype, again suggesting that the ER regulatory sequences drive expression close to the level of the endogenous CLV1 gene (Figures 5A and 5B). The ER:CB1 construct, which provided significantly poorer rescue of clv1-1 compared with the ER:CLV1 construct (Figure 3A), provided nearly complete rescue of clv1-11 (Figures 5A and 5B). This finding suggests that the C-terminal domain of CLV1 contains no motifs specific for CLV1 signaling.
Figure 5.
The Chimeric Receptors CB2, CB2m, CB3, and CB4 Are Dominant Negative.
(A) The relative size of the fifth whorl compared with the number of carpels in clv1-11 and clv1-11 transformed with ER:CLV1, ER:CB1, ER:CB2, ER:CB2m, ER:CB3, ER:CB4, empty vector, and control transgenes. The number of independent transgenic lines and the number of T3 plants analyzed are shown in the table at bottom. Ten fully expanded siliques of 30-day-old plants were counted for the number of carpels and dissected for the size of the fifth whorl before desiccation. The vertical and horizontal bars represent standard errors for the fifth whorl size and the number of carpels, respectively. Note that the almost complete rescue of the clv1-11 phenotype is observed for ER:CLV1 and ER:CB1. On the other hand, the chimeric receptors ER:CB2, ER:CB2m, ER:CB3, and ER:CB4 enhance the phenotypes of clv1-11 plants.
(B) Comparison of the silique shapes of clv1-11 plants transformed with ER:CLV1, ER:CB2, ER:CB2m, ER:CB3, empty vector, and control transgenes. Note that the shape of the ER:CLV1 siliques is close to that of wild-type siliques and that the siliques of ER:CB2, ER:CB2m, and ER:CB3 transgenic plants are club shaped, reflecting a more severe phenotype than that seen in the clv1-11 and control lines. Bar = 2 mm.
The ER:CB2, ER:CB2m, ER:CB3, and ER:CB4 transgenes enhanced the clv1-11 mutant phenotype. This enhancement included all aspects of the Clv1− phenotype: mean carpel number, extent of fifth-whorl growth, overall silique morphology, and size of the shoot apical meristem (Figures 5A and 5B). Both the wild-type and mutant versions of the BRI1 kinase domains in ER:CB2 and ER:CB2m, respectively, were equally effective in enhancing the Clv1− phenotype. The ER:CB3 construct contained just the extracellular domain of CLV1 and retained dominant-negative effects. The ER:CB4 construct, in which only the kinase domain of CLV1 was replaced by the BRI1 kinase domain, enhanced the clv1-11 phenotype as well, suggesting that neither the juxtamembrane domain nor the C-terminal tail domain is involved in the induction of the dominant-negative effect. Each dominant-negative transgene gave rise to a similar level of Clv1− phenotype enhancement, suggesting similar target(s) and mechanisms of action. Thus, constructs containing a CLV1 extracellular domain and a BRI1 kinase domain exhibited a dominant-negative effect.
We did not observe the induction of any phenotypes outside of the meristem with any of the chimeric receptors with the exception of Bri1− phenotypes. As we observed for these transgenes in the clv1-1 background, the chimeric constructs induced cosuppression of BRI1 in ∼20% of the lines analyzed. These plants did not show clv1-11 suppression and were comparable phenotypically to clv1-11 (data not shown).
Precise CLV1 Expression Is Not Essential for Its Function
CLV1 normally is expressed specifically within the internal cell layers of the shoot meristem, predominantly the L3 layer (Clark et al., 1997; Fletcher et al., 1999; Trotochaud et al., 1999). CLV3 is expressed in overlying, minimally overlapping cells in the L1, L2, and topmost L3 layers (Fletcher et al., 1999). Remarkably, driving CLV1 expression under the control of the ER regulatory elements, which drive endogenous and transgene expression in all layers of the shoot meristem (Yokoyama et al., 1998), in clv1-1 and clv1-11 plants had no impact on meristem development beyond rescuing the Clv1− phenotype. When ER:CLV1 was transformed into wild-type plants, no effect on shoot or flower meristem development was observed (data not shown).
CLV1 Can Function Outside of the Meristem
The elongation of internodes and pedicels is important in determining the overall aerial structure of plants. Each flower meristem gives rise to flower organs that are separated from the inflorescence stem by a pedicel. The pedicel elongates with the development of floral organs. Mutant screens for altered inflorescence branching patterns have revealed ER (Torii et al., 1996; Yokoyama et al., 1998; Lease et al., 2001a) and other genes (Lease et al., 2001b; Venglat et al., 2002). The commonly used Arabidopsis Ler ecotype harbors the er mutation, resulting in an altered inflorescence branching pattern caused by a reduction in the length of internodes and pedicels, the growth of clustered flowers at the apex of the inflorescence, and shorter, wider, and blunt-tipped siliques. A precise analysis of stems and pedicels of er mutant plants indicated that the short stem/pedicel phenotype is attributable mainly to a decrease in cell number (Yokoyama et al., 1998).
We initially observed that the size of siliques and pedicels of the clv1-1 er and clv1-11 er plants transformed with the ER:CLV1 construct was increased compared with that in clv1-1 er and clv1-11 er, respectively (data not shown). Because the ER:CLV1 construct also suppressed the meristem defects of clv1-1 and clv1-11 mutants, we could not exclude the possibility that the increase in pedicel and silique size in clv1-1 er ER:CLV1 and clv1-11 er ER:CLV1 transgenic plants was the indirect consequence of a restoration of normal meristem function. To eliminate transgene-induced changes in meristem size, the ER:CLV1 construct was introduced into Ler plants and the size of the siliques and pedicels was scored. The pedicels of Ler ER:CLV1 plants were 27% longer than the pedicels of Ler plants transformed with the control construct (wild-type pedicel, 7.74 ± 0.05 mm, n = 185; ER:CLV1 pedicel, 9.82 ± 0.03 mm, n = 166). Interestingly, the length of the gynoecium was not affected significantly by the ER:CLV1 construct (wild-type silique, 12.96 ± 0.05 mm, n = 185; ER:CLV1 silique, 12.42 ± 0.07 mm, n = 166). Thus, the expression of CLV1 under the control of the ER promoter can partially rescue the pedicel but not the gynoecium phenotypes of er plants, indicating that CLV1 can function outside of the meristem.
Ecotype Genetic Variation Affects Phenotypes of clv1 Mutants
During backcrosses of two of the clv1 null alleles from the Ws-2 background (clv1-12 and clv1-13) into Ler, we detected a significant effect of ecotype background on the severity of Clv1− phenotypes (Table 2). Note that in the analysis described below, we detected a significant frequency of a “valveless” phenotype among some of the genotypes tested, as indicated in Table 2. This phenotype was first described in detail for clv2 mutants, which often develop replum tissue in the place of carpels, such that the flowers can develop one or even zero carpels (Kayes and Clark, 1998). In flowers with the valveless phenotype, carpel number is no longer an accurate reflection of flower meristem defects. Thus, we also calculated the number of carpels per flower from mutant plants using only those flowers that did not exhibit a valveless phenotype. These measurements are described below.
Table 2.
Ecotype Background and er Modify clv1 and clv3 Phenotypes
| Plant | Ecotypea | No. of Siliques Analyzed |
No. of Carpels per Flower (mean ± SE) |
Valveless Siliques (%) |
No. of Carpels Excluding Valveless Siliques (mean ± SE)b |
Relative Size of the Fifth Whorl (% ± SE) |
|---|---|---|---|---|---|---|
| clv1-4 ER | Unk.c/Ws-2 | 330 | 3.58 ± 0.04 | 51 | 4.15 ± 0.02 | >80 |
| clv1-4 er123 | Unk.c/Ws-2 | 250 | 3.71 ± 0.06 | 54 | 4.39 ± 0.04 | >80 |
| clv1-4 er | Unk.c/Ler | 100 | 5.2 ± 0.09 | 15 | 5.35 ± 0.08 | >80 |
| clv1-12 ER | Ws-2 | 260 | 2.57 ± 0.05 | 0 | 2.57 ± 0.05 | 0 |
| clv1-12 er123 | Ws-2 | 160 | 2.83 ± 0.06 | 0 | 2.83 ± 0.06 | 1.25 ± 0.62 |
| clv1-12 ER | Ws-2/Col | 240 | 2.77 ± 0.05 | 0 | 2.77 ± 0.05 | 1.93 ± 0.54 |
| clv1-12 er106 | Ws-2/Col | 210 | 2.81 ± 0.05 | 0 | 2.81 ± 0.05 | 0 |
| clv1-12 er | Ws-2/Ler | 280 | 3.12 ± 0.04 | 0 | 3.12 ± 0.04 | 12.13 ± 1.15 |
| clv1-13 ER | Ws-2 | 260 | 2.64 ± 0.05 | 0 | 2.64 ± 0.05 | 0 |
| clv1-13 ER | Ws-2/Col | 200 | 2.71 ± 0.05 | 0 | 2.71 ± 0.05 | 0.25 ± 0.20 |
| clv1-13 er106 | Ws-2/Col | 170 | 2.66 ± 0.06 | 0 | 2.66 ± 0.06 | 1.23 ± 0.51 |
| clv1-13 er | Ws-2/Ler | 270 | 3.28 ± 0.04 | 0 | 3.28 ± 0.04 | 6.25 ± 0.97 |
| clv3-2 er | Ler | 230 | 5.29 ± 0.05 | 0 | 5.29 ± 0.05 | >80 |
| clv3-8 ER | Unk.d | 117 | 4.16 ± 0.11 | 50 | 5.10 ± 0.08 | >80 |
| clv3-3 er | Ws-2/Ler | 285 | 2.57 ± 0.04 | 0 | 2.57 ± 0.04 | 0 |
| clv3-3 ER | Ws-2 | 274 | 2.78 ± 0.05 | 0 | 2.78 ± 0.05 | 0 |
Populations with two ecotypes indicated refer to a segregating F2 population of a cross between the two ecotypes.
Mean number of carpels per flower excluding flowers exhibiting the valveless phenotype.
Unknown, Estland or Limbaugh.
Unknown.
clv1-12 displayed a mean carpel number of 2.57 ± 0.05 per flower in the Ws-2 ER background and a mean carpel number of 3.12 ± 0.04 in the first backcross into the Ler background (Table 2), revealing a highly significant effect of the Ler background on the clv1-12 phenotype (Table 3). Similarly, the mean carpel number of clv1-13 mutants increased from 2.64 ± 0.05 to 3.28 ± 0.04 in the same backcross (Table 2), which again is highly significant based on Student's t test (Table 3).
Table 3.
Statistical Tests of Phenotypic Variation
| Reference Plants | Compared with | Student's t Test P Value | Significantly Different? |
|---|---|---|---|
| clv1-12 ER (Ws-2) | clv1-12 er123 (Ws-2) | 0.00081 | Yes |
| clv1-12 er (Ws-2/Ler) | 4.13 E−16 | Yes | |
| clv1-12 ER (Ws-2/Col) | 0.00163 | Yes | |
| clv1-12 ER (Ws-2/Col) | clv1-12 er106 (Ws-2/Col) | 0.59704 | No |
| clv1-13 ER (Ws-2/Col) | clv1-13 er106 (Ws-2/Col) | 0.58964 | No |
| clv1-4 ER (Unk.a/Ws-2) | clv1-4 er123 (Unk.a/Ws-2) | 8.69 E−5 | Yes |
| clv1-4 er (Unk.a/Ler) | 1.48 E−39 | Yes | |
| clv3-3 ER (Ws-2) | clv3-3 er (Ws-2/Ler) | 0.00119 | Yes |
| clv3-8 ER (Unk.)b | clv3-2 er (Ler) | 0.09590 | No |
Unknown, Estland or Limbaugh.
Unknown.
To determine if this enhancement of clv1 null phenotypes was the result of ecotype variation, due to the er mutation, and/or specific for clv1, we analyzed clv1 and clv3 mutants in a number of backcrosses (Tables 2 and 3). clv1-12 crossed to the er-123 allele within the same Ws-2 background displayed a lesser, but statistically significant, effect compared with crosses into Ler. This finding indicates that there is both a specific but minor effect from the er mutation and a larger effect of the Landsberg background. We observed a significant phenotypic enhancement when crossing clv1-12 in the Ws-2 ER background to the Col ER background, again revealing an ecotype effect (Tables 2 and 3). However, there was no detectable difference between crossing clv1-12 to the Col ER or the Col er-106 background, suggesting that the specific effect of er is minor and/or masked by the Col background. clv1-13 displayed results similar to clv1-12 (Tables 2 and 3).
clv1-4, which is in an uncharacterized ecotype listed by McKelvie (1962) as either Estland or Limbaugh, exhibited effects of both er or a linked factor and ecotype backgrounds. clv1-4 er-123 and clv1-4 ER plants analyzed from a segregating mixed ecotype population revealed a minor, but statistically significant, enhancement of the Clv1− phenotype in plants homozygous for er (Tables 2 and 3). A much stronger Clv1− enhancement was observed in clv1-4 crossed to the Ler background (Tables 2 and 3).
clv3 mutants, on the other hand, displayed different effects of ecotype backgrounds and the er mutation. clv3-2 and clv3-8 are presumed null alleles that have phenotypes similar to that of clv1-4 (Clark et al., 1995). We observed no significant differences between the clv3-2 allele from the Ler background and the clv3-8 allele from an unknown ER background (Tables 2 and 3). The weak clv3-3 allele has a phenotype similar to that of clv1 null alleles (Fletcher et al., 1999). We observed a significant effect of crossing clv3-3 ER from the Ws-2 background to Ler. However, the phenotype of clv3-3 crossed to Ler was suppressed compared with the phenotype of clv3-3 in the Ws-2 ER background, which is the opposite of our observation for clv1 null alleles. Thus, ecotypes to a major degree, and the er mutation to a minor degree, have a specific effect on the phenotype of clv1 mutants. Pursuing the genes responsible for the ecotype differences could prove useful in identifying additional factors that interact with CLV1 and/or other redundant receptor kinases.
DISCUSSION
Previous models of CLV1 signaling did not explain differences in the observed effects of clv1 alleles. Specifically, the observation that alleles with absent kinase domains (clv1-6 and clv1-7) exhibited weaker phenotypes than those with missense mutations within the LRR and/or kinase domains could not be explained easily. Alleles of clv2 and clv3 displayed no such disparity between lesion and phenotypic effect (Clark et al., 1995; Kayes and Clark, 1998; Fletcher et al., 1999; Jeong et al., 1999). This work provides several lines of evidence suggesting that the stronger clv1 alleles are dominant negative and likely interfere with additional receptor kinase(s) within the meristems (Figure 6). By assessing the dominant-negative effects of CLV1/BRI1 chimeric receptors, we provide evidence defining receptor interactions and show that CLV1 and BRI1 kinase domains are not interchangeable. Furthermore, our results indicate that CLV1 can act outside of the meristem to regulate pedicel length in the absence of ER function. Finally, ectopic expression of CLV1 within the meristem does not disrupt normal meristem development.
Figure 6.
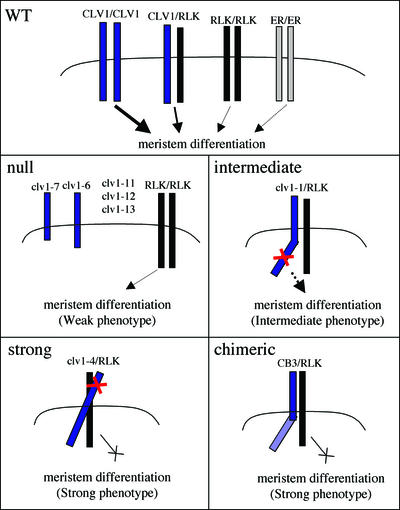
Model of Dominant-Negative Receptor Action.
A speculative model for the role of CLV1, an additional unknown receptor kinase with functional overlap (RLK), and ER in regulating meristem development. Scenarios for wild-type plants (WT), clv1 null mutants, intermediate clv1 mutants, strong clv1 mutants, and the dominant-negative chimeric receptors are indicated. The model predicts receptor interactions between CLV1 and RLK, which can lead to interference with RLK function in the presence of clv1 dominant-negative isoforms and in the presence of chimeric receptors. The thickness of the arrows suggests the relative strength of signaling, and lesions in clv1 proteins are indicated with a red X.
Unknown Receptor Kinase(s) May Share Functions with CLV1
Several lines of evidence support the existence of an additional receptor kinase(s) that has functional overlap with CLV1 (Figure 6). The first such indication came from analyses of three new null alleles of clv1, each of which displayed weak phenotypes comparable to those of clv1-6 and clv1-7 mutants. This finding suggested that all of the intermediate and strong mutant clv1 alleles are dominant negative. Consistent with this hypothesis, cosuppression of the expression of the intermediate clv1-1 allele in transgenic plants partially rescued the mutant clv1-1 phenotype, such that the cosuppressed plants appeared phenotypically similar to clv1 null mutants.
Many different types of proteins could be inactivated by clv1 dominant-negative protein isoforms, including ligands, other receptor kinases, and cytoplasmic signaling factors. Previous genetic analyses of clv3 mutants are most consistent with a receptor kinase(s) being the target of mutant clv1 interference (Figure 6). The strongest dominant-negative alleles of clv1 approach the phenotype of clv3 null alleles, and clv3 null alleles are epistatic to weak, intermediate, and strong clv1 alleles (Clark et al., 1993, 1995). Thus, CLV3 retains some function in clv1 mutants, especially in clv1 null mutants. Because CLV3 is secreted and required to activate CLV1 (Trotochaud et al., 1999; Rojo et al., 2002), CLV3 must activate another target in clv1 null mutants, presumably a receptor kinase. Furthermore, the hypothesis that the clv1 dominant-negative isoforms sequester the putative CLV3 ligand is not consistent with studies on a likely feedback loop between WUS and CLV3 (Brand et al., 2000; Schoof et al., 2000; Lenhard et al., 2002). The net result of this loop is to increase CLV3 expression in mutant plants in which CLV signaling is diminished. Indeed, CLV3 expression as detected by RNA in situ hybridization is increased in clv1 mutant backgrounds (Brand et al., 2000). Thus, CLV3 is unlikely to be rate limiting in the clv1 dominant-negative backgrounds.
If an additional receptor kinase functions in overlap with CLV1, why has this receptor not been identified in genetic screens? One possible explanation is that the unknown receptor has a somewhat lesser role in CLV3 signaling, such that mutant alleles might not exhibit a phenotype. clv1 null alleles display fairly subtle deviations from the wild type in some ecotype backgrounds, so that a receptor with lower expression levels or lesser signaling effectiveness compared with CLV1 might not display any significant phenotype. Part of the reason for this might lie in the CLV3/WUS feedback loop. A second possible explanation is that the unknown receptor also might function in additional developmental pathways, such that mutants would display pleiotropic phenotypes. Finally, the unknown receptor might be represented by more than one gene, such that inactivation of any one gene would not exhibit a phenotype. Note that most of our genetic screens have been performed in the clv1-1 mutant background, in which the activity of any receptors with functional overlap would be expected to be reduced as a result of the dominant-negative character of this allele. Ideally, genetic screens should be performed in the clv1 null mutant background, in which the activity of these hypothetical receptors would be most important phenotypically.
Potential Mechanisms by Which Dominant-Negative clv1 Alleles Act
Most of the clv1 alleles identified to date are dominant negative. The severity of the phenotypes of these mutant proteins varies depending on the location of the lesion within the CLV1 protein. The mutations that give an intermediate phenotype are clustered in the activation loop of the kinase domain. The mutations that give a strong phenotype are found in the fourth, fifth, and ninth LRRs in the predicted extracellular domain (Figure 6).
We were unable to find any example in the literature of dominant-negative mutations within the extracellular domains of animal receptor kinases. Missense mutations in the ligand binding domain of the receptor-Tyr kinase torso and deletion of the Toll extracellular domain are gain-of-function mutations (Schneider et al., 1991; Li et al., 2002b). The dominant-negative mutations in CLV1 are localized in LRRs four, five, and nine and are situated in or close to the predicted β-strand of the respective LRRs. Interestingly, these mutants exhibit a stronger phenotype than the dominant-negative mutations located in the cytoplasmic domain. Their phenotypes approach that of clv3 null mutants. The fact that the phenotype is stronger with the LRR-located mutations than with the mutations in the cytoplasmic domain suggests that the blockage of the signaling pathway by the mutations in the cytoplasmic domain is not complete (Figure 6).
For lesions in the kinase domain, we noted several differences and similarities when comparing clv1 alleles with dominant-negative mutations in animals. Deletion of all or part of the cytoplasmic domain of the receptor-Tyr kinase epidermal growth factor receptors or other receptor-Tyr kinases results in dominant-negative mutations (Kashles et al., 1991; Chen et al., 1993; Prager et al., 1994; Guichard et al., 2002). The two CLV1 alleles that mimic this kind of mutation, clv1-6 and clv1-7, exhibit a weak phenotype similar to that of null alleles. Several explanations for this difference can be suggested. The truncated clv1-6 and clv1-7 receptors could be unstable at the plasma membrane or mislocalized. If stable at the plasma membrane, the clv1-6 and clv1-7 mutant proteins would appear unable to interfere with receptors present in the meristem that have functional overlap.
Most of the dominant-negative mutations in protein kinases (TGF-β receptor, MEKK1, Alk6, IGF-1R, and IRAK-4) or in receptor-Tyr kinases (IGF-1R and IRAK-4) in animals are clustered at the ATP binding site of the kinase domain, corresponding to domain II of CLV1 (Kong et al., 1994; Sanchez et al., 1994; Zou and Niswander, 1996; Kalebic et al., 1998; Li et al., 2002a). It is of interest that no clv1 alleles have been identified that contain mutations within key catalytic residues of the kinase domain, suggesting that some catalytic activity may be required within the full-length CLV1 for receptor interaction and, hence, dominant-negative properties. We have noted that when expressed as kinase domains in Escherichia coli, clv1-1, clv1-2, and clv1-9 retain some autophosphorylation activity (data not shown). Interestingly, the mutations in clv1-1, clv1-2, and clv1-9 lie within or close to the activation loop in the kinase domain (Torii and Clark, 2000). Some mutations in animals are comparable to these: the EGFRDN3 mutation is located in domain VIb, and the bmpr2 mutation is located between domains VIb and VII (Zou and Niswander, 1996; Guichard et al., 2002; Rudarakanchana et al., 2002). Studies on bmpr2 showed that this dominant-negative mutant is not expressed at the plasma membrane but instead is retained in the endoplasmic reticulum (Rudarakanchana et al., 2002).
The phenotypes observed in clv1-1 plants suggest that the mutant protein is able to interfere with the signaling of other receptors with functional overlap. This interference could occur by clv1 proteins interacting directly with the other receptors and blocking their function (Figure 6). Consistent with the idea of a direct interaction between receptors, the clv1-1 allele was described previously as incompletely dominant (Clark et al., 1995, 1996). Moreover, the hypothesis that dominant-negative clv1 isoforms interfere with the function of other receptors is consistent with the extent of Clv1− rescue when ER:CLV1 and ER:CB1 were transformed into clv1-1 and clv1-11 plants. Both of these constructs are nearly able to completely rescue the clv1-11 null phenotype, suggesting that in the absence of dominant-negative protein isoforms, both wild-type CLV1 and CB1 are effective transducers of CLV3 signal. However, in the presence of clv1-1, both CLV1 and CB1 are significantly less effective at signaling, as measured by the partial Clv1− rescue each provides. The phenotype of clv1-1 ER:CLV1 plants is similar to that of clv1-1/+ plants, consistent with ER elements driving expression within the meristem at levels similar to those in the endogenous CLV1 gene. clv1-1 ER:CB1 plants are nearly as severe as clv1 null alleles, indicating that CB1 function is particularly sensitive to clv1-1 interference.
We previously examined clv1 protein accumulation in various clv1 mutant plants (Trotochaud et al., 1999). These studies revealed that clv1 dominant-negative isoforms accumulate within the meristem; however, we were unable to conclude that these proteins accumulate normally, because the proportion of meristem cells per sample was affected dramatically by the Clv1− phenotype. The various clv1 isoforms were identified as components of protein complexes, some of them similar to CLV1 complexes observed in wild-type plants, although the extracellular domain isoforms were found in a novel complex of an apparently monomeric size. However, it is difficult to draw clear conclusions about the mechanism of clv1 dominant-negative action from this approach for several reasons. First, most of the components of the complexes are unknown. Second, it is not certain that complexes from different genotypes of approximately the same size are in fact the same protein complexes. Third, interactions responsible for clv1 dominant-negative properties might not be stable under our isolation methods and thus might not be represented in our analysis. The identification of other receptor(s) that have functional overlap with CLV1 would be critical for using complex formation to assess the mechanism of dominant-negative clv1 function and potential receptor interactions.
Dominant-Negative Chimeric Receptors
Although the chimeric CB1 receptor kinase was functional, especially in a clv1 null background, the proteins encoded by the chimeric constructs ER:CB2, ER:CB2m, ER:CB3, and ER:CB4 appear to act as dominant-negative proteins. These constructs transformed into clv1-1 and clv1-11 plants enhanced the mutant phenotype, with transgenic plants displaying an increased number of carpels per flower and an enlarged fifth whorl compared with clv1-1 and clv1-11 plants. Thus, expression of the extracellular domain of CLV1 with a heterologous kinase domain in transgenic plants was able to block receptor signaling within the meristem in a dominant manner.
One important aspect of the chimeric receptors is that they did not require overexpression to achieve their dominant-negative character. Expression of wild-type CLV1 under the control of the ER regulatory elements in clv1-1 and clv1-11 plants is consistent with ER-driven expression being close to, but less than, the expression level of the endogenous CLV1 gene. The same ER regulatory sequences driving ER:CB2, ER:CB2m, ER:CB3, and ER:CB4 expression in clv1-11 plants give rise to phenotypes stronger than those seen in clv1-1, clv1-2, and clv1-9 plants. Indeed, the phenotype of these chimeric receptors is similar to that of the strong clv1-4 allele.
Each of the receptors ER:CB2, ER:CB2m, ER:CB3, and ER:CB4 resulted in nearly identical phenotypes when transformed into clv1-11. This finding suggests a common mechanism for these receptors in interfering with signaling within the meristem. This indicates that kinase activity is not required for interference, because the putative kinase-active CB2 and kinase-inactive CB2m were similar in severity. It also indicates that the key to the dominant-negative function is likely the substitution of the BRI1 kinase domain, because this feature is common to all four constructs. Thus, although the kinase domains of the plant LRR-RLKs show strong sequence similarity (the kinase domains of CLV1 and BRI1 show 63% similarity), they are not interchangeable.
Natural Variation Affects Meristem Development
We have observed that the phenotypes of various clv1 alleles are affected significantly by natural variation between different Arabidopsis ecotypes. In fact, in every case, crossing clv1 alleles from one ecotype to another resulted in significant modification of the mutant phenotype. The strongest Clv1− phenotype enhancement was observed in crosses to the Ler background. A significant, albeit lesser, portion of this enhancement appeared to be the consequence of the er mutation within this ecotype. This finding suggests some overlap in function between CLV1 and ER within the shoot meristem. The activity of these receptors clearly is not equivalent in meristem regulation, based on driving CLV1 expression under the control of the ER regulatory elements. The effect of the er mutation could reflect a low level of nonspecific crosstalk between the CLV1 and ER signaling pathways. In the Ler backcrosses, we conclude that the majority of the phenotypic enhancement is the result of non-er factors. Furthermore, other ecotype backcrosses in which the wild-type ER allele was fixed also resulted in significant phenotypic variation. Importantly, in at least some cases, this variation appeared to be specific for CLV1. The genes responsible for this natural variation make good candidates for CLV1-interacting factors.
Ectopic CLV1 Expression Affects Pedicel, but Not Meristem, Development
The ER regulatory elements have been shown to match endogenous ER patterns and to drive expression throughout the meristem as well as in developing organ primordia (Yokoyama et al., 1998). The putative ligand for CLV1, CLV3, and the target of CLV1 signaling, WUS, both are expressed only within specific cell layers of the shoot meristem, with the exception that WUS also is expressed in ovule primordia (Gross-Hardt et al., 2002). Thus, it is of interest to determine if CLV1 placed in ectopic locations at the physiologically relevant levels provided by ER regulatory elements can exert any effect on signaling or development. Any phenotypic effect of CLV1 ectopic expression would suggest that CLV1 is capable of interacting with additional signaling systems. We observed that pedicel length was affected significantly by the ER:CLV1 transgene in Ler plants. Because pedicel length was shortened by the er mutation, the ER:CLV1 transgene suppressed the Er− pedicel phenotype. Given the overlap in ER and CLV1 function detected within the meristem, one possible explanation for this finding is that CLV1 replaces ER function within the developing pedicels. The ER:CLV1 suppression of er phenotypes does not extend to the Er− gynoecium phenotypes. Lease et al. (2001b) recently described agb1, a mutant allele of the heterotrimeric G-protein β-subunit, which exhibits phenotypes similar to those of er. They concluded that AGB1 may function with ER to regulate aspects of silique development but may function in parallel in other aspects of development. Thus, ER may use alternate signaling mechanisms in different tissues. This finding may explain the differential effect of CLV1 on Er− phenotypes. However, many pathways affect organ elongation, and there is no way at this time to conclude that CLV1 acts within the ER pathway in pedicels.
Surprisingly, one site where ectopic CLV1 expression did not affect development was in the shoot meristem. Here, the location of CLV1 expression matched well the adjacent expression of the putative ligand, CLV3, and the expression of the target, WUS, in a subdomain of CLV1 expression (Clark et al., 1997; Mayer et al., 1998; Fletcher et al., 1999). One could predict that ectopic CLV1 expression within the L1 and L2 layers of the shoot meristem would block CLV3 signal generated in the L1 and L2 layers from reaching L3, where WUS normally is repressed by CLV1 action. However, we observed no such effect, suggesting either that the CLV3/WUS feedback loop overcomes any ligand sequestration by CLV1 or that the L1 and L2 layers lack the factors necessary for the presumed CLV1–CLV3 interaction.
METHODS
clv1 and clv3 Alleles
The clv3-3 allele (line CS6421) of Arabidopsis thaliana was identified in ecotype Wassilewskija-2 (Ws-2) and obtained from the ABRC at Ohio State University (Columbus). clv3-3 was backcrossed to Landsberg erecta (Ler) to isolate the clv3-3 er double mutant. The clv3-2 allele has been described previously (Clark et al., 1995; Fletcher et al., 1999). The clv3-8 allele (line CS3604) was isolated by G. Redei and obtained from the ABRC. No ecotype information was available for clv3-8. PCR analysis of this allele revealed a chromosomal breakpoint between the 5′ end of intron 2 and the 3′ end of exon 3 (data not shown), similar to the breakpoint position in clv3-2. The T-DNA clv1-12 and clv1-13 insertion lines were identified by screening the Arabidopsis Knockout Facility α population (http://www.biotech.wisc.edu/Arabidopsis/). Products were amplified by PCR with CLV1-specific (5′-GTTTCCGATTCCGGCCGGGATTGAACCGGT-3′ and 5′-AACTAACCTGGGGCGATGTATCCATAA-GGG-3′) and JL-202 (5′-CATTTTATAATAACGCTGCGGACATCTAC-3′; left border specific) primers and sequenced to determine the site of the T-DNA insertion. These insertions are available as lines CS3954 and CS3955 from the ABRC. These two lines, originally in the Ws-2 background, were backcrossed to Ler, Ws-2 er-123 (Lease et al., 2001a), and Columbia er-106 (Lease et al., 2001a) for a comparative analysis of the effect of the mutation in both backgrounds.
The F2 population was screened for the Clv1− and Er− phenotypes and allowed to self-pollinate. Phenotypic analyses were performed on the F3 generation. The clv1-11 line (background Ler) was generated by crossing the enhancer trap–containing T-DNA line DsE2 (CS8047) donated to the Nottingham Stock Centre by Rob Martienssen (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) to the Ac transposase T-DNA line Ac2 (CS8044). Unlinked transposition of the Ds element was screened for in the F2 population by growing seeds on naphthalene acetamide– and kanamycin-containing plates (Sundaresan et al., 1995). Doubly resistant plants were transferred to soil and allowed to self-pollinate. Phenotypic analysis of the F3 segregating population of line clv1-11 showed the clv1 phenotype as a recessive mutation. DNA samples were taken from two plants that displayed the mutation and one that did not, and thermal asymmetric interlaced PCR was performed. Sequencing of the bands amplified in each case revealed the same sequence with an eight-base duplication of endogenous sequence, typical of Ds insertions. The sequence indicated the insertion site within the CLV1 gene, between nucleotides 753 and 754 of the cDNA (Figure 1), which translates to amino acids 227 and 228 of the protein. Seeds from the selfed F3 plants showing the mutation were collected, and all subsequent analysis was performed on these homozygous F4 seeds.
We had reported previously that the clv1-10 allele, which was isolated as an intragenic enhancer of clv1-1, contained a second mutation within the kinase domain coding sequence (Pogany et al., 1998). Subsequent analysis could not replicate this result. Resequencing of the CLV1 coding sequence from the clv1-10 genome revealed instead that the clv1-10 allele contains a second lesion within the Leu-rich repeat domain coding sequence, as indicated in Figure 1.
Seeds were sown on a 1:1:1 mix of topsoil:perlite:vermiculite and allowed to imbibe for 5 days at 4°C. Plants were grown at 22°C under ∼800 foot-candles of continuous cool-white fluorescent light. The transgenic constructs were introduced into Agrobacterium tumefaciens strain GV3101 by the heat-shock method and transformed into clv1-1, clv1-11, or Ler by vacuum infiltration (5 min at 25 inches of Hg) (Bechtold et al., 1993; Bechtold and Pelletier, 1998). Transgenic seedlings were selected by resistance to Basta herbicide (Hoechst Schering AgrEvo, Wilmington, DE) diluted 1:1000. The plants were sprayed 10 days after germination and then three times per week for at least 2 weeks.
CLV1 cDNA, CLV1/BRI1, and CLV1/BRI1m Expression Constructs
For the chimeric receptor construct ER:CB2, a FauI restriction site was inserted in the CLV1 and BRI1 cDNA sequences in the first kinase domain by site-directed mutagenesis. Four oligonucleotides were used: FclvFauI (5′-CGGTAAAGGCGGGGCTGGAATTGTC-3′) and RclvFauI (5′-GACAATTCCAGCCCCGCCTTTACCG-3′) for CLV1 and FbriFauI (5′-GATTGGTTCTGGCGGGTTTGGAGATG-3′) and RbriFauI (5′-CATCTCCAAACCCGCCAGAACCAATC-3′) for BRI1. This FauI site was used to clone the 5′ CLV1 fragment in frame with the 3′ BRI1 fragment to give the amino acid sequence 5′-LKEENIIGSGGFGD-3′. A PstI-EcoRV fragment of CB2 was replaced by the mutated bri1-101 fragment (XhoI site at 3340 bp mutated) in CB2m. For the CB3 construct, an AseI site was created in the transmembrane domains of CLV1 cDNA and BRI1 cDNA. Four oligonucleotides were used: AseITM_CLV1f (5′-ATCACCGGATTA-ATCCTAATCA-3′) and AseITM_CLV1r (5′-GATTAGGATTAATCCGGTGAT-3′) for insertion of the AseI site in CLV1 cDNA and AseITM_BRI1f (5′-ATATTTGGATTAATCCTTGTTGG-3′) and AseITM_BRI1r (5′-CCA-ACAAGGATTAATCCAAATAT-3′) for insertion of the AseI site in BRI1 cDNA. The SpeI-AseI fragment from the CLV1 cDNA was ligated to the AseI-EcoRV fragment from BRI1 cDNA.
For the CB4 construct, the kinase domain of CB2 was used as a template for PCR amplification with the oligonucleotides FbriPstI (5′-CGG-AAGATTGCGATAGGATCAG-3′) and RClvBriXIKD (5′-GTGCACAAC-TTCCCTCATTGTCGGTCGTCTCC-3′). This first PCR amplified the kinase domain of BRI1 until domain XI of the kinase domain and added the first nucleotides of the C-terminal tail domain of CLV1 at the 3′ end of the sequence. A second PCR was performed on the CLV1 cDNA cloned in pBluescript II KS+ (Stratagene) with the oligonucleotides FBriClvXIKD (5′-GGAGACGACCGACAATGAGGGAAGTTGTGCAC-3′) and the T3 primer. This second PCR amplified the C-terminal tail domain of CLV1 with a few nucleotides belonging to domain XI of the BRI1 kinase domain at the 5′ end of the sequence. A third PCR was performed using as a template the two fragments mentioned above with the oligonucleotides FbriPstI and T3. This fragment was digested with PstI-EcoRV and ligated in the PstI-EcoRV–digested CB2 construct to replace the C-terminal tail domain of BRI1 with the CLV1 C-terminal tail domain. This fragment was sequenced after cloning.
For the CB1 construct, the same strategy was used. The first PCR amplification was performed on the CLV1 cDNA with the primers KSQ.1 (5′-CCGCCACCGTCACATAG-3′) and RevBriClvXIKD (5′-GGCCATGAC-TTGTACCATCGTAGGCCTTGC-3′). A second PCR was performed on the BRI1 cDNA cloned into pBluescript II KS+ (Stratagene) with the oligonucleotides ForBriClvXIKD (5′-GCAAGGCCTACGATGGTACAAGTC-ATGGCC-3′) and T7. These two fragments were used as templates in a third PCR with primers KSQ.1 and T7. The SpeI-NsiI fragment from the CLV1 cDNA was ligated to a NsiI-EcoRV fragment from this third PCR, generating a chimeric kinase domain in which the C-terminal tail domain of CLV1 was replaced with the C-terminal tail domain of BRI1. As mentioned above, the construct was sequenced after cloning. The fragment, including the ER promoter (−1678 bp), the CLV1 cDNA or chimeric constructs, and the E9 terminator, was subcloned into the pCB302 vector (Xiang et al., 1999). Fragments including only the ER promoter and the E9 terminator inserted into the pCB302 vector and the pCB302 empty vector were used as controls.
Scanning Electron Microscopy Analysis
Inflorescence tips of 15- or 30-day-old plants were fixed in 4% glutaraldehyde and 25 mM NaPO4 overnight at 4°C. Tissues were washed in 25 mM NaPO4 and incubated for several days in 1% osmium tetroxide and 25 mM NaPO4 at 4°C. Samples then were dehydrated sequentially through an ethanol series. Samples were dried with a critical point drier with liquid CO2, dissected to remove obstructive organs and expose the shoot apical meristem, coated with gold, and observed with a scanning electron microscope (S3200N; Hitachi, Tokyo, Japan). Images were collected digitally.
RNA Extraction and Reverse Transcriptase–PCR
Total RNA was isolated from 100 mg of inflorescences from approximately 30-day-old plants using the Trizol reagent (Life Technologies/Gibco BRL). One microgram of total RNA was reverse transcribed from an oligo(dT15) primer in a 25-μL reverse transcription reaction as recommended by the manufacturer (Moloney murine leukemia virus reverse transcriptase; Promega). One microliter of the first-strand cDNA reaction was used as a template for PCR amplification with specific oligonucleotides for CLV1 (forward, 5′-CCGCCACCGTCACATAG-3′ or 5′-CGG-CGTTGTTCTCACCGTCA-3′ [upstream of the CLV1 intron as a control for genomic DNA contamination]; reverse, 5′-ATGATCACCACATTT-CGCATC-3′ or 5′-CCAACAGGTTTCTTCCCAGCT-3′) and CLV1/BRI1 (forward, 5′-CGGCGTTGTTCTCACCGTCA-3′; reverse, 5′-CTGATCCTATCGCAATCTTCCG-3′). As an internal control, primers for the Arabidopsis βATPase genes were used. The PCR products were loaded on ethidium bromide–stained agarose gels. Each band was quantified with the Gel Doc 2000 Gel Documentation System (Bio-Rad) using Quantity One software and normalized to the controls. First, the background of the no-DNA PCR sample was removed for the clv1 and transgene bands. Then, the values were adjusted on the loading controls (βATPase). The values obtained for clv1-1 were normalized to the clv1-1 controls, which were assigned the value of 100. For the ER:CLV1, ER:CB2, ER:CB2m, and ER:CB3 transgenes, the highest ER:CLV1-expressing line was used to normalize the values of the other lines.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank Joshua Stomel for determining the correct sequence of the clv1-10 allele, Brody DeYoung for the sequence of the clv1-7 allele, Keiko Torii for isolating the ER promoter, Sang-Kee Song for generating the clv3-3 er double mutants, and Erik Colegrove for assistance in planting. This work was supported by National Institutes of Health Grant 1R01GM62962-01A1 to (S.E.C.). A.D. was supported in part by Organogenesis Postdoctoral Fellowship (University of Michigan, Ann Arbor). Isolation of clv1-12 and clv1-13 insertion alleles was supported by the National Science Foundation Grant DBI-9975808 to (F.E.X.).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010504.
References
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199. [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Brand, U., Fletcher, J.C., Hobe, M., Meyerowitz, E.M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. [DOI] [PubMed] [Google Scholar]
- Brand, U., Grunewald, M., Hobe, M., and Simon, R. (2002). Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 129, 565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R.H., Ebner, R., and Derynck, R. (1993). Inactivation of the type II receptor reveals two receptor pathways for the diverse TGF-beta activities. Science 260, 1335–1338. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Jacobsen, S.E., Levin, J.Z., and Meyerowitz, E.M. (1996). The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122, 1567–1575. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Running, M.P., and Meyerowitz, E.M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397–418. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Running, M.P., and Meyerowitz, E.M. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057–2067. [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. [DOI] [PubMed] [Google Scholar]
- Fletcher, J.C., Brand, U., Running, M.P., Simon, R., and Meyerowitz, E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. [DOI] [PubMed] [Google Scholar]
- Friedrichsen, D.M., Joazeiro, C.A., Li, J., Hunter, T., and Chory, J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123, 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Hardt, R., Lenhard, M., and Laux, T. (2002). WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev. 16, 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard, A., Srinivasan, S., Zimm, G., and Bier, E. (2002). A screen for dominant mutations applied to components in the Drosophila EGF-R pathway. Proc. Natl. Acad. Sci. USA 99, 3752–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Z., Wang, Z.Y., Li, J., Zhu, Q., Lamb, C., Ronald, P., and Chory, J. (2000). Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288, 2360–2363. [DOI] [PubMed] [Google Scholar]
- Jeong, S., Trotochaud, A.E., and Clark, S.E. (1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11, 1925–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalebic, T., Blakesley, V., Slade, C., Plasschaert, S., Leroith, D., and Helman, L.J. (1998). Expression of a kinase-deficient IGF-I-R suppresses tumorigenicity of rhabdomyosarcoma cells constitutively expressing a wild type IGF-I-R. Int. J. Cancer 76, 223–227. [DOI] [PubMed] [Google Scholar]
- Kashles, O., Yarden, Y., Fischer, R., Ullrich, A., and Schlessinger, J. (1991). A dominant negative mutation suppresses the function of normal epidermal growth factor receptors by heterodimerization. Mol. Cell. Biol. 11, 1454–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayes, J.M., and Clark, S.E. (1998). CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125, 3843–3851. [DOI] [PubMed] [Google Scholar]
- Kong, G., Penn, R., and Benovic, J.L. (1994). A beta-adrenergic receptor kinase dominant negative mutant attenuates desensitization of the beta 2-adrenergic receptor. J. Biol. Chem. 269, 13084–13087. [PubMed] [Google Scholar]
- Laux, T., Mayer, K.F., Berger, J., and Jurgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96. [DOI] [PubMed] [Google Scholar]
- Lease, K.A., Lau, N.Y., Schuster, R.A., Torii, K.U., and Walker, J.C. (2001. a). Receptor serine/threonine protein kinases in signalling: Analysis of the erecta receptor-like kinase of Arabidopsis thaliana. New Phytol. 151, 133–143. [DOI] [PubMed] [Google Scholar]
- Lease, K.A., Wen, J., Li, J., Doke, J.T., Liscum, E., and Walker, J.C. (2001. b). A mutant Arabidopsis heterotrimeric G-protein β subunit affects leaf, flower, and fruit development. Plant Cell 13, 2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard, M., Jurgens, G., and Laux, T. (2002). The WUSCHEL and SHOOTMERISTEMLESS genes fulfill complementary roles in Arabidopsis shoot meristem regulation. Development 129, 3195–3206. [DOI] [PubMed] [Google Scholar]
- Leyser, H.M.O., and Furner, I.J. (1992). Characterizations of three shoot apical meristem mutants of Arabidopsis thaliana. Development 116, 397–403. [Google Scholar]
- Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- Li, S., Strelow, A., Fontana, E.J., and Wesche, H. (2002. a). IRAK-4: A novel member of the IRAK family with the properties of an IRAK-kinase. Proc. Natl. Acad. Sci. USA 99, 5567–5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.X., Agaisse, H., Mathey-Prevot, B., and Perrimon, N. (2002. b). Differential requirement for STAT by gain-of-function and wild-type receptor tyrosine kinase Torso in Drosophila. Development 129, 4241–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, K.F., Schoof, H., Haecker, A., Lenhard, M., Jurgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. [DOI] [PubMed] [Google Scholar]
- McKelvie, A.D. (1962). A list of mutant genes in Arabidopsis thaliana. Radiat. Bot. 1, 233–241. [Google Scholar]
- Medford, J.I., Behringer, F.J., Callos, J.D., and Feldmann, K.A. (1992). Normal and abnormal development in the Arabidopsis vegetative shoot apex. Plant Cell 4, 631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany, J.A., Simon, E.J., Katzman, R.B., de Guzman, B.M., Yu, L.P., Trotochaud, A.E., and Clark, S.E. (1998). Identifying novel regulators of shoot meristem development. J. Plant Res. 111, 307–313. [Google Scholar]
- Prager, D., Li, H.L., Asa, S., and Melmed, S. (1994). Dominant negative inhibition of tumorigenesis in vivo by human insulin-like growth factor I receptor mutant. Proc. Natl. Acad. Sci. USA 91, 2181–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo, E., Sharma, V.K., Kovaleva, V., Raikhel, N.V., and Fletcher, J.C. (2002). CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 14, 969–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudarakanchana, N., Flanagan, J.A., Chen, H., Upton, P.D., Machado, R., Patel, D., Trembath, R.C., and Morrell, N.W. (2002). Functional analysis of bone morphogenetic protein type II receptor mutations underlying primary pulmonary hypertension. Hum. Mol. Genet. 11, 1517–1525. [DOI] [PubMed] [Google Scholar]
- Sanchez, I., Hughes, R.T., Mayer, B.J., Yee, K., Woodgett, J.R., Avruch, J., Kyriakis, J.M., and Zon, L.I. (1994). Role of SAPK/ERK kinase-1 in the stress-activated pathway regulating transcription factor c-Jun. Nature 372, 794–798. [DOI] [PubMed] [Google Scholar]
- Schneider, D.S., Hudson, K.L., Lin, T.Y., and Anderson, K.V. (1991). Dominant and recessive mutations define functional domains of Toll, a transmembrane protein required for dorsal-ventral polarity in the Drosophila embryo. Genes Dev. 5, 797–807. [DOI] [PubMed] [Google Scholar]
- Schoof, H., Lenhard, M., Haecker, A., Mayer, K.F., Jurgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. [DOI] [PubMed] [Google Scholar]
- Shiu, S.H., and Bleecker, A.B. (2001). Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci. STKE 2001, RE22. [DOI] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9, 1797–1810. [DOI] [PubMed] [Google Scholar]
- Torii, K.U., and Clark, S.E. (2000). Receptor-like kinases in plant development. In Advances in Botanical Research, M. Kries and J.C. Walker, eds (San Diego, CA: Academic Press), pp. 226–269.
- Torii, K.U., Mitsukawa, N., Oosumi, T., Matsuura, Y., Yokoyama, R., Whittier, R.F., and Komeda, Y. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8, 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud, A.E., Hao, T., Guang, W., Yang, Z., and Clark, S.E. (1999). The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 11, 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venglat, S.P., Dumonceaux, T., Rozwadowski, K., Parnell, L., Babi, V., Martienssen, R., Selvarag, G., and Datla, R. (2002). The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G.L., et al. (1998). Xa21D encodes a receptor-like molecule with a leucine-rich repeat domain that determines race-specific recognition and is subject to adaptive evolution. Plant Cell 10, 765–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, C., Han, P., Lutziger, I., Wang, K., and Oliver, D.J. (1999). A mini binary vector series for plant transformation. Plant Mol. Biol. 40, 711–717. [DOI] [PubMed] [Google Scholar]
- Yokoyama, R., Takahashi, T., Kato, A., Torii, K.U., and Komeda, Y. (1998). The Arabidopsis ERECTA gene is expressed in the shoot apical meristem and organ primordia. Plant J. 15, 301–310. [DOI] [PubMed] [Google Scholar]
- Zou, H., and Niswander, L. (1996). Requirement for BMP signaling in interdigital apoptosis and scale formation. Science 272, 738–741. [DOI] [PubMed] [Google Scholar]