Abstract
The RNA polymerase (pol) III-transcribed (e.g. tRNA and 5S rRNA) genes of traditionally studied organisms rely on gene-internal promoters that precisely position the initiation factor, TFIIIB, on the upstream promoter-less DNA. This is accomplished by the ability of the TFIIIB subunit, TFIIB-related factor (Brf1), to make stable protein–protein interactions with TATA-binding protein (TBP) and place it on the promoter-less upstream DNA. Unlike traditional model organisms, Schizosaccharomyces pombe tRNA and 5S rRNA genes contain upstream TATA promoters that are required to program functional pol III initiation complexes. In this study we demonstrate that S.pombe (Sp)Brf does not form stable interactions with TBP in the absence of DNA using approaches that do reveal stable association of TBP and S.cerevisiae (Sc)Brf1. Gel mobility analyses demonstrate that a TBP–TATA DNA complex can recruit SpBrf to a Pol III promoter. Consistent with this, overproduction of SpBrf in S.pombe increases the expression of a TATA-dependent, but not a TATA-less, suppressor tRNA gene. Since previous whole genome analysis also revealed TATA elements upstream of tRNA genes in Arabidopsis, this pathway may be more widespread than appreciated previously.
INTRODUCTION
In eukaryotes, Pol I synthesizes large ribosomal rRNA (35S-45S depending on the species), Pol II synthesizes mRNAs and some small nuclear (sn)RNAs (e.g. U1, U2), and Pol III synthesizes mostly tRNAs and 5S rRNA, as well as U6 snRNA and few other non-protein coding transcripts (1). Pols I, II and III all require TATA-binding protein (TBP) or a closely related protein for initiation even though some of the target genes do not contain the sequence-specific TBP-binding site, TATAAA (or closely related sequences, referred to as a TATA box or element) (2,3). In the absence of a TATA element, other factors bring TBP to the promoter via protein–protein interactions (reviewed in 4,5). A second highly conserved initiation factor, TFIIB, exhibits very weak interactions with TBP in the absence of DNA, but recognizes the TBP–TATA DNA complex with substantially higher affinity (6,7). TFIIB and TFIIB-related factors (e.g. Brf1) are multifunctional proteins of central importance that interact with several other factors, help determine the start site of transcription, participate in promoter melting and select the appropriate Pol for transcription initiation (8–13). TFIIB is used by Pol II, Brf1 is used by Pol III for tRNA and 5S rRNA (TATA-less) genes, and a distinct protein, Brf2, is used by Pol III for human U6 and related type 3 genes that contain TATA elements (14–16).
The classical Pol III promoters that characterize the hundreds-to-thousands of tRNA and 5S rRNA genes in the traditionally studied eukaryotes use internal (i.e. downstream of transcription start site) promoter elements to initiate the process of transcription complex assembly. The assembly factor TFIIIC binds to the tRNA internal control elements and initiates protein–protein interactions that result in the deposition of TBP onto the TATA-less upstream DNA (17–20). Thus, direct association with Brf1 allows TBP to bind the upstream TATA-less DNA, even if it is comprised of only G and C residues (16,20,21). In addition to the TATA-less pathway of TBP recruitment, this TBP–Brf1 arrangement can also support TFIIIC-independent transcription by Pol III, in vitro, via direct recruitment of TBP by the TATA elements of the single U6 gene and the four TATA-containing tRNA genes (of a total of 274 tRNA genes) that can be found in the Saccharomyces cerevisiae genome (22–24).
Characterization of the Pol III transcription system in the fission yeast S.pombe has revealed, among other things, that all of its Pol III-dependent genes appear to contain upstream TATA sequences as one element of their multipartite promoters (25–27, reviewed in 28). Upstream TATAs were shown to be required for functional tRNA expression in S.pombe and in vitro, and for 5S rRNA synthesis in vivo (27). It is important to note for the purposes of this discussion that although a small number of TATA-containing tRNA genes from traditional organisms are known to exist, those that have been studied in great detail represent rare exceptions which in most cases characterize highly specialized tRNA genes (see discussion in 27). In contrast to this, all tRNA and 5S genes appear to contain upstream TATA elements in S.pombe (27). In the present study we examine the TBP-binding characteristics of SpBrf that are associated with the requirement for TATA elements in S.pombe.
Although a prior study demonstrated that SpBrf co-purified with SpPol III and SpTFIIIC, while SpTBP did not, it did not assess the potential for interaction between TBP and Brf (27). Because this potential would appear to be an issue of central importance to the requirement for upstream TATA elements, it was examined in detail. Here we show that SpBrf and SpTBP do not stably associate in vitro nor are they found associated in extracts, as monitored by co-immunoprecipitation (IP), but that they can form a complex with TATA-containing DNA. The results indicate an innate insufficiency of SpBrf to bring TBP to the upstream DNA and that this represents a novel characteristic of the Pol III system in S.pombe. While similar insufficiency would be detrimental in organisms with TATA-less Pol III genes, in S.pombe this is balanced by the ubiquitous TATA elements that are precisely positioned upstream of the Pol III-transcribed genes in this yeast. Since a whole genome survey of Arabidopsis thaliana also revealed TATA elements upstream of its tRNA genes, the Pol III transcription complex assembly pathway described here may be more widespread in nature than appreciated previously, and also suggests that it may represent an ancient eukaryotic Pol III transcription system.
MATERIALS AND METHODS
Yeast strains
The yeast strains used are listed in Table 1. Suppression was monitored on plates containing EMM lacking uracil and containing 30 mg/l adenine, with or without thiamin (5 mg/l).
Table 1. Schizosaccharomyces pombe strains used in this study.
| Strain | Comments | Source |
|---|---|---|
| ySD5 (h+, ade6-704, leu1-32, ura4–, his3-237) | Recipient of Brf1-HA allele | This report |
| yMW2 (h+, ade6-704, leu1-32, his3-237, ura4–, Δbrf1::[brf1+-HA-ura4+]) | IP of SpBrf1-HA | This report |
| yAS78 (hS–, ade6-704, leu1-32::[tRNAMSer7T-leu1+], ura4–) | SpBrf1 overexpression | This report |
| yAS50 (hS–, ade6-704, leu1-32, ura4–) | Control strain | 27 |
| yTM2a (hS–, ade6-704, leu1-32::[tRNAMSer7T-leu1+], ura4–/pRep93X-spBrf1(ura4+)) | Suppression +/– thiamin | This report |
Construction of a C-terminal 3HA and 6His tagging vector
For epitope tagging of SpBrf, a vector designated p3HAHis was constructed using synthetic DNA 5′-pGATCCGTCGACCATCATCATCATCATCATGCCTACCCATACGACGTTCCAGACTACGCTGCCTACCCATATGATGTGCCTGATTACGCCGCCTATCCATACGATGTCCCAGACTACGCTTAGCCC (Lofstrand Labs Inc., Gaithersburg) containing sequence encoding six histidine residues (bold) and three repeats of the Influenza hemagglutinin (HA) epitope, YPYDVPDYA (underline) with 5′ BamHI and SalI sites and a 3′ SmaI site cloned into the BamHI/SmaI sites of pREP93X (29).
Construction of a S.pombe strain expressing epitope-tagged brf-6His-3HA (brf-HA)
The chromosomal copy of brf1+ was replaced with epitope tagged brf1-HA by a one-step knock-in procedure (30). First, the 3′-flanking region of the S.pombe brf gene was amplified by PCR using primers 5′-GACTGAGTCGACCACTGAGTAATTATTAGTATCCATC and 5′-GACTGAGTCGACTCT GCTCAATTCTATCCAGAAAGG containing SalI sites (underlined) and cloned into the SalI site of pBluescript II SK(+)-Ura4+ (a gift from T. Kokubo) to create pUraFl. Secondly, a 3′-segment of the coding region of the S.pombe brf1 gene (1074 bp) was PCR amplified from genomic DNA using primers 5′-GACTGACCATGGGTTTGCCGGATATCTAAGACGTCT and 5′-GACTGAGTCGACCGACTGCTCTTCGTCAAATAACAT and cloned into the NcoI/SalI sites of p3HAHis to create pHaBrf. A fragment containing the 3′-segment of brf1, the 6His-3HA tag, and the nmt1 terminator was amplified from pHaBrf using primers 5′-GTCGCTCTCTAGACTCACTTTCTGACTTATAGTCGC and 5′-GAGTTAGACTC GAATTCGAGCTCG and cloned into the XbaI/EcoRI sites of pUraFl to create pCBrf.
For yeast transformation, a contiguous fragment containing the 3′ segment of brf1, the 6His-3HA sequence, the nmt1 terminator, ura4+ and the 3′-flanking region of brf1 was obtained by digestion of pCBrf with XbaI and XhoI followed by PCR using primers 5′-TCTGCTCAATTCTATCCAGAAAGGAAAGG and 5′-GTTTGCCGGATATCTAAGACGTCTCATATG. The resulting fragment (expected size, 5009 bp) was gel-purified and used to transform ySD5 to a ura+ phenotype. Multiple stable ura4+ transformants were selected and characterized for ploidy, correct genomic modification by PCR and Southern blotting, and for expression of tagged Brf1 by western blotting analysis; yMW2 (also known as yMW1-2) is the transformant used in this study.
Purification of S.pombe TBP (SpTBP)
Recombinant SpTBP containing a N-terminal 6His-tag was expressed and purified under native conditions using the pET system (Novagen) (26). Cells were grown to an OD600 of 0.6 in LB broth containing 25 ug/ml kanamycin at 25°C. SpTBP was then induced with IPTG at a final concentration of 0.5 mM and cells were harvested after 8 h at 25°C. Cells were suspended in Buffer A [25 mM HEPES–KOH, pH 7.6, 12.5 mM MgCl2, 10% glycerol, 0.05% Nonidet P-40, 100 mM KCl, 7 mM β-mercaptoethanol (β-ME), 0.1 mM phenylmethylsulfonyl fluoride (PMSF)], containing 200 ug/ml lysozyme, and sonicated five times for 45 s. The pellet was removed after centrifugation at 12 000 g for 30 min. The extract (20 ml, ∼20 mg/ml) was loaded onto a 5 ml heparin HiTrap column (Amersham) equilibrated in Buffer A. After wash with Buffer A, SpTBP was eluted with a linear gradient of 0.2–0.6 M KCl in Buffer A and followed by western blotting. Fractions containing SpTBP were pooled and loaded onto a 0.5 ml Ni–NTA agarose (Qiagen) column equilibrated in Buffer A containing 10 mM imidazole. After washing with Buffer A containing 300 mM NaCl and 10 mM imidazole, SpTBP was eluted in Buffer A containing 150 mM imidazole and dialyzed into 25 mM HEPES–KOH, pH 7.6, 12.5 mM MgCl2, 10% glycerol, 100 mM KCl, 1 mM DTT and 0.1 mM PMSF. The concentration of SpTBP was determined by SDS–PAGE using bovine serum albumin as a standard.
Recombinant S.pombe Brf1p (SpBrf)
Recombinant S.pombe Brf1p (SpBrf) was purified after expression as a C-terminal 6His-tagged protein under denaturing conditions using the pET system (Novagen), similar to that described for purification of ScBrf1 (31). A sense primer BRF1 (5′-CTAGCGCCATGGGATGTCCAAATTGCGGTT, NcoI) and antisense primer BRF2 (5′-GAGCTCGAGTTACGACTGCTCTTCGTCAAATA, XhoI) were used to amplify S.pombe cDNA (26) and cloned into the NcoI/XhoI sites of pET28b (pET28b-Brf). Cells were suspended in Buffer B (20 mM Tris–Cl, pH 7.6, 500 mM NaCl, 0.1 mM PMSF, 7 mM β-ME and 10% glycerol) and sonicated five times for 45 s. Inclusion bodies were collected by centrifugation at 12 000 g for 30 min and solubilized in Buffer B containing 6 M guanidine–HCl for 1 h at 25°C. Insoluble material was removed by centrifugation at 18 000 r.p.m. for 30 min and the supernatant was incubated with 1 ml of Ni–NTA agarose beads for 1 h at 25°C. The beads were washed twice with Buffer B containing 10 mM imidazole and SpBrf was eluted with Buffer B containing 6 M urea and 300 mM imidazole and step dialyzed into Buffer D (20 mM Tris–HCl, pH 7.6, 100 mM NaCl, 0.1 mM PMSF, 1 mM DTT, 10% glycerol and 10 mM ZnSO4) containing 3, 1.5, 0.7 and then 0 M urea. SpBrf was quantitated by SDS–PAGE using bovine serum albumin as a protein standard.
Schizosaccharomyces pombe B” (SpBdp1) was expressed in bacteria and purified
A sense primer, SPB1UPN (TAGAATTCCCATatgtcaaggtttgcccctaaatttacagcccg, NdeI) and an antisense primer SPB2UPN (GATCGGATCCattaaaccacttgcccgactacctcg, BamHI) were used for PCR amplification using a S.pombe cDNA library as template and cloned into the NdeI/BamHI sites of pET28a. SpBdp1 was expressed as an N-terminal 6His fusion protein and purified under native conditions. Cells were grown to an OD600 of 0.6 in LB broth containing 25 μg/ml kanamycin at 25°C. Synthesis of SpBdp1 was induced with IPTG at a final concentration of 0.5 mM and cells were harvested after 8 h at 25°C. The pellet was suspended in Buffer A (50 mM Tris–HCl, pH 7.9, 12.5 mM MgCl2, 10% glycerol, 0.05% Nonidet P-40, 100 mM NaCl, 7 mM β-ME and 0.1 mM PMSF containing 200 μg/ml lysozyme, and sonicated for 45 s five times. The pellet was removed by centrifugation at 12 000 g for 30 min and the extract (20 ml, ∼20 mg/ml) was loaded onto a 0.5 ml Ni–NTA agarose (Qiagen) column equilibrated in Buffer A containing 10 mM imidazole. The column was washed with Buffer A containing 300 mM NaCl and 10 mM imidazole. SpBdp1 was eluted with Buffer A containing 150 mM imidazole. Fractions containing SpBdp1 were pooled and dialyzed into 50 mM Tris–HCl, pH 7.9, 12.5 mM MgCl2, 10% glycerol, 100 mM NaCl, 1 mM DTT and 0.1 mM PMSF and loaded onto a 1 ml DEAE column (Amersham) equilibrated in Buffer A. The column was washed with Buffer A. SpBdp1 was eluted with a linear gradient from 0.1 to 1.0 M NaCl in buffer A. The fractions containing SpBdp1 were pooled and the concentration of SpBdp1 was determined by SDS–PAGE using bovine serum albumin as a standard.
Schizosaccharomyces pombe Pol III (SpPol III) was immunoaffinity purified as described previously (27). Saccharomyces cerevisiae (sc)Pol III, as well as recombinant ScTBP, ScBrf1 and scB” (Bdp1), were kindly provided by G. Kassavetis and E. P. Geiduschek (UC, San Diego, CA).
Co-immunoprecipitation
Schizosaccharomyces pombe extracts at ∼20 mg/ml were prepared from yAS50 or yMW2 as described (26). One milliliter of extract was incubated with 2 µl pre-immune serum, 2 µl anti-SpTBP, 2 µl affinity-purified anti-SpBrf or 20 µl agarose-immobilized mouse anti-HA monoclonal Ab (12CA5; Boehringer) for 1 h at 4°C. For co-IP with anti-SpTBP or anti-SpBrf, 20 µl of protein A–Sepharose beads (Amersham Pharmacia Biotech) was subsequently added and the mixture was incubated for 1 h at 4°C. The beads were collected by centrifugation at 1000 g for 5 min and washed three times with 1 ml NET-2 (150 mM NaCl, 50 mM Tris–HCl, pH 7.5). Bound material was eluted with 20 µl of 1.5 × SDS loading buffer for 5 min at 94°C and analyzed by western blotting using affinity-purified anti-SpTBP Ab (1:1000 dilution), affinity-purified anti-SpBrf Ab (1:2000 dilution), anti-pTR6 serum (1:5000 dilution) and anti-pTAF130 serum (1:5000 dilution). Affinity-purified anti-SpBrf and anti-SpTBP were described previously (26,27). Anti-pTR6 and anti-pTAF130 sera were kindly provided by T. Kokubo (Nara Institute of Science and Technology, Japan). Blots were developed using the ECL system (Amersham Pharmacia Biotech).
In vitro protein–protein interactions
To monitor interactions between SpTBP and ScBrf1 or SpBrf, SpTBP (15 ng) was incubated with SpBrf or ScBrf1 (60 ng) in the presence of 1 µl of anti-SpTBP Ab in NET-2 buffer in a total volume of 100 ul for 1 h at 4°C. Twenty μl of protein A–Sepharose (Amersham Pharmacia Biotech) was added and the mixture was incubated for 1 h at 4°C. The beads were washed three times with 1 ml NET-2. The bound proteins were eluted with 20 μl of 1.5 × SDS loading buffer for 5 min at 94°C. Inputs (1/13th volume) and eluates (1/8th volume) were analyzed by western blotting using affinity-purified anti-SpTBP Ab (1:2000 dilution) or mouse anti-His6 Ab (1:2000 dilution; Boehringer). For interactions between ScTBP and ScBrf1 or SpBrf, ScTBP (10 ng) was incubated with SpBrf or ScBrf1 (60 ng) in the presence of 1 ul of anti-ScTBP Ab (kindly provided by P. A. Weil, Vanderbilt University School of Medicine, Nashville, TN). Inputs (1/13th) and eluates (1/8th) were analyzed by western blotting using affinity-purified anti-SpTBP (1:10 000 dilution) or mouse anti-His6 Ab (1:2000 dilution; Boehringer).
DNA binding assays
The probe was derived from the S.cerevisiae U6 gene (SNR6). Complementary oligonucleotides corresponding to position –45 to –8 of SNR6 were annealed in TE containing 300 mM NaCl, purified from a 12% native polyacrylamide gel and labeled with polynucleotide kinase and [γ-32P]ATP (Lofstrand Labs Inc.). DNA binding assays were performed in buffer containing 40 mM Tris–Cl, pH 8.0, 60 mM NaCl, 7 mM MgCl2, 6% glycerol, 1 mM DTT, 100 ug/ml BSA, 0.1% Tween-20, 10 ng/l of poly(dG–dC)poly(dG–dC), 1 ng DNA probe (∼5000 d.p.m.), TBP (1–2.6 ng, calibrated to give comparable levels of complex formation) and varying amounts of Brf1 as indicated. Reaction mixtures were incubated for 35 min at 25°C. The protein–DNA complexes were analyzed on 4% native polyacrylamide gels (InVitrogen, Carlsbad, CA) containing 25 mM Tris–Cl, pH 8.3, 0.19 M glycine, 0.2 mM MgOAc, 2.5% glycerol and 0.5 mM DTT. The electrode buffer was 25 mM Tris–Cl, pH 8.3, 0.19 M glycine, and 0.2 mM MgOAc. Electrophoresis was performed at 140 V for 35 min at 4°C. The gel was dried and exposed to a FUJIFILM screen (Fuji) and analyzed by PhosphorImager.
RESULTS
SpTBP is not stably associated with SpBrf in extracts of S.pombe
It was demonstrated previously that while SpBrf was readily detected in association with immunoaffinity-purified SpPol III and spTFIIIC complexes, SpTBP could not be detected in either complex (27). Moreover, the SpPol III complex also contained SpBdp1 (formerly known as spB”; 32), suggesting that SpTBP might not form stable association with SpBrf even if the other known component of TFIIIB, Bdp1, was also present (27). As one approach to more closely examine the potential for interaction between SpTBP and SpBrf, we modified the S.pombe brf1 gene to encode a SpBrf fusion protein with a C-terminal tag containing three HA epitopes, in a haploid strain designated yMW2. The structure of the integrated construct was confirmed by PCR and Southern blotting (data not shown), which indicated that the single copy of brf1 was replaced by brf1-HA as summarized in Figure 1A. This brf1 gene modification caused no appreciable difference in growth rate of yMW2 at 32 or 37°C as compared with the control strain, yAS50, in the appropriate growth conditions (not shown).
Figure 1.
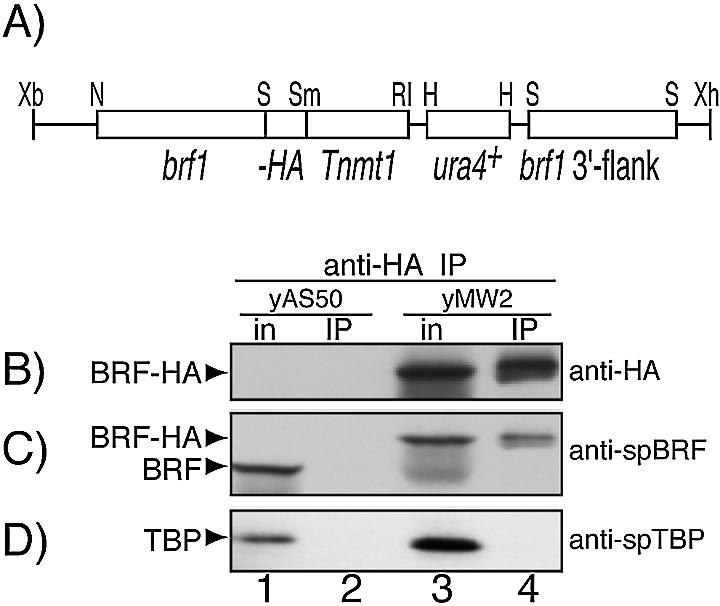
(A) The brf1-HA locus in strain, yMW2, which expresses HA epitope-tagged SpBrf. (B–D) Extracts prepared from the control strain yAS50 and experimental strain yMW2 were subjected to IP with anti-HA agarose beads. Input extracts (in) and immunoprecipitates were analyzed by western blotting using the following antibodies (Abs) for detection: (B) anti-HA, (C) anti-SpBrf and (D) anti-SpTBP, as indicated on the right. Detected proteins are identified by the arrowheads on the left. The mobility of SpBrf-HA is slower than endogenous SpBrf due to three HA epitopes and a 6His tag.
Extracts prepared from yMW2 and yAS50 were subjected to IP by anti-HA followed by western blotting using anti-HA, anti-SpBrf and anti-SpTBP for detection (Fig. 1B–D). The inputs (lanes 1 and 3) and IPs (lanes 2 and 4) were examined. Anti-HA detected a protein of the expected size in yMW2 (Fig. 1B, lane 3) but not yAS50 (lane 1), as expected. Anti-SpBrf recognized endogenous SpBrf in yAS50 (Fig. 1C, lane 1) and the slower migrating SpBrf-HA in yMW2 (lane 3). Most significantly, although SpTBP was readily detectable in yAS50 and yMW2, it was not co-immunoprecipitated with SpBrf-HA (Fig. 1D, lane 4).
It was possible that although SpBrf-HA supported normal growth, the HA tag might interfere with interaction with SpTBP. To address this, we performed co-IP using anti-SpBrf antibodies (Abs) and examined them for the presence of SpTBP (Fig. 2A and B). Anti-SpBrf specifically immunoprecipitated SpBrf and SpBrf-HA from yAS50 and yMW2 (Fig. 2A, lanes 3 and 6) while pre-immune serum did not (lanes 2 and 5). Again, while SpTBP was readily detectable in the extract (Fig. 2B, lanes 1 and 4), it was not co-immunoprecipitated with either SpBrf or SpBrf-HA (Fig. 2B, lanes 3 and 6).
Figure 2.
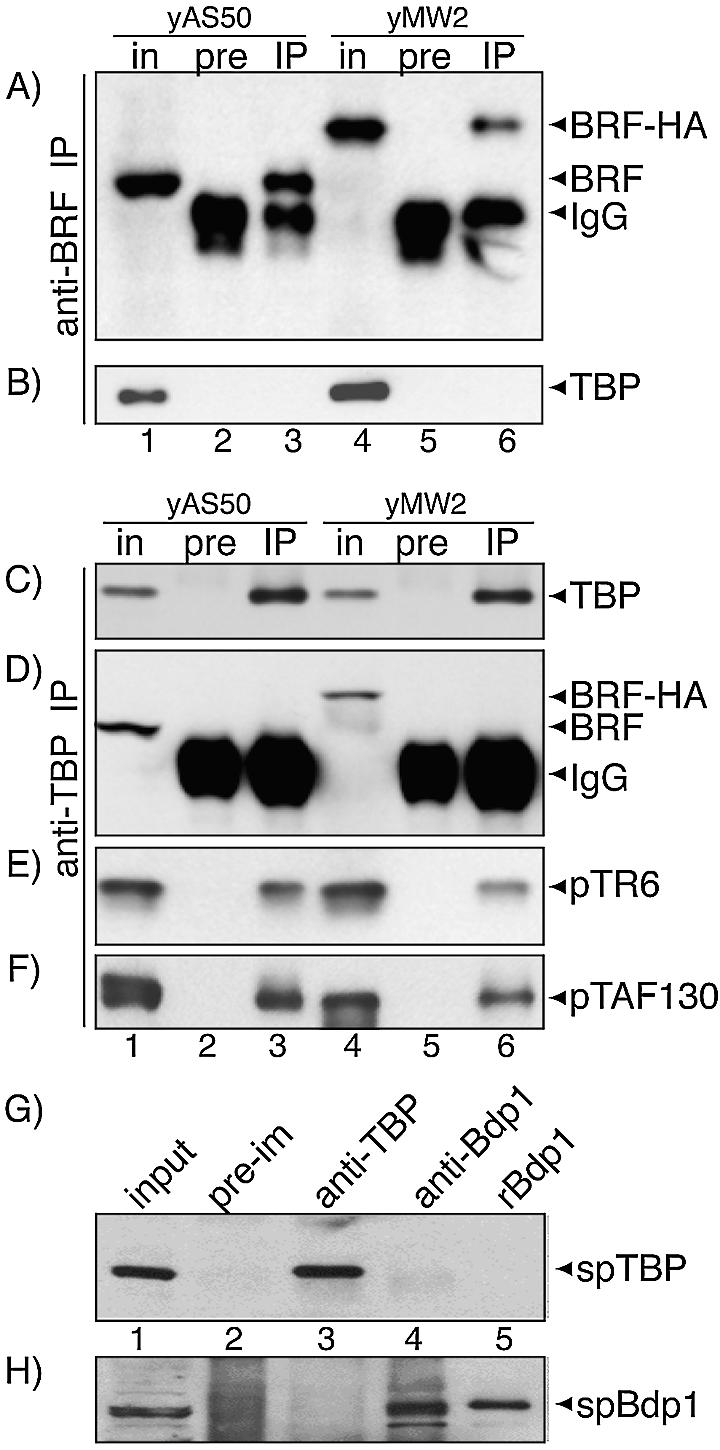
(A–F) SpTBP and SpBrf are not associated in S.pombe extract, whereas SpTBP and the Pol II TAFs, pTR6 and pTAF130 are. yAS50 and yMW2 extracts were analyzed directly (in) or first subjected to IP with pre-immune (pre), or anti-SpBrf Ab (IP) and analyzed by western blotting using the following Ab for detection: (A) anti-SpBrf, (B) anti-SpTBP. (C–F) was performed as in (A and B) except that anti-SpTBP was used for IP. Input extracts (in), pre-immune (pre) or anti-SpTBP (IP) were analyzed by western blotting using the following Ab for detection: (C) anti-SpTBP, (D) anti-SpBrf, (E) anti-pTR6 and (F) anti-pTAF130. (G and H) SpBdp1 and SpTBP are not associated in S.pombe extract. This experiment was performed as above. Input extract (in), pre-immune (pre), anti-SpTBP (αTBP) and anti-SpBdp1 (αB”) (lanes 1–4) were analyzed by western blotting using the following Ab for detection: (G) anti-SpTBP, (H) anti-SpBdp1. Lane 5 contained purified recombinant SpBdp1 as marker. Detected proteins are indicated by arrowheads on the right.
In a reciprocal approach, we examined anti-SpTBP IPs for the presence of SpBrf and SpBrf-HA (Fig. 2C and D). Anti-SpTBP Abs immunoprecipitated SpTBP while pre-immune Abs did not (Fig. 2C, lanes 3 and 6). While SpBrf and SpBrf-HA were readily detectable in the input (Fig. 2D, lanes 1 and 4, respectively), neither was co-immunoprecipitated with SpTBP (Fig. 2D, lanes 3 and 6). Two previously characterized TAFIIs referred to as pTR6 and TAF130 (33,34), were readily detectable in the anti-SpTBP co-IP as expected (Fig. 2E and F). This indicated that the deficiencies of SpBrf and SpBrf-HA to associate with SpTBP were not due to a general failure of the co-IP protocol. Moreover, since TAFIIs are less abundant than Brf1 in some yeast (35), the lack of detectable SpBrf in the TBP co-IP would appear to be significant. These data demonstrate that TBP and Brf1 are not stably associated in extracts of S.pombe.
We also asked if TBP and Bdp1 are associated in S.pombe by reciprocal IP and western blotting. While the corresponding antisera efficiently precipitated the cognate polypeptide, in both cases the other protein was not detected in the IP (Fig. 2G and H). This suggested that TBP and Bdp1 are not stably associated in extracts of S.pombe.
Insufficiency to stably associate with SpTBP in the absence of DNA is inherent to SpBrf
Because the data suggested the possibility that Brf1 might be innately deficient in recognition of free TBP, we examined the potential for purified recombinant TBP and Brf1 proteins to interact in vitro. Protein mixtures were subjected to anti-TBP co-IP, and the inputs (In) and immunoprecipited material were examined by western blotting. We included ScTBP and ScBrf1 with each other as positive controls and with the reciprocal S.pombe proteins, to provide additional information. The experiments in Figure 3A and B differed only with regard to whether SpTBP or ScTBP (Fig. 3A and B, respectively) and the corresponding affinity-purified Abs for IP were used.
Figure 3.
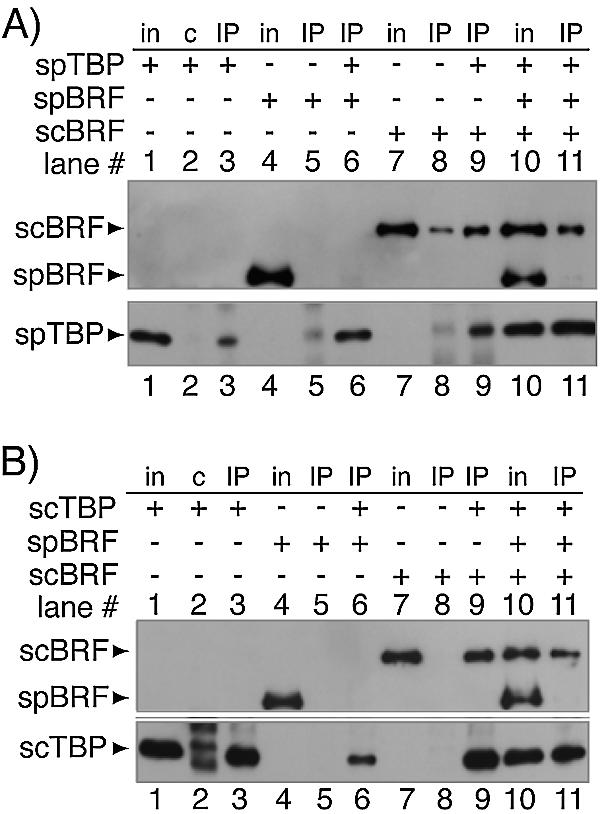
Recombinant SpBrf does not form a stable association with recombinant TBP in the absence of DNA, while recombinant ScBrf1 does. Mixtures of the indicated proteins were incubated and subjected to IP with anti-SpTBP Ab (A) or anti-ScTBP Ab (B). After the IPs were washed, the bound material was eluted and examined by western blotting using anti-6His Ab (upper panels, A and B), anti-SpTBP Ab (lower panel, A) or anti-ScTBP (lower panel, B). The panels in (A and B) differ only as to whether SpTBP or ScTBP were used, respectively. Input proteins (in) and IPs with pre-immune (c) or IPs with anti-ScTBP Ab (IP) were analyzed. Proteins detected by western blotting are indicated on the left.
The most relevant finding in Figure 3A and B is that while ScBrf1 was co-immunoprecipited with TBP (lanes 9), SpBrf was not (lanes 6). To confirm this and exclude the possibility that our SpBrf preparation might contain an inhibitor that would generally block TBP–Brf1 interaction, co-IP experiments were performed with SpBrf, ScBrf1 and TBP (Fig. 3A and B, lanes 10 and 11). ScBrf1 but not SpBrf could be co-immunoprecipitated with either SpTBP or ScTBP (Fig. 3A and B, lanes 11). This indicated that SpBrf competes poorly if at all for TBP binding and suggests that it is innately limited in the ability to stably associate with TBP. Consistent with the above results, we noted that a small amount of ScBrf1 was immunoprecipitated by anti-SpTBP in the absence of exogenous TBP (Fig. 3A, lane 8, upper panel). Upon close examination, this appeared to be due to the presence of TBP, as a contaminant, in the anti-SpTBP Ab preparation (Fig. 3A, lower panel, lane 8) that probably leeched from the TBP-matrix during affinity purification of our anti-SpTBP Abs. However, it should be noted that while similar low level TBP was also present in lane 5 (Fig. 3A, lower panel), in this case, SpBrf was not co-immunoprecipitated (Fig. 3A, lane 5, upper panel). The cumulative data suggest that either ScTBP or SpTBP could associate with ScBrf1 but that neither could stably associate with SpBrf. Interaction between SpTBP and ScBrf1 was not unexpected since SpTBP could fully complement the null mutation in the ScTBP gene, SPT15, in S.cerevisiae (36). The data in Figure 3 supported the results obtained in S.pombe extract (Figs 1 and 2). We conclude that the inability to detect association between SpTBP and SpBrf reflects an innate insufficiency of Brf1 to stably interact with TBP in the absence of DNA.
Recognition of TBP–TATA DNA by SpBrf
To test the ability of SpBrf to associate with a TBP–TATA DNA complex, we performed EMSA with both ScTBP (Fig. 4A) and SpTBP (Fig. 4B). TBP formed a complex with the TATA-containing probe (Fig. 4, filled arrowheads, lanes 2) that was competed by an excess of unlabeled TATA-containing DNA but not with DNA that contained mutations of the TATA sequence, demonstrating that the interaction was specific (not shown). Addition of either ScBrf1 or SpBrf led to complexes of slower mobility (open arrowheads, lanes 3–12), similar to what has been reported for ScBrf1 (37,38). We note that the DNA–TBP–Brf complex can be further super-shifted by recombinant B” (data not shown), suggesting that the low mobility band contains TBP and Brf. In contrast to the ability of full length SpBrf to associate with the TBP–TATA complex, a purified recombinant fragment (SpBrf amino acids 1–383) (20) that corresponds to the TFIIB-homologous region of SpBrf plus the Brf-homologous region 1, but lacks Brf homologous regions 2 and 3, failed to associate to any notable degree (data not shown).
Figure 4.
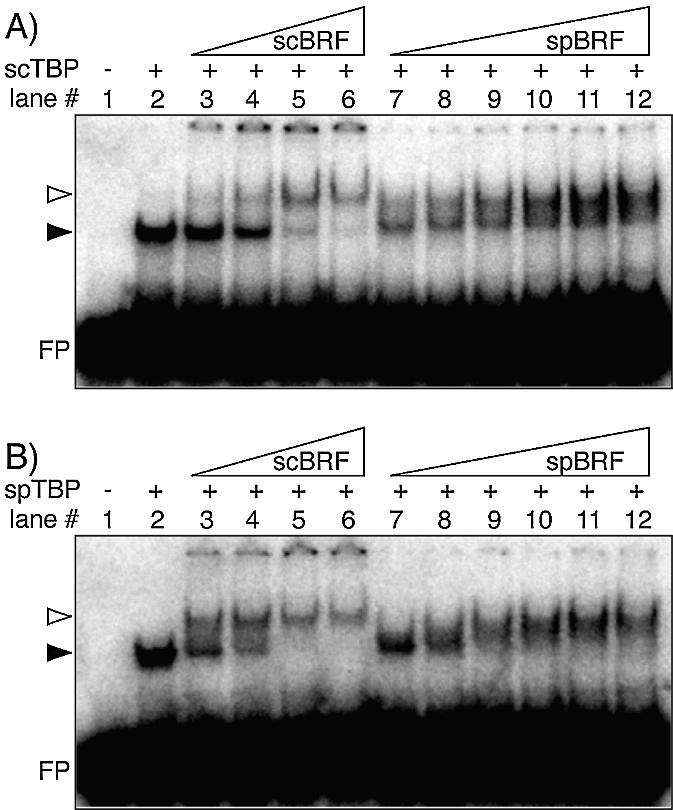
SpBrf can recognize a TBP–TATA DNA complex. Interactions between recombinant TBP, TATA DNA and Brf1 were examined by EMSA. (A and B) differ only as to the source of TBP; ScTBP (A) or SpTBP (B), as indicated. For each, varying concentrations of ScBrf1 (12–120 ng, lanes 3–6) and SpBrf (4–120 ng, lanes 7–12) were examined. The protein–DNA complexes are indicated by arrows.
We noted that in some experiments, ScBrf1 led to a more distinct TBP–DNA–Brf1 complex than did SpBrf, the latter of which produced more of a smear (e.g. compare lanes 6 and 12). This suggested that SpBrf may form a less stable complex with TBP–DNA as compared with ScBrf1, although this was not examined exhaustively. In any case, the positive EMSA results revealed that SpBrf is capable of specific recognition of a TBP–TATA DNA complex.
Overproduction of SpBrf increases expression from a TATA-dependent tRNA gene in S.pombe
We characterized previously a system to examine tRNA expression in S.pombe in which Pol III-mediated expression of the opal suppressor tRNASerUGAM gene can be monitored by suppression of ade6-704 and a resulting decrease in red pigment (25). In this system, increased levels of tRNASerUGAM transcripts lead to increased suppression (25), dependent on the TATA element located upstream of the tRNASerUGAM gene (27). For the present study, we developed a modified version of this suppression system to serve as a reporter for SpBrf function. We developed strain yAS78, which harbors the TATA-dependent allele, tRNASerUGAM-7T, and produces an attenuated suppressor that results in partial suppression (25). In this system, overproduction of SpBrf might lead to increased transcription of tRNASerUGAM-7T and suppression of yAS78. For this purpose, SpBrf DNA was cloned into the S.pombe expression vector, pRep93X, whose promoter is repressed by the addition of exogenous thiamin (29,39).
Transformation of yAS78 with pRep93X-SpBrf led to strain yTM2a (yYH1), which exhibited increased suppression in the absence of thiamin but not in the presence of thiamin (Fig. 5A). Concordant with this, western blotting demonstrated overproduction of SpBrfp in the absence of thiamin and repression in the presence of thiamin as expected (Fig. 5B). As a control for loading, we also probed the western blot for spLa protein (Fig. 5C). The results established that thiamin-sensitive overproduction of SpBrf was specific and that it could stimulate tRNASerUGA M-mediated suppression. Attempts to activate suppression by overproduction of SpBrfp in strains carrying a tRNASerUGA gene with a mutated TATA sequence (GGATCC) (27) were unsuccessful (not shown). The results indicate that SpBrf stimulates tRNA expression from TATA-containing but not TATA-less tRNASerUGAM genes. In addition to demonstrating its Pol III-related activity, the results also indicate that the in vivo level of SpBrfp may be a critical determinant of tRNASerUGAM expression in S.pombe.
Figure 5.
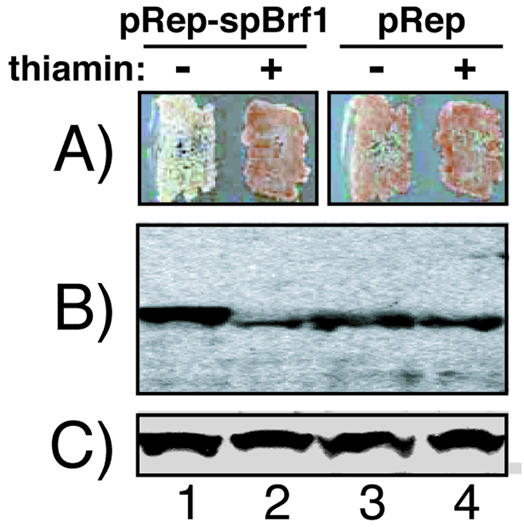
Overproduction of SpBrf increases expression from a TATA-dependent tRNA gene in S.pombe. (A) Schizosaccharomyces pombe strain yAS78 transformed with pRep93X-SpBrf (lanes 1 and 2) or empty pRep93X vector (lanes 3 and 4) were grown on media containing limiting adenine (30 mg/l) in the absence or presence of thiamin as indicated. The cells shown in (A) were grown in liquid media in the absence or presence of thiamin and extracts were analyzed by western blotting using anti-SpBrf Ab (B) or anti-spLa Ab (C). Note that pRep93X-SpBrf expresses FLAG epitope-tagged SpBrf (28) whose mobility is only slightly slower (lane 1) than endogenous SpBrf (lanes 2–4) in this gel.
DISCUSSION
A conclusion of this work is that SpBrf is innately insufficient to associate with TBP in the absence of DNA. Nonetheless SpBrf can be recruited to a TBP–TATA complex and function in TATA-dependent transcription by Pol III. These findings contribute significantly to our understanding of Pol III transcription in S.pombe as it can account for the need for the TATA sequences that appear upstream of all tRNA and 5S rRNA genes in this yeast (27). These results reflect a concerted diversion of two components of a bipartite transcription control mechanism in S.pombe, the cis-acting element (TATA) and the trans-acting factors that recognize it (TBP), that distinguish it from other model systems. That the upstream TATA elements appear to functionally compensate for the inability of SpBrf to associate with TBP in S.pombe provides a natural species-wide illustration of the compensatory function of cis elements and trans-acting factors in Pol III transcription initiation. In S.cerevisiae, the vast majority of tRNA and 5S rRNA genes do not contain TATA elements, and an important function of Brf1 in this organism, that is lacking in S.pombe, is to bring TBP to the upstream DNA (16). From these observations it seems reasonable to infer that differences in the ability to associate with TBP in the absence of DNA is a species-specific characteristic of Brf1 that primarily reflects a compensatory mechanism to accommodate the Pol III promoters in these yeasts.
It is interesting to note that historically, TATA elements were first found upstream of mRNA genes and considered as hallmarks of Pol II transcription. Recognition that U6 snRNA transcription required an upstream promoter established a role for TATA elements in Pol III transcription and led to the realization that all three eukaryotic Pols require TBP for initiation. Since an upstream TATA was shown to function in rRNA synthesis by Pol I in S.pombe (27), it would since appear that all Pol I and Pol III-transcribed genes are TATA-containing in this yeast, as are most Pol II-transcribed genes. Therefore, it is noteworthy that the only known TATA-less promoters in fission yeast are those encoding (a subset of) mRNAs (40).
The expression level of SpBrf is a critical determinant of TATA-dependent tRNASerUGAM transcription in S.pombe
Overproduction of plasmid-encoded SpBrf led to increased suppression of ade6-704 by the suppressor gene, tRNASerUGAM (Fig. 5). Overproduction of SpBrf could not stimulate suppression from a tRNASerUGAM gene whose upstream TATA element had been altered by site-directed mutagenesis. In addition to providing evidence that our SpBrf construct is active, the results also indicate that the in vivo level of SpBrfp may be a critical determinant of tRNASerUGAM transcription in vivo. We note that in this regard SpBrf may be similar to ScBrf1 (41).
We wish to emphasize that while our results fail to show SpBrf–TBP interaction in vitro, and in dilute extracts, the possibility that these proteins may interact in the absence of DNA in vivo cannot be excluded. However, in this regard we note that the SpBrf overexpression studies indicate that although the intranuclear concentration of SpBrf is increased sufficiently to stimulate the TATA-containing tRNA suppressor gene, the TATA-less tRNA gene remains inactive. These results suggest that SpBrf cannot recruit TBP to the TATA-less promoter even when overexpressed.
Species-specificity and evolutionary divergence of SpBrf
The Brf1 proteins of human, S.cerevisiae and S.pombe are most homologous in their TFIIB-related N-terminal domains, and are less conserved in their C-terminal domains (28). The N-terminal domain of Brf1 is homologous to the TFIIB core region, containing a predicted zinc-ribbon and two TFIIB repeats, while the C-terminal half is Brf1-specific, containing three conserved tracts referred to as Brf-homologous regions 1–3 that are not present in TFIIB (42–46). While both halves of ScBrf1 interact with TBP–DNA, the C-terminal domain, especially Brf-homologous region 2, constitutes principal ScTBP interaction sites and is important for TFIIIB assembly (31,46). The C-terminal one-third of SpBrf is shorter than it is in ScBrf1 and HsBrf1, by 96 and 175 amino acids, respectively (29). The previously noted sequence tracts adjacent to the Brf-homology regions that are lacking in SpBrf relative to ScBrf1 and HsBrf1 (29) are likely to contribute to the species-specific interactions of Brf1 with TBP in the absence of DNA.
It has been noted that TFIIB forms a complex with TBP–DNA even though it only weakly associates with TBP in the absence of DNA (47,48). Multiple variables, including sequences surrounding the TATA sequence as well as species-specific regions of TBP, can determine the nature of the TFIIB–TBP–TATA interaction (49–51). As noted in the Results section above, the Brf-specific region of SpBrf is important for stable association with a TBP–TATA DNA complex since truncation of SpBrf beyond amino acids 383 abrogates the association. More detailed future analyses may reveal specific regions of SpBrf that are important for the recognition of TBP–TATA DNA.
It is noteworthy that although ScBrf1 can interact with SpTBP, data not shown indicate that its overproduction does not lead to increased tRNA expression in our suppression assay. Rather, ScBrf1 produced a dominant negative effect on suppression (not shown), perhaps because it squelches SpTBP. The results suggest that although ScBrf1 can interact with SpTBP, it cannot function with the Pol III machinery in S.pombe and suggests that the species-specific activities of Brf1 extend beyond its ability to interact with TBP.
It should be emphasized that this work focused primarily on one function of Brf1, its potential for TBP recruitment, and not other functions in Pol III transcription (16). In this regard we note that since SpBrf is found associated with TFIIIC in S.pombe, it probably also functions to bridge TBP and TFIIIC, similar to S.cerevisiae, to provide direction to Pol III initiation (26).
Compensatory divergence of Pol III promoters and their trans-acting factors
As alluded to above, the variety of Pol III promoters in S.cerevisiae appears more complex than in S.pombe since the former contains both TATA-less (most) and TATA-containing (i.e. U6 and four tRNA) Pol III-dependent genes. While in S.cerevisiae, both TATA-less and TATA-containing promoters are accommodated by a single TFIIIB entity, the situation is more complex in higher eukaryotes. Most tRNA genes in Drosophila melanogaster are TATA-less (27,52) and TBP-related factor (TRF), rather than TBP, directs transcription of TATA-less tRNA genes by Pol III (53). In this case, TRF stably associates with Brf1 (53). TBP-dependent tRNA transcription has also been observed in D.melanogaster (54,55), but for an unusual, TATA-containing tRNA gene (see discussion in 27). In the human Pol III system Brf1 and Brf2 are differentially used at the TATA-less and TATA-containing promoters represented by tRNA and U6 promoters, respectively (14,15). Thus, different mechanisms of compensation balance the cis-acting elements with the trans-acting factors to accommodate TATA-less Pol III transcription in various eukaryotes. In so far as one can assume that a TATA-dependent transcription system was established before the divergence of archaeal and eucaryal lineages (3,6,56,57), S.pombe may reflect a simplified eucaryal system that, for some reason, retained its TATA elements and therefore did not develop a TATA-less system of Pol III transcription and the need for a TBP-recruitment function of Brf.
Acknowledgments
ACKNOWLEDGEMENTS
We thank E. P. Geiduschek, G. Kassavetis, T. Kokubo and P. A. Weil for kindly providing reagents, A. Sakulich for creating yAS78 and R. V. Intine for critical reading of this manuscript.
REFERENCES
- 1.Paule M.R. and White,R.J. (2000) Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res., 28, 1283–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hernandez N. (1993) TBP, a universal eukaryotic transcription factor? Genes Dev., 7, 1291–1308. [DOI] [PubMed] [Google Scholar]
- 3.Soppa J. (1999) Transcription initiation in Archaea: facts, factors and future aspects. Mol. Microbiol., 31, 1295–1305. [DOI] [PubMed] [Google Scholar]
- 4.Albright S.R. and Tjian,R. (2000) TAFs revisited: more data reveal new twists and confirm old ideas. Gene, 242, 1–13. [DOI] [PubMed] [Google Scholar]
- 5.Pugh B.F. (2000) Control of gene expression through regulation of the TATA-binding protein. Gene, 255, 1–14. [DOI] [PubMed] [Google Scholar]
- 6.Langer D., Hain,J., Thuriaux,P. and Zillig,W. (1995) Transcription in archaea: similarity to that in eucarya. Proc. Natl Acad. Sci. USA, 92, 5768–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagrange T., Kapanidis,A.N., Tang,H., Reinberg,D. and Ebright,R.H. (1998) New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev., 12, 34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kassavetis G., Letts,G. and Geiduschek,E.P. (2001) The RNA polymerase III transcription initiation factor TFIIIB participates in two steps of promoter opening. EMBO J., 20, 2823–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kassavetis G.A., Braun,B.R., Nguyen,L.H. and Geiduschek,E.P. (1990) S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell, 60, 235–245. [DOI] [PubMed] [Google Scholar]
- 10.Reinberg D., Orphanides,G., Ebright,R., Akoulitchev,S., Carcamo,J., Cho,H., Cortes,P., Drapkin,R., Flores,O., Ha,I., Inostroza,J.A., Kim,S., Kim,T.K., Kumar,P., Lagrange,T., LeRoy,G., Lu,H., Ma,D.M., Maldonado,E., Merino,A., Mermelstein,F., Olave,I., Sheldon,M., Shiekhattar,R., Zawel,L. et al. (1998) The RNA polymerase II general transcription factors: past, present and future. Cold Spring Harb. Symp. Quant. Biol., 63, 83–103. [DOI] [PubMed] [Google Scholar]
- 11.Pan G. and Greenblatt,J. (1994) Initiation of transcription by RNA polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. J. Biol. Chem., 269, 30101–30104. [PubMed] [Google Scholar]
- 12.Hawkes N.A. and Roberts,S.G. (1999) The role of human TFIIB in transcription start site selection in vitro and in vivo. J. Biol. Chem., 274, 14337–14343. [DOI] [PubMed] [Google Scholar]
- 13.Cho E.J. and Buratowski,S. (1999) Evidence that transcription factor IIB is required for a post-assembly step in transcription initiation. J. Biol. Chem., 274, 25807–25813. [DOI] [PubMed] [Google Scholar]
- 14.Teichmann M., Wang,Z. and Roeder,R.G. (2000) A stable complex of a novel transcription factor IIB-related factor, human TFIIIB50 and associated proteins mediate selective transcription by RNA polymerase III of genes with upstream promoter elements [In Process Citation]. Proc. Natl Acad. Sci. USA, 97, 14200–14205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schramm L., Pendergrast,P.S., Sun,Y. and Hernandez,N. (2000) Different human TFIIIB activities direct RNA polymerase III transcription from TATA-containing and TATA-less promoters [In Process Citation]. Genes Dev., 14, 2650–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geiduschek E.P. and Kassavetis,G.A. (2001) The RNA polymerase III transcription apparatus. J. Mol. Biol., 310, 1–26. [DOI] [PubMed] [Google Scholar]
- 17.Chedin S., Ferri,M.L., Peyroche,G., Andrau,J.C., Jourdain,S., Lefebvre,O., Werner,M., Carles,C. and Sentenac,A. (1998) The yeast RNA polymerase III transcription machinery: a paradigm for eukaryotic gene activation. Cold Spring Harb. Symp. Quant. Biol., 63, 381–389. [DOI] [PubMed] [Google Scholar]
- 18.Kassavetis G.A., Bardeleben,C., Bartholomew,B., Braun,B.R., Joazeiro,C.A.P., Pisano,M. and Geiduschek,E.P. (1994) In Conaway,R.C. and Conaway,J.W. (eds), Transcription: Mechanisms and Regulation. Raven Press, Ltd, New York, NY, pp. 107–126.
- 19.Willis I.M. (1993) RNA polymerase III genes, factors and transcriptional specificity. Eur. J. Biochem., 212, 1–11. [DOI] [PubMed] [Google Scholar]
- 20.Poon D. and Weil,P.A. (1993) Immunopurification of yeast TATA-binding protein and associated factors. Presence of transcription factor IIIB transcriptional activity. J. Biol. Chem., 268, 15325–15338. [PubMed] [Google Scholar]
- 21.Jaozeiro C.A.P., Kassavetis,G.A. and Geiduschek,E.P. (1996) Alternative outcomes in assembly of promoter complexes: the roles of TBP and a flexible linker in placing TFIIIB on tRNA genes. Genes Dev., 10, 725–739. [DOI] [PubMed] [Google Scholar]
- 22.Dieci G., Percudani,R., Giuliodori,S., Bottarelli,L. and Ottonello,S. (2000) TFIIIC-independent in vitro transcription of yeast tRNA genes. J. Mol. Biol., 299, 601–613. [DOI] [PubMed] [Google Scholar]
- 23.Whitehall S.K., Kassavetis,G.A. and Geiduschek,E.P. (1995) The symmetry of the yeast U6 RNA gene’s TATA box and the orientation of the TATA-binding protein in yeast TFIIIB. Genes Dev., 9, 2974–2985. [DOI] [PubMed] [Google Scholar]
- 24.Jaozeiro C.A.P., Kassavetis,G.A. and Geiduschek,E.P. (1994) Identical components of yeast transcription factor IIIB are required and sufficient for transcription of TATA box-containing and TATA-less genes. Mol. Cell. Biol., 14, 2798–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamada M., Sakulich,A.L., Koduru,S.B. and Maraia,R. (2000) Transcription termination by RNA polymerase III in fission yeast: a genetic and biochemically-tractable model system. J. Biol. Chem., 275, 29076–29081. [DOI] [PubMed] [Google Scholar]
- 26.Huang Y., Hamada,M. and Maraia,R.J. (2000) Isolation and cloning of four subunits of a fission yeast TFIIIC complex that includes an ortholog of the human regulatory protein TFIIICbeta. J. Biol. Chem., 275, 31480–31487. [DOI] [PubMed] [Google Scholar]
- 27.Hamada M., Huang,Y., Lowe,T.M. and Maraia,R.J. (2001) Widespread use of TATA elements in the core promoters for RNA Polymerases III, II and I in fission yeast. Mol. Cell. Biol., 21, 6870–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y. and Maraia,R.J. (2001) Comparison of the RNA polymerase III transcription machinery in S. pombe, S. cerevisiae and humans (Review). Nucleic Acids Res., 29, 2675–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Y., Hamada,M., Patel,J. and Maraia,R.J. (2001) Construction of FLAG and histidine tagging vectors for Schizosaccharomyces pombe. Yeast, 18, 463–468. [DOI] [PubMed] [Google Scholar]
- 30.Rothstein R.J. (1983) One-step gene disruption in yeast. Methods Enzymol., 101, 202–211. [DOI] [PubMed] [Google Scholar]
- 31.Kassavetis G.A., Kumar,A., Ramirez,E. and Geiduschek,E.P. (1998) Functional and structural organization of Brf, the TFIIB-related component of the RNA polymerase III transcription initiation complex. Mol. Cell. Biol., 18, 5587–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willis I.M. (2002) A universal nomenclature for subunits of the RNA polymerase III transcription initiation factor TFIIIB. Genes Dev., 16, 1337–1338. [DOI] [PubMed] [Google Scholar]
- 33.Shibuya T., Tsuneyoshi,S., Azad,A.K., Urushiyama,S., Ohshima,Y. and Tani,T. (1999) Characterization of the ptr6(+) gene in fission yeast: a possible involvement of a transcriptional coactivator TAF in nucleocytoplasmic transport of mRNA. Genetics, 152, 869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitsuzawa H., Seino,H., Yamao,F. and Ishihama,A. (2001) Two WD repeat-containing TATA-binding protein-associated factors in fission yeast that suppress defects in the anaphase-promoting complex. J. Biol. Chem., 276, 17117–17124. [DOI] [PubMed] [Google Scholar]
- 35.Lee T.I. and Young,R.A. (1998) Regulation of gene expression by TBP-associated proteins. Genes Dev., 12, 1398–1408. [DOI] [PubMed] [Google Scholar]
- 36.Fikes J.D., Becker,D.M., Winston,F. and Guarente,L. (1990) Striking conservation of TFIID in Schizosaccharomyces pombe and Saccharomyces cerevisiae. Nature, 346, 291–294. [DOI] [PubMed] [Google Scholar]
- 37.Colbert T., Lee,S., Schimmack,G. and Hahn,S. (1998) Architecture of protein and DNA contacts within the TFIIIB-DNA complex. Mol. Cell. Biol., 18, 1682–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kassavetis G.A., Joazeiro,C.A.P., Pisano,M., Geiduschek,E.P., Colbert,T., Hahn,S. and Blanco,J.A. (1992) The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell, 71, 1055–1064. [DOI] [PubMed] [Google Scholar]
- 39.Maundrell K. (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene, 123, 127–130. [DOI] [PubMed] [Google Scholar]
- 40.Sakurai H. and Ishihama,A. (2001) Transcription organization and mRNA levels of the genes for all 12 subunits of the fission yeast RNA polymerase II. Genes Cells, 6, 25–36. [DOI] [PubMed] [Google Scholar]
- 41.Sethy-Coraci I., Moir,R.D., Lopez-de-Leon,A. and Willis,I.M. (1998) A differential response of wild type and mutant promoters to TFIIIB70 overexpression in vivo and in vitro. Nucleic Acids Res., 26, 2344–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andrau J.C., Sentenac,A. and Werner,M. (1999) Mutagenesis of yeast TFIIIB70 reveals C-terminal residues critical for interaction with TBP and C34. J. Mol. Biol., 288, 511–520. [DOI] [PubMed] [Google Scholar]
- 43.Hahn S. and Roberts,S. (2000) The zinc ribbon domains of the general transcription factors TFIIB and Brf: conserved functional surfaces but different roles in transcription initiation. Genes Dev., 14, 719–730. [PMC free article] [PubMed] [Google Scholar]
- 44.Khoo B., Brophy,B. and Jackson,S.P. (1994) Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev., 8, 2879–2890. [DOI] [PubMed] [Google Scholar]
- 45.Wang Z. and Roeder,R.G. (1995) Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB and high-mobility-group protein 2-related domains. Proc. Natl Acad. Sci. USA, 89, 7026–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kassavetis G., Bardeleben,C., Kumar,A., Ramirez,E. and Geduschek,E.P. (1997) Domains of the Brf component of RNA polymerase III transcription factor IIIB (TFIIIB): functions in assembly ofTFIIIB-DNA complexes and recruitment of RNA polymerase to the promoter. Mol. Cell. Biol., 17, 5299–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barberis A., Muller,C.W., Harrison,S.C. and Ptashne,M. (1993) Delineation of two functional regions of transcription factor TFIIB. Proc. Natl Acad. Sci. USA, 90, 5628–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stargell L.A. and Struhl,K. (1996) A new class of activation-defective TATA-binding protein mutants: evidence for two steps of transcriptional activation in vivo. Mol. Cell. Biol., 16, 4456–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Imbalzano A.N., Zaret,K.S. and Kingston,R.E. (1994) Transcription factor (TF) IIB and TFIIA can independently increase the affinity of the TATA-binding protein for DNA. J. Biol. Chem., 269, 8280–8286. [PubMed] [Google Scholar]
- 50.Wolner B.S. and Gralla,J.D. (2001) TATA-flanking sequences influence the rate and stability of TATA-binding protein and TFIIB binding. J. Biol. Chem., 276, 6260–6266. [DOI] [PubMed] [Google Scholar]
- 51.Zhao X. and Herr,W. (2002) A regulated two-step mechanism of TBP binding to DNA. A solvent-exposed surface of TBP inhibits TATA box recognition. Cell, 108, 615–627. [DOI] [PubMed] [Google Scholar]
- 52.Dingermann T., Burke,D.J., Sharp,S., Schaack,J. and Soll,D. (1982) The 5-flanking sequences of Drosophila tRNAArg genes control their in vitro transcription in a Drosophila cell extract. J. Biol. Chem., 257, 14738–14744. [PubMed] [Google Scholar]
- 53.Takada S., Lis,J.T., Zhou,S. and Tjian,R. (2000) A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell, 101, 459–469. [DOI] [PubMed] [Google Scholar]
- 54.Ouyang C., Martinez,M.J., Young,L.S. and Sprague,K.U. (2000) TATA-binding protein-TATA interaction is a key determinant of differential transcription of silkworm constitutive and silk gland-specific tRNA(Ala) genes. Mol. Cell. Biol., 20, 1329–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trivedi A., Young,L.S., Ouyang,C., Johnson,D.L. and Sprague,K.U. (1999) A TATA element is required for tRNA promoter activity and confers TATA-binding protein responsiveness in Drosophila Schneider-2 cells. J. Biol. Chem., 274, 11369–11375. [DOI] [PubMed] [Google Scholar]
- 56.Hausner W., Frey,G. and Thomm,M. (1991) Control regions of an archaeal gene. A TATA box and an initiator element promote cell-free transcription of the tRNA(Val) gene of Methanococcus vannielii. J. Mol. Biol., 222, 495–508. [DOI] [PubMed] [Google Scholar]
- 57.Woese C.R. (2002) On the evolution of cells. Proc. Natl Acad. Sci. USA, 99, 8742–8747. [DOI] [PMC free article] [PubMed] [Google Scholar]


