Abstract
The hsp70/hsc70-associating protein Hap46 of human origin, also called BAG-1M (Bcl-2-associated athanogene 1), has been characterized previously as a DNA binding protein, which is able to stimulate transcription. By use of in vitro assays we now show that Hap46-mediated transcriptional activation can occur from linearized as well as from supercoiled circular DNA and does not require the presence of a transcription promoter. Accordingly, we observed no preferential binding of Hap46 to overlapping DNA fragments covering the sequence of the cytomegalovirus (CMV) early promoter, thus suggesting non-specific binding. The C-terminal deletion variant Hap46ΔC47, which is unable to associate with hsp70/hsc70 molecular chaperones, produced greatly diminished effects on transcription, indicating a significant involvement of hsp70/hsc70 chaperones but not an absolute requirement. In contrast, deletion of the acidic hexarepeat region, as in variant Hap46Δ12-62, did not disturb transcriptional stimulation. While full-length Hap46 readily formed complexes with a series of structurally unrelated transcription factors, variant Hap46ΔC47 proved incapable of doing so. Together these data suggest that transcriptional stimulation is a major biological activity of Hap46 and point to involvement of hsp70/hsc70 molecular chaperones in transcription in concert with Hap46, thus providing a link between hsp70/hsc70 molecular chaperones and components of the transcription machinery.
INTRODUCTION
The hsp70/hsc70-associating protein Hap46/BAG-1M (Bcl-2-associated athanogene 1) of 46 kDa apparent molecular weight has been described as a multifunctional factor affecting a great variety of cellular functions. These are thought to be mediated by specific target proteins, each of which specifies the respective response and somehow interacts with Hap46/BAG-1M (1–13). Such a multitude of effects and interactions subsequently became much easier to understand when it was realized that Hap46/BAG-1M directly interacts with the ATP binding domains of chaperones of the 70 kDa heat shock protein (hsp70) family (5,14–16). These hsp70 chaperones employ their substrate binding domains to promiscuously complex a great variety of unrelated protein structures, in particular if these contain hydrophobic areas or are partially misfolded (for reviews, see 17–19). In this way, hsp70 and its constitutively expressed counterpart hsc70 are able to form ternary complexes with Hap46/BAG-1M and various other components, as has been demonstrated in vitro for several purified model proteins (5,6). Chaperones of the hsp70/hsc70 type, working as interaction mediators, may thus explain the multitude of Hap46/BAG-1M-association partners. Subse quently, Hap46/BAG-1M was observed to directly interact with DNA (20,21) through its highly basic N-terminal portion (compare with Fig. 1) resulting in transcriptional stimulation in vitro. Hap46/BAG-1M was found to similarly enhance transcription in intact cells if these had been exposed previously to heat shock conditions (20).
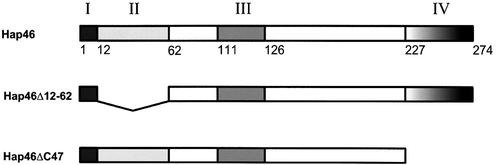
Figure 1.
Domain structure of Hap46. I, DNA binding region; II, acidic hexarepeat region (consensus sequence Ser-Glu-Glu-X-Thr-Arg); III, ubiquitin-like region. IV, hsp70/hsc70 binding region. Deletion variants are shown schematically in relation to the domain structure of Hap46.
Taken together, these observations give rise to the idea that hsp70 and hsc70 might be involved in interactions between Hap46 and the transcriptional apparatus of the cell. In the present study we show that Hap46 can initiate in vitro transcription independent of any promoter probably by guiding transcription complexes to the DNA. Moreover, we provide evidence for the participation of hsp70s in Hap46-stimulated transcription and show that a series of transcription factors requires the presence of hsp70/hsc70 chaperones for associating with Hap46.
MATERIALS AND METHODS
Proteins
Fusion constructs of glutathione S-transferase (GST) with full-length Hap46 and deletion variant Hap46ΔC47 (Fig. 1) were described previously (20). A cDNA fragment encoding Hap46Δ12-62 (compare with Fig. 1) was generated by inverse PCR with primers 5′-CCGGGTCGAGCGGCGCCGGGTTTT-3′ and 5′-GTGACCAGGGAGGAAATGGCGGCA-3′, inserted into BamHI and EcoRI sites of pGEX-2T (Amersham Pharmacia), and verified by sequencing. Proteins were expressed and purified as before (20) and GST portions were removed by thrombin cleavage for in vitro transcription and gel-shift experiments. As judged by SDS–PAGE and Coomassie Blue or silver staining, proteins were at least 95% pure. As experienced by others (13), deletion variant Hap46Δ12-64 is less stable than Hap46ΔC47 and wild type Hap46. Bovine hsc70 was purified from brain (22).
COS-7 cells were cultured at 37°C in RPMI 1640 medium supplemented with 5% fetal calf serum and transiently transfected with plasmid pHEGO, encoding the human estrogen receptor, by use of reagent DOTAP (Roche Molecular Biochemicals), as before (20). Cells were harvested 30 h later and extracts prepared in 50 mM Tris–HCl, pH 8.0, containing 150 mM NaCl, 0.1% Nonidet P-40, and a protease inhibitor cocktail. The estrogen receptor was highly purified by standard co-immunoprecipitation using monoclonal antibody H222 (450 µg/ml; Abbott Laboratories), specific for this receptor, coupled via rabbit anti-mouse IgG (Sigma) onto protein G–Sepharose (Amersham Pharmacia) and extensively washed with the above extract preparation buffer.
Protein interaction experiments
Reticulocyte lysate was used in a coupled transcription– translation system (TNT, Promega) to produce 35S-labeled proteins from T3, T7 or SP6 promoter-containing plasmids pSV2Wrec (mouse glucocorticoid receptor), pHEGO (human estrogen receptor), pCREBΔα (human cAMP response element binding protein CREB), pCbfA1 (human core binding factor subunit A1, also called Runx2) and pTβ-STOP-GAX (human growth arrest homeobox transcription factor). Standard 50 µl transcription–translation reaction products were used for interaction experiments after prior treatment with 1 U calf intestinal phosphatase (Promega) and 50 U DNase I (Roche Molecular Biochemicals) for 30 min at 37°C. Interactions with GST–Hap46 fusion proteins attached to GSH–Sepharose (Amersham Pharmacia) were assayed in 20 mM HEPES buffer, pH 7.4, 100 mM KCl, 10 mM MgCl2, 0.1% Nonidet P-40, 1 mM DTT, as before (5). Matrices were washed extensively with 0.1% Nonidet P-40 in HEPES buffered saline, pH 7.4. Bound proteins were eluted with SDS sample buffer and analyzed by SDS–PAGE and autoradiography or immunoblotting, as before (5). Hsc70 was detected by monoclonal antibody N27F3-4 (Stressgen), diluted 1:1000, in combination with a peroxidase-conjugated second antibody and enhanced chemiluminescence reagent (Amersham).
The specific immuno-matrix (see above) carrying the purified estrogen receptor (10 µl slurry per assay) was pre-blocked with 20% calf serum in the above HEPES buffer and incubated overnight at 8°C with GST–Hap46 fusion protein (5 µg) and hsc70 included or not. After extensive washing, bound proteins were eluted with SDS buffer, as above. Analysis was by SDS–PAGE and immunoblotting with GST-specific antibody Z-5 (sc-459; Santa Cruz), diluted 1:1000, for detection of GST–Hap46. After stripping the membrane, hsc70 was visualized by antibody N27F3-4, as above.
Electrophoretic mobility-shift assays
For plasmid mobility-shift assays we routinely used plasmid pcDNA3/CAT (Invitrogen) in 20 mM HEPES, pH 7.4, 100 mM KCl, 10 mM MgCl2, 1 mM DTT and 4% glycerol. Hap46 protein (5 µg) was incubated in a total volume of 25 µl for 30 min at 25°C with DNA (200 ng) and hsc70 (10 µg), as indicated. Analysis was in standard 0.8% agarose gels and DNA bands were visualized by ethidium bromide staining.
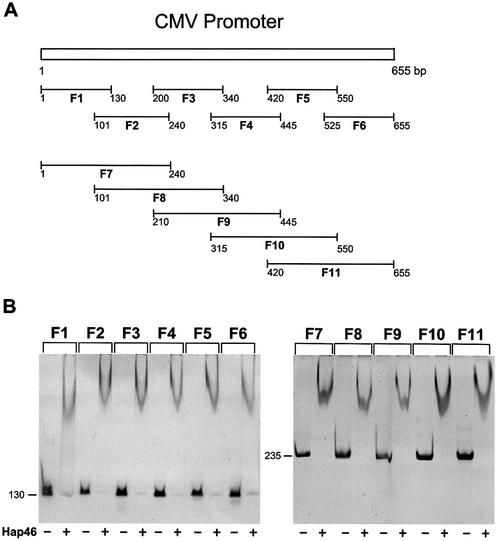
For the analysis of Hap46 binding to the cytomegalovirus (CMV) early promoter we produced a set of 11 overlapping PCR fragments, called F1 to F11 (compare with Fig. 4A). The following primers were used: 5′-CGATGTACGGGCCAGATATAC-3′ and 5′-CGGGCCATTTACCGTAAGTTA-3′ (F1), 5′-GCGTTACATAACTTACGGTAAATGGCC-3′ and 5′-CCGTAAATAGTCCACCCATTGACG-3′ (F2), 5′-CCATTGACGTCAATGGGTGGA-3′ and 5′-CTGGGCATAATGCCAGGCGGG-3′ (F3), 5′-ATGGCCCGCCTGGCATTA TGC-3′ and 5′-CTATCCACGCCCATTGATGTA-3′ (F4), 5′-GCAGTACATCAATGGCGCTGG-3′ and 5′-GGAGTTGTTACGACATTTTGG-3′ (F5), 5′-CTTTCCAAAATGTCGTAACAA-3′ and 5′-AATTTCGATAAGCCAGTAAGC-3′ (F6) using plasmid pcDNA3/CAT as template. To generate fragments F7 to F11 the same primers were used according to the scheme depicted in Figure 4A. Fragments were purified using the Wizard PCR purification system (Promega). Standard mobility-shift assays were carried out in 6% polyacrylamide gels as before (20) routinely using 100 ng DNA and 3 µg Hap46 protein. DNA bands were visualized by staining with ethidium bromide. In this assay system we find that Hap46·DNA complexes migrate as curved bands (20).
Figure 4.
Binding of Hap46 to fragments of the CMV promoter. (A) The CMV promoter sequence of 655 bp was divided by PCR into six portions of ∼130 bp each and five portions of ∼235 bp, as detailed in Materials and Methods. (B) The above DNA fragments were incubated with Hap46 and analyzed by gel electrophoresis, as described in Materials and Methods. Ethidium stained bands are shown in dark.
In vitro transcription
Transcription assays with HeLa nuclear extract (Promega; 50 µg protein/25 µl assay) and ATP, CTP, UTP (final concentrations 400 µM each), as well as GTP (16 µM) and 5 µCi [32P]GTP (3000 Ci/mmol, ICN) were carried out in 20 mM HEPES buffer, pH 7.9, 100 mM KCl, 3 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT and 20% glycerol for 60 min at 30°C. Full-length Hap46 or Hap46 deletion variants (5–10 µg/25 µl assay) were added, as indicated. Control experiments showed that this amount of Hap46 per assay was close to maximum in terms of transcriptional stimulation (data not shown). Incorporated radioactivity was quantitated by scintillation counting and RNA was analyzed on 4–6% denaturing polyacrylamide gels. Routinely, a CMV promoter-containing 1485 bp fragment of pcDNA3/CAT, generated by digestion with NruI and BamHI, served as template. In some experiments (compare with Fig. 2A), plasmid pcDNA3/CAT was used without linearization. Promoter-less prokaryotic fragments of 517 and 396 bp length were obtained by HinfI digestion of plasmid pGEM-4Z (Promega). Promoter-less templates of eukaryotic origin were the 2.3 kb glucocorticoid receptor cDNA, produced by BglII/XbaI digestion of plasmid pSV2Wrec (23), and the 0.9 kb Hap46 cDNA, generated by HindIII/EcoRI from plasmid pcDNA3–Hap46 (20).
Figure 2.
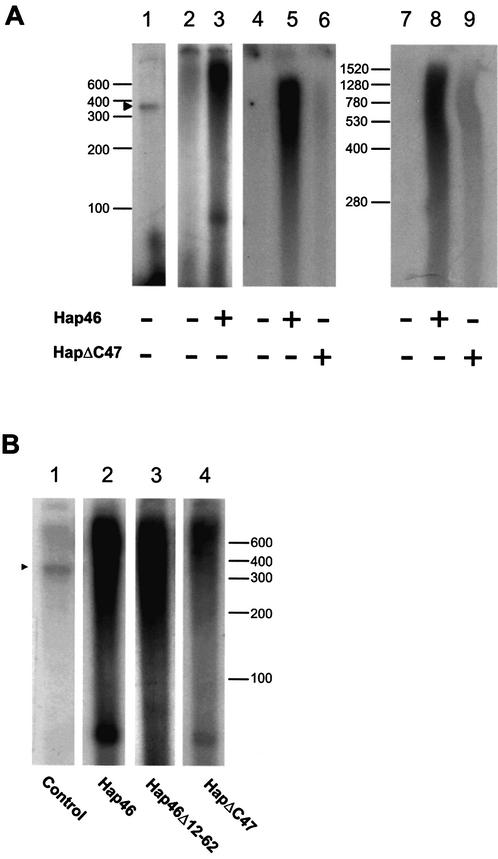
Hap46 stimulates transcription from circular and linearized DNA. (A) In the experiments of lanes 2 and 3, plasmid pcDNA3/CAT was used for transcription assays with HeLa nuclear extracts, as described in Materials and Methods, either in the absence or presence of full-length Hap46, as indicated. In the experiments of lanes 4–9, prokaryotic DNA of 517 bp (lanes 4–6) and eukaryotic DNA of 2.3 kb (lanes 7–9) were similarly used in the absence (lanes 4 and 7) or presence of full-length Hap46 (lanes 5 and 8) or Hap46ΔC47 (lanes 6 and 9), as described in Materials and Methods. The same result was obtained with other prokaryotic or eukaryotic DNAs of 396 and 0.9 kb, respectively (not shown). As control, a 1485 bp template containing the CMV promoter was used with no Hap46 added (lane 1). (B) The same linear CMV promoter-containing template was used in transcription assays either in the absence (lane 1) or presence of full-length Hap46 (lane 2), Hap46Δ12-62 (lane 3) and Hap46ΔC47 (lane 4). Labeled RNA was analyzed by gel electrophoresis and autoradiography. (A) Lanes 7–9 were analyzed on 4% polyacrylamide gels, all others were run on 6% gels. Positions of RNA size markers are shown along margins. Arrowheads mark the position of the CMV promoter-specific transcript of 363 nt.
RESULTS
Hap46 stimulates in vitro transcription from linear and circular DNA
Previous in vitro transcription experiments using HeLa cell nuclear extracts and linearized DNA fragments resulted in significant enhancement of RNA synthesis upon addition of Hap46 (20). We now show that Hap46 can produce similar effects if the template is provided in the form of supercoiled circular DNA (Fig. 2A, lane 3 versus 2). Direct counting of incorporated radioactivity revealed a 6.5-fold stimulation by Hap46.
Concomitant with this transcriptional activation, Hap46 associates with supercoiled circular DNA, as evidenced by retardation of electrophoretic mobility in agarose gels (Fig. 3, lane 2 versus 1). In this experiment the plasmid DNA was present as a mixture of monomers and dimers (lower and upper bands in lane 1, respectively) and both forms became upshifted (Fig. 3, lane 2). Binding of Hap46 to plasmid DNA was obtained with several constructs independent of whether they contained eukaryotic sequences (data not shown), suggesting that Hap46 binding occurs independent of nucleotide sequences. Upon including purified hsc70 in the incubation of Hap46 with supercoiled circular DNA, we observed a supershift in electrophoretic mobility of both DNA bands (Fig. 3, lane 3), suggesting that Hap46 can simultaneously associate with DNA and hsc70.
Figure 3.
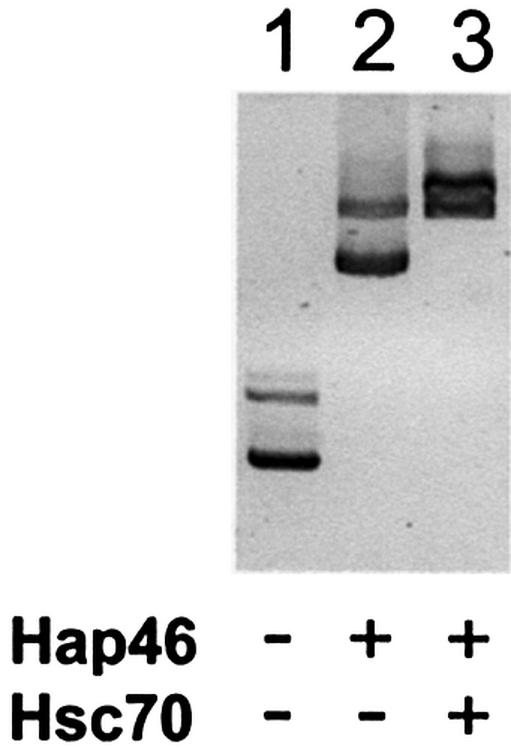
Hap46 binds to circular DNA. Plasmid pcDNA3/CAT was incubated with full-length Hap46 and hsc70, as indicated, and submitted to electrophoretic moblity-shift assays in 0.8% agarose gels, as described in Materials and Methods. Ethidium stained bands are shown here in dark.
To further characterize the stimulatory effect of Hap46 on in vitro transcription, we carried out a series of experiments with several linear DNA fragments, which are devoid of promoter elements. Figure 2A shows examples of this using prokaryotic (lanes 4–6) and eukaryotic sequences (lanes 7–9). Upon addition of Hap46, we observed significant transcriptional activity (Fig. 2A, lanes 5 and 8) relative to controls without Hap46 (lanes 4 and 7). In the absence of Hap46, standard transcription experiments employing a DNA fragment containing the CMV promoter yielded the expected distinct transcripts (Fig. 2A, lane 1), whereas all RNA patterns obtained in the presence of Hap46 were quite diffuse.
Maximum transcriptional stimulation requires the region of Hap46 involved in binding to hsp70/hsc70 chaperones
Figure 2A also shows in vitro transcription experiments with variant Hap46ΔC47 from which the 47 C-terminal amino acid residues were deleted (compare with Fig. 1). In the presence of this variant protein we observed greatly decreased stimulation of RNA synthesis, compared with full-length Hap46 (Fig. 2A, lanes 6 and 9 versus 5 and 8). As hsp70s are highly abundant in extracts from HeLa cell nuclei (5,24), which were used here for in vitro transcription and since deletion variant Hap46ΔC47 is unable to associate with these molecular chaperones (see below and Fig. 5A), proteins of the hsp70 family are suggested to be significantly involved in Hap46-mediated transcriptional stimulation.
Figure 5.
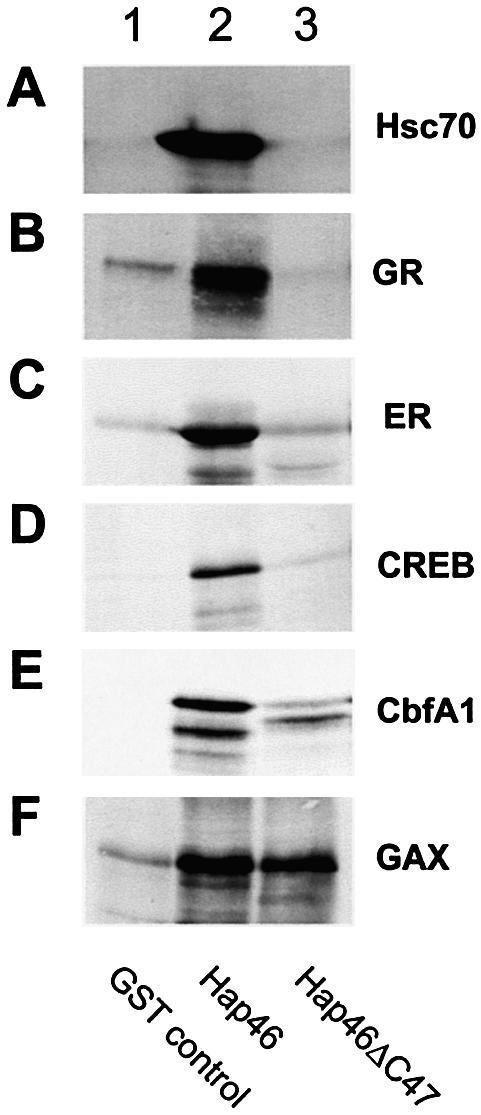
Interaction of Hap46 with transcription factors. Fusion proteins of GST with full-length Hap46 (lanes 2) or Hap46ΔC47 (lanes 3) were attached to GSH–Sepharose and used as affinity matrices for in vitro expressed radiolabeled transcription factors, as described in Materials and Methods. For controls, plain GSH–Sepharose was used (lanes 1). Rabbit reticulocyte lysate was used as such in the experiment of (A). Eluates were analyzed by SDS–PAGE followed either by autoradiographic detection of radiolabeled bands or, in the case of hsc70 (A), by specific immunoblotting.
The above result was confirmed in separate assays with a CMV promoter-containing construct and full-length Hap46 as well as deletion variants Hap46Δ12-62 and Hap46ΔC47 (compare with Fig. 1). Variant Hap46Δ12-62, from which the acidic hexapeptide repeat region had been deleted, was able to activate transcription and its stimulatory effect was slightly stronger than that of full-length Hap46 (Fig. 2B, lane 3 versus 2). Hap46ΔC47 also produced transcriptional stimulation, although at 70–85% lower levels as compared with full-length Hap46 (Fig. 2B, lane 4 versus 2). Nevertheless, such activation occurring independent of hsp70/hsc70 chaperones may still be of biological relevance. It remains uncertain, however, whether a previously reported increase in the expression of a reporter gene upon transfecting a Hap46ΔC47 construct into cells (20) relates to the activation observed in the present in vitro studies. Aside from difficulties in quantitatively controlling such transfections, measurements of chloramphenicol acetyltransferase activity in transfected cells were shown to be unreliable in subsequent analyses. Following publication of the previous paper (20), we obtained only minimal transcriptional stimulation upon transfection of various cell types with a Hap46ΔC47–cDNA construct in seven of seven independent experiments (data not shown). This was indeed the reason why we subsequently modified the procedure to using in vitro transcription assays in which we observed stimulation by Hap46ΔC47 in all of the 10 experiments at the low levels described above. Moreover, full-length Hap46 and Hap46ΔC47 yielded the respective increases in transcriptional activity whether the DNA template was in linear or supercoiled circular form and whether it contained promoter elements.
Hap46 binds unspecifically to the CMV early promoter
The above experiments demonstrate that DNA fragments devoid of promoter sequences are able to support Hap46-stimulated transcription (compare with Fig. 2A). Further more, we observed binding of Hap46 to DNA fragments independent of origin and whether they contained eukaryotic promoter elements (20). While these data do not support the notion of sequence or promoter-specific DNA binding a recent publication suggested specificity for the CMV promoter, which was supposed to contain a specific binding site for Hap46/BAG-1M since the protein bound only to one small fragment of the CMV promoter (21). As this observation was not substantiated by published data and is in clear contrast to all our results, we generated from the CMV promoter two sets of consecutive PCR fragments of ∼130 and 235 bp overlapping as schematically depicted in Figure 4A. These were then individually tested for interaction with Hap46 and all fragments were found to be shifted in electrophoretic mobility assays (Fig. 4B). As no significant differences were detectable amongst fragments of similar lengths, we conclude that Hap46 does not exert sequence-specific binding to the CMV promoter.
Hap46 interacts with several transcription factors through hsp70/hsc70 chaperones
Even though the above in vitro experiments (compared with Fig. 2A and B) provide no evidence for specificity in transcriptional stimulation by Hap46, previous experiments using intact cells clearly showed increased expression of several genes upon overexpression of Hap46 or the somewhat larger isoform Hap50 (20,25). Thus, we surveyed a series of transcription factors for interactions with Hap46, which—in principle—might either require the presence of a hsp70/hsc70 chaperone or could occur independent of hsp70s. For protein interaction experiments we used full-length Hap46 and Hap46ΔC47 fused to GST and attached to GSH–Sepharose. As expected from previous data (16,21,26), Hap46ΔC47 was unable to retain hsc70 while full-length Hap46 readily interacted (Fig. 5A, lane 3 versus 2). Hsc70 is an abundant protein in extracts of all cell types, including rabbit reticulocyte lysate.
Transcription factors were in vitro expressed in the reticulocyte lysate system and assayed with the above full-length Hap46 and Hap46ΔC47 affinity matrices. To eliminate indirect interactions through simultaneous binding of Hap46 and the various transcription factors to DNA, we pretreated all protein samples with DNase I. The data in Figure 5 show that the glucocorticoid receptor (Fig. 5B), estrogen receptor (Fig. 5C), the cAMP response element binding protein CREB (Fig. 5D) and core binding factor CbfA1, also called Runx2, (Fig. 5E) were retained by full-length Hap46 (lanes 2), but not by variant Hap46ΔC47 (lanes 3). In control experiments using only GST attached to GSH–Sepharose, we obtained background signals (Fig. 5, lanes 1). In another control the Hap46ΔN10 deletion variant (20) showed perfect interaction with the above transcription factors in the presence of hsc70 (data not shown). However, when we investigated the homeodomain-containing transcription factor Gax we found that full-length Hap46 and Hap46ΔC47-specific matrices retained this protein to a comparable extent (Fig. 5F).
In the reverse approach we used the highly purified human estrogen receptor attached to a specific immuno-matrix and examined for retention of full-length Hap46. As shown in Figure 6, the amount of Hap46 paralleled that of hsc70 present in the incubations, again demonstrating involvement of this molecular chaperone.
Figure 6.
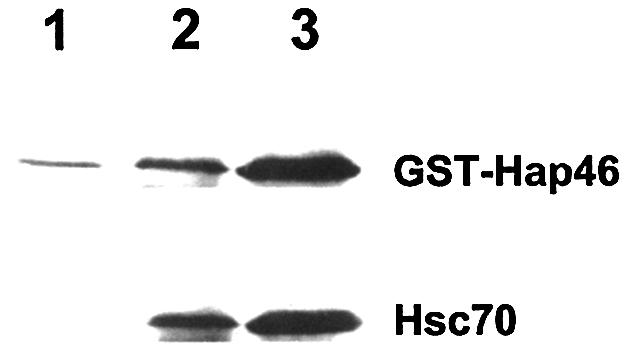
Interaction of Hap46 and hsc70 with the estrogen receptor. The estrogen receptor attached to a specific immunomatrix was incubated with the GST–Hap46 fusion protein, as described in Materials and Methods. The experiment of lane 2 contained minimal amounts of endogenous hsc70 co-purified with the receptor, while further hsc70 (5 µg) was added in the experiment of lane 3. As control (lane 1), the plain immunomatrix was used without estrogen receptor but with hsc70 (5 µg) added.
DISCUSSION
The data presented here further strengthen the view that Hap46/BAG-1M is a multifunctional protein, which is able to interact with DNA and with hsp70/hsc70 chaperones. To achieve these distinct biochemical activities, Hap46/BAG-1M employs separate regions of the polypeptide chain: N-terminal sequences are required for binding to DNA (20,21) while the C-terminal portion is involved in interactions with hsp70s (compare with Fig. 5A) (16,21,26). In this way, Hap46 can act as a molecular link between such divergent components as DNA and hsp70/hsc70 molecular chaperones. Simultaneous interactions then result in multi-component complexes of the type DNA·Hap46·hsc70 as evidenced by the DNA super-shift shown in Figure 3. Such complexes in themselves are well suited for further interactions through hsp70/hsc70 components. The fact that we observe rather large shifts in agarose gels (compare with Fig. 3) implies that multiple copies of Hap46 can simultaneously associate with DNA, possibly in a cooperative manner. Evidence for such multimeric binding was indeed obtained when increasing amounts of Hap46 were incubated with short DNA fragments, resulting in a progressive upshift of complexes (20). Interestingly, Hap46/BAG-1M similarly interacts with sequences of eukaryotic and prokaryotic origin and appears to act as non-specific DNA binding protein (20). This became very evident when we analyzed its binding to a series of overlapping DNA fragments covering the CMV early promoter. Since all these fragments were upshifted upon incubation with Hap46/BAG-1M (compare with Fig. 4B) sequence-specific interaction with the CMV promoter, as suggested by others (21), is certainly not prevalent. However, we cannot exclude limited preferences for some as yet unidentified clusters of nucleotides.
When we examined Hap46 for the ability to associate with a series of transcription factors, which were selected to belong to structurally different protein families, we discerned two distinct types of interactions. Steroid hormone receptors, CREB and the core binding factor CbfA1/Runx2 were found to require the presence of hsc70 for forming complexes (compare with Fig. 5B–E), i.e. the potential for interaction was lost upon deletion of the C-terminal portion as in variant Hap46ΔC47 which renders Hap46 unable to associate with hsp70/hsc70 chaperones (compare with Fig. 5A). These data clearly show that association of Hap46 with these transcription factors—and possibly many more—only occurs in the presence of hsp70 or hsc70 and when Hap46 itself is able to interact with these molecular chaperones. In contrast, the homeodomain-containing transcription factor Gax, which is involved in the regulation of cell viability (27), was found to associate with Hap46 independent of hsp70/hsc70 chaperones (compare with Fig. 5F). This suggests that some specific transcription factors may be able to directly interact with Hap46/BAG-1M, a finding that requires further investigation.
The above mentioned transcription factor CbfA1/Runx2 is involved in mammalian osteogenic development from mesenchymal progenitor cells and in osteoblast function (28,29) and was found to be mutated in patients affected by the skeletal disorder dysplasia cleidocranialis (for a review, see 30). Our observation of CbfA1/Runx2 interacting with Hap46/BAG-1M may thus suggest some involvement of the latter in osteogenesis and bone metabolism; however, future studies will have to establish whether Hap46/BAG-1M participates in bone homeostasis. Similarly, Hap46/BAG-1M may affect some of the manifold biological actions of transcription factor CREB (for a review, see 31), which transmits the effects of intracellular cAMP-levels on gene expression.
We hypothesize that the major biological significance of Hap46/BAG-1M is the stimulatory effect on transcriptional activity. Such transcriptional stimulation occurs in intact cells under specific conditions, most notably heat stress, when Hap46/BAG-1M becomes translocated to the cell nucleus (20) together with hsp70 (24). In fact, heat-induced nuclear accumulation of hsp70 and Hap46/BAG-1M may well contribute to the long-known transcriptional activation of a set of genes under such stress conditions. Transcriptional stimulation is more obvious under much less complex circumstances, which are effective in in vitro assays as used here. In the presence of Hap46/BAG-1M the majority of RNA products are much longer than the run-off transcript of 363 nt when a template is used that bears the CMV promoter, and there is no preference for transcripts of defined lengths (compare with Fig. 2). This might either be due to end-initiation followed by premature termination or alternatively Hap46/BAG-1M-stimulated transcription may start at any site. We greatly favor the latter possibility because Hap46/BAG-1M itself is a strong transcriptional activator, it does not require a template, which contains promoter sequences (compare with Fig. 2A), and binds to any type of DNA fragment provided (compare with Figs 3 and 4) (20). Involvement of hsp70/hsc70 chaperones is evident from the fact that the C-terminal deletion variant Hap46ΔC47 produced greatly diminished transcriptional stimulation (compare with Fig. 2A and B). While hsp70/hsc70 chaperones are certainly the major direct binding partners of Hap46/BAG-1M which employ the C-terminal domain, the existence of some other proteins of minor cellular abundance involved in nuclear events and interacting with the very same domain cannot—in principle—be excluded; however, such proteins still need to be identified. We thus infer that hsp70/hsc70 chaperones function to coordinate Hap46 into complexes with transcription factors and possibly other components of the transcriptional machinery, resulting in activation. The observation, however, that Hap46ΔC47 is still able to stimulate transcription to a limited extent independent of hsp70/hsc70 chaperones (compare with Fig. 2A and B) suggests that there must exist ways for Hap46/BAG-1M to function without associating with hsp70 or hsc70 possibly involving direct interactions with constituents of the transcriptional machinery. It is important to note that hsp70s not only function to associate with incorrectly folded polypeptide chains (see Introduction) but are required for the assembly of multimeric protein structures. This is most obvious in the case of steroid hormone receptor complexes (for reviews, see 32,33) but is also evident for heteromeric complexes of several protein kinases involved in cell proliferation (for a review, see 34) and for the assembly of other multiprotein complexes (35,36). It thus appears likely that a major function of Hap46 in concert with hsp70s is the chaperoning of components of the transcriptional apparatus to DNA, perhaps similar to the recently discussed action of the SWI/SNF complex on nucleosomal states (37). Several molecules of Hap46 bound to DNA may then recruit and mobilize—mainly through hsp70/hsc70 chaperones—various components involved in transcription. This probably includes the formation of bridging complexes with enhancer elements remotely located on the DNA as, for example, steroid hormone receptors which bind to specific response elements on the DNA and interact with Hap46 via hsp70s (compare with Fig. 5B and C).
Several alternative forms of Hap/BAG-1 have been described (38–41) and are expressed in a stage- and site-specific fashion during mouse development (42), most notably a protein isoform of apparent molecular weight 50 kDa, called Hap50 or BAG-1L. It carries an N-terminal extension of 71 amino acid residues and includes the cluster of basic amino acids which was shown previously to be involved in DNA binding (20). Upon overexpression in cells, this large isoform exerts transcriptional effects similar to those described for Hap46/BAG-1M (20), but independent of heat stress (25) and, in fact, it is nuclearly localized independent of heat stress (25,39,41,43,44). It is this Hap50/BAG-1L isoform which has been reported to functionally cooperate with the vitamin D receptor (45,46) as well as the androgen receptor (43) whereby such cooperation requires the C-terminal hsp70/hsc70- interaction domain. In contrast to Hap46/BAG-1M and Hap50/BAG-1L, shorter isoforms do not contain the N-terminally located positively charged amino acid clusters and consequently are inert towards DNA (21). They function as regulators of hsp70/hsc70 chaperones in protein folding reactions (47,48). Interestingly, Hap46/BAG-1M takes an intermediary position in that it may act both as transcriptional activator based on the present studies and previous reports (20,21) and as hsp70/hsc70 chaperone cofactor in cytosolic protein folding (5,14,49,50), depending on physiological conditions.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr M. Zeiner for valuable discussions during the course of these studies, M. Siebert for technical assistance, B. Segnitz for purifying hsc70 from bovine brain, and Ph. Henrich for help with the figures. We are very grateful to Drs A. Alonso, P. Angel, P. Chambon, M. Danielsen and A. Kuhn for providing plasmids. This investigation was supported by the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Zeiner M. and Gehring,U. (1995) A protein that interacts with members of the nuclear hormone receptor family: identification and cDNA cloning. Proc. Natl Acad. Sci. USA, 92, 11465–11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takayama S., Sato,T., Krajewski,S., Kochel,K., Irie,S., Millan,J.A. and Reed,J.C. (1995) Cloning and functional analysis of BAG-1: a novel Bcl-2-binding protein with anti-cell death activity. Cell, 80, 279–284. [DOI] [PubMed] [Google Scholar]
- 3.Bardelli A., Longati,P., Albero,D., Goruppi,S., Schneider,C., Ponzetto,C. and Comoglio,P.M. (1996) HGF receptor associates with the anti-apoptotic protein Bag-1 and prevents cell death. EMBO J., 15, 6205–6212. [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H.-G., Takayama,S., Rapp,U.R. and Reed,J.C. (1996) Bcl-2 interacting protein, BAG-1, binds to and activates the kinase Raf-1. Proc. Natl Acad. Sci. USA, 93, 7063–7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeiner M., Gebauer,M. and Gehring,U. (1997) Mammalian protein RAP46: an interaction partner and modulator of 70 kDa heat shock proteins. EMBO J., 16, 5483–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bimston D., Song,J., Winchester,D., Takayama,S., Reed,J.C. and Morimoto,R.I. (1998) BAG-1, a negative regulator of Hsp70 chaperone activity, uncouples nucleotide hydrolysis from substrate release. EMBO J., 17, 6871–6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Froesch B.A., Takayama,S. and Reed,J.C. (1998) BAG-1L protein enhances androgen receptor function. J. Biol. Chem., 273, 11660–11666. [DOI] [PubMed] [Google Scholar]
- 8.Kullmann M., Schneikert,J., Moll,J., Heck,S., Zeiner,M., Gehring,U. and Cato,A.C.B. (1998) RAP46 is a negative regulator of glucocorticoid receptor action and hormone induced apoptosis. J. Biol. Chem., 273, 14620–14625. [DOI] [PubMed] [Google Scholar]
- 9.Liu R., Takayama,S., Zheng,Y., Froesch,B., Chen,G., Zhang,X., Reed,J.C. and Zhang,X.-K. (1998) Interaction of BAG-1 with retinoic acid receptor and its inhibition of retinoic acid-induced apoptosis in cancer cells. J. Biol. Chem., 273, 16985–16992. [DOI] [PubMed] [Google Scholar]
- 10.Matsuzawa S., Takayama,S., Froesch,B.A., Zapata,J.M. and Reed,J.C. (1999) p53-inducible human homologue of Drosophila seven in absentia (Siah) inhibits cell growth: suppression by BAG-1. EMBO J., 17, 2736–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naishiro Y., Adachi,M., Okuda,H., Yawata,A., Mitaka,T., Takayama,S., Reed,J.C., Hinoda,Y. and Imai,K. (1999) BAG-1 accelerates cell motility of human gastric cancer cells. Oncogene, 18, 3244–3251. [DOI] [PubMed] [Google Scholar]
- 12.Schneikert J., Hübner,S., Martin,E. and Cato,A.C.B. (1999) A nuclear action of the eukaryotic cochaperone RAP46 in downregulation of glucocorticoid receptor activity. J. Cell Biol., 146, 929–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lüders J., Demand,J. and Höhfeld,J. (2000) The ubiquitin-related BAG-1 provides a link between the molecular chaperones hsc70/hsp70 and the proteasome. J. Biol. Chem., 275, 4613–4617. [DOI] [PubMed] [Google Scholar]
- 14.Gebauer M., Zeiner,M. and Gehring,U. (1997) Proteins interacting with the molecular chaperone hsp70/hsc70: physical associations and effects on refolding activity. FEBS Lett., 417, 109–113. [DOI] [PubMed] [Google Scholar]
- 15.Höhfeld J. and Jentsch,S. (1997) GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J., 16, 6209–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takayama S, Bimston,D.N., Matsuzawa,S., Freeman,B.C., Aime-Sempe,C., Xie,Z., Morimoto,R.I. and Reed,J.C. (1997) BAG-1 modulates the chaperone activity of hsp70/hsc70. EMBO J., 16, 4887–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellis R.J. and Hartl,F.U. (1999) Principles of protein folding in the cellular environment. Curr. Opin. Struct. Biol., 9, 102–110. [DOI] [PubMed] [Google Scholar]
- 18.Fink A.L. (1999) Chaperone-mediated protein folding. Physiol. Rev., 79, 425–449. [DOI] [PubMed] [Google Scholar]
- 19.Mayer M.P., Brehmer,D., Gassler,C.S. and Bukau,B. (2001) Hsp70 chaperone machines. Adv. Protein Chem., 59, 1–44. [DOI] [PubMed] [Google Scholar]
- 20.Zeiner M., Niyaz,Y. and Gehring,U. (1999) The hsp70-associating protein Hap46 binds to DNA and stimulates transcription. Proc. Natl Acad. Sci. USA, 96, 10194–10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi N., Sasaki,R., Takahashi,J., Takayama,S., Reed,J.C. and Andoh,T. (2001) BAG-1M, an isoform of of Bcl-2-interacting protein BAG-1, enhances gene expression driven by CMV promoter. Biochem. Biophys. Res. Commun., 286, 807–814. [DOI] [PubMed] [Google Scholar]
- 22.Chappell T.G., Konforti,B.B., Schmid,S.L. and Rothman,J.E. (1987) The ATPase core of a clathrin uncoating protein. J. Biol. Chem., 262, 746–751. [PubMed] [Google Scholar]
- 23.Danielsen M., Northrop,J.P. and Ringold,G.M. (1986) The mouse glucocorticoid receptor: mapping of functional domains by cloning, sequencing and expression of wild-type and mutant receptor proteins. EMBO J., 5, 2513–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Velazquez J.M. and Lindquist,S. (1984) Hsp70: nuclear concentration during environmental stress and cytoplasmatic storage during recovery. Cell, 36, 655–662. [DOI] [PubMed] [Google Scholar]
- 25.Niyaz Y., Zeiner,M. and Gehring,U. (2001) Transcriptional activation by the human Hsp70-associating protein Hap50. J. Cell Sci., 114, 1839–1845. [DOI] [PubMed] [Google Scholar]
- 26.Takayama S., Xie,Z. and Reed,J.C. (1999) An evolutionarily conserved family of hsp70/hsc70 molecular chaperone regulators. J. Biol. Chem., 274, 781–786. [DOI] [PubMed] [Google Scholar]
- 27.Perlman H., Luo,Z., Krasinski,K., Le Roux,A., Mahfoudi,A., Smith,R.C., Branellec,D. and Walsh,K. (1999) Adenovirus-mediated delivery of Gax transcription factor to rat carotid arteries inhibits smooth muscle proliferation and induces apoptosis. Gene Ther., 6, 758–763. [DOI] [PubMed] [Google Scholar]
- 28.Ducy P., Zhang,R., Geoffroy,V. Ridall,A.L. and Karsenty,G. (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell, 89, 747–754. [DOI] [PubMed] [Google Scholar]
- 29.Karsenty G., Ducy,P., Starbuck,M., Priemel,M., Shen,J., Geoffroy,V. and Amling,M. (1999) Cbfa1 as a regulator of osteoblast differentiation and function. Bone, 25, 107–108. [DOI] [PubMed] [Google Scholar]
- 30.Rodan G.A. and Harada,S. (1997) The missing bone. Cell, 89, 677–680. [DOI] [PubMed] [Google Scholar]
- 31.Quinn P.G. (2002) Mechanisms of basal and kinase-inducible transcription activation by CREB. Prog. Nucleic Acid Res. Mol. Biol., 72, 296–305. [DOI] [PubMed] [Google Scholar]
- 32.Gehring U. (1998) Steroid hormone receptors and heat shock proteins. In Litwack,G. (ed.), Vitamins and Hormones. Academic Press, San Diego, Vol. 54, pp. 167–205. [DOI] [PubMed]
- 33.Cheung J. and Smith,D.F. (2000) Molecular chaperone interactions with steroid receptors: an update. Mol. Endocrinol., 14, 939–946. [DOI] [PubMed] [Google Scholar]
- 34.Helmbrecht K., Zeise,E. and Rensing,L. (2000) Chaperones in cell cycle regulation and mitogenic signal transduction: a review. Cell Prolif., 33, 341–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marshall O.J. and Harley,V.R. (2001) Identification of an interaction between SOX9 and HSP70. FEBS Lett., 496, 75–80. [DOI] [PubMed] [Google Scholar]
- 36.Maheswaran S., Englert,C., Zheng,G., Lee,S.B., Wong,J., Harkin,D.P., Bean,J., Ezzell,R., Garvin,A.J., McCluskey,R.T., DeCaprio,J.A. and Haber,D.A. (1998) Inhibition of cellular proliferation by the Wilms tumor suppressor WT1 requires association with the inducible chaperone Hsp70. Genes Dev., 12, 1108–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narlikar G.J., Phelan,M.L. and Kingston,R.E. (2001) Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol. Cell, 8, 1219–1230. [DOI] [PubMed] [Google Scholar]
- 38.Packham G., Brimmel,M. and Cleveland,J.L. (1997) Mammalian cells express two differently localized Bag-1 isoforms generated by alternative translation initiation. Biochem. J., 328, 807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takayama S., Krajewski,S., Krajewska,M., Kitada,S., Zapata,J.M., Kochel,K., Knee,D., Scudiero,D., Tudor,G., Miller,G.J., Miyashita,T., Yamada,M. and Reed,J.C. (1998) Expression and location of Hsp70/Hsc-binding anti-apoptotic protein BAG-1 and its variants in normal tissues and tumor cell lines. Cancer Res., 58, 3116–3131. [PubMed] [Google Scholar]
- 40.Yang X., Chernenko,G., Hao,Y., Ding,Z., Pater,M.M., Pater,A. and Tang,S.-C. (1998) Human BAG-1/RAP46 protein is generated as four isoforms by alternative translation initiation and overexpressed in cancer cells. Oncogene, 17, 981–989. [DOI] [PubMed] [Google Scholar]
- 41.Brimmell M., Burns,J.S., Munson,P., McDonald,L., O’Hare,M.J., Lakhani,S.R. and Packham,G. (1999) High level expression of differentially localized BAG-1 isoforms in some oestrogen receptor-positive human breast cancers. Br. J. Cancer, 81, 1042–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crocoll A., Blum,M. and Cato,A.C. (2000) Isoform-specific expression of BAG-1 in mouse development. Mech. Dev., 91, 355–359. [DOI] [PubMed] [Google Scholar]
- 43.Knee D.A., Froesch,B.A., Nuber,U., Takayama,S. and Reed,J.C. (2001) Structure-function analysis of Bag1 proteins. Effects on androgen receptor transcriptional activity. J. Biol. Chem., 276, 12718–12724. [DOI] [PubMed] [Google Scholar]
- 44.Hague A., Packham,G., Huntley,S., Shefford,K. and Evenson,J.W. (2002) Deregulated Bag-1 protein expression in human oral squamous cell carcinomas and lymph node metastases. J. Pathol., 197, 60–71. [DOI] [PubMed] [Google Scholar]
- 45.Guzey M., Takayama,S. and Reed,J.C. (2000) BAG-1L enhances trans-activation function of the vitamin D receptor. J. Biol. Chem., 275, 40749–40756. [DOI] [PubMed] [Google Scholar]
- 46.Witcher M., Yang,X., Pater,A. and Tang,S.-C. (2001) BAG-1 p50 isoform interacts with the vitamin D receptor and its cellular overexpression inhibits the vitamin D pathway. Exp. Cell Res., 265, 167–173. [DOI] [PubMed] [Google Scholar]
- 47.Lüders J., Demand,J., Papp,O. and Höhfeld,J. (2000) Distinct isoforms of the cofactor BAG-1 differentially affect hsc70 chaperone function. J. Biol. Chem., 275, 14817–14823. [DOI] [PubMed] [Google Scholar]
- 48.Nollen E.A.A., Brunsting,J.F., Song,J., Kampinga,H.H. and Morimoto,R.I. (2000) Bag-1 functions in vivo as a negative regulator of hsp70 chaperone activity. Mol. Cell. Biol., 20, 1083–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gebauer M., Zeiner,M. and Gehring,U. (1998) Interference between proteins Hap46 and Hop/p60, which bind to different domains of the molecular chaperone hsp70/hsc70. Mol. Cell. Biol., 18, 6238–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gässler C.S., Wiederkehr,T., Brehmer,D., Bukau,B. and Mayer,M.P. (2001) Bag-1M accelerates nucleotide release for human Hsc70 and Hsp70 and can act concentration-dependent as positive and negative cofactor. J. Biol. Chem., 276, 32538–32544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.