Abstract
Ensemble and single-molecule fluorescence measurements of 2′-(4-hydroxyphenyl)-5-[5-(4-methylpiperazine-1-yl) benzimidazo-2-yl]-benzimidazole (H-258)– calf thymus (CT) DNA complexes at various [H-258]/[DNA bp] ratios were performed to elucidate the binding of H-258 with DNA. Upon binding to double-stranded CT DNA (CT ds DNA) at a [H-258]/[DNA bp] ratio of 0.05 the relative fluorescence quantum yield, Φf, of H-258 increases from 0.02 to 0.58. The fluorescence decay can be fitted almost by a mono-exponential model with a lifetime of ∼3.6 ns. This indicates that H-258 binds almost quantitatively in the minor groove of DNA at low [H-258]/[DNA bp] ratios. With increasing [H-258]/[DNA bp] ratios, e.g. 0.15 and 0.20, the fluorescence quantum yield of H-258 decreases to 0.28 and 0.19, respectively. Fitting of the fluorescence decays measured for higher [H-258]/[DNA bp] ratios reveals the presence of additional shorter fluorescence lifetime components in the range of 0.5–2.0 ns. Our results suggest that H-258 partially intercalates in G:C sequences at higher [H-258]/[DNA bp] ratios reflected by a lifetime component of 1.5–2 ns. In addition, stacking or adsorption of H-258 molecules on DNA occurs at higher [H-258]/[DNA bp] ratios. These molecules exhibit a short fluorescence lifetime of ∼500 ps and are more exposed to the aqueous environment. Fluorescence transients of the intensity and lifetime of single H-258 CT ds DNA demonstrate that weakly (unspecific) bound H-258 molecules exhibit a shorter fluorescence lifetime and a strongly reduced photostability.
INTRODUCTION
Hoechst 33258 [H-258: 2′-(4-hydroxyphenyl)-5-[5-(4-methylpiperazine-1-yl) benzimidazo-2-yl]-bezimidazole] is the parent compound of the benzimidazole family (Fig. 1). H-258 and its ethoxy analog Hoechst 33342 (H-342) are commercially available DNA-binding compounds widely used as fluorescent cytological stains for DNA (1). Because of its double-strand DNA-binding affinity (2,3), H-258 can affect transcription and translation (4), and can block topo isomerase I (5) as well as helicase activity (6). Accordingly, this molecule can be very useful for the design of new drugs for gene regulation.
Figure 1.
Molecular structure of the bis-benzimidazole derivatives H-258 and H-342 at pH 7.0.
It has been shown that H-258 and its derivatives offer protection against radiation-induced strand breaks (7,8). Strand breakage is among the most important lesions leading to radiation-induced cell death because it disrupts the continuity and integrity of the double helix (9). Therefore, the role of H-258 and its derivatives as potential radio-protectors has been the subject of continuous investigation (10). Radiation-induced strand breakage is caused by direct ionization of DNA molecules and by reactions between DNA and radicals generated in the surrounding media by the absorption of energy—among which the hydroxyl radical (·OH) being the most effective. The hydroxyl radical-induced DNA radicals are the precursors of DNA-strand breaks (9,11). Pulse radiolysis studies showed that the protection of H-258 has two components: scavenging of OH-radicals by the DNA-bound ligand and additional protection due to efficient quenching of OH-radical-induced DNA-radicals (12–14).
Although H-258 possesses several pKa values, it becomes a mono-cationic species at pH 7.0 (Fig. 1) (15–18). In A:T-rich sequences of the minor groove, the electrostatic potential is highly negative (19). X-ray crystallographic and NMR studies using oligonucleotides with various sequences showed that at pH 7.0, H-258 preferentially binds in the minor groove at A:T-rich sequences in B-DNA at the edges of four to five contiguous base pairs (bp) (20–31) with nanomolar affinity (32). Quantitative DNAse I footprinting studies revealed that among various A:T sequences, AATT represents the most favored binding site of H-258 (33). It has long been suggested (34–36) that at least two different binding modes for H-258 with DNA–A:T-specific strong binding with an intense increase in fluorescence, and at a high [H-258]/[DNA bp] ratio (≥0.1) an unspecific weaker binding, have to be considered. Absorption and fluorescence titration studies using various polynucleotides and natural DNA [e.g. double-stranded calf thymus DNA (CT ds DNA)–42% G:C] suggested that unspecific binding between H-258 and DNA might occur due to the following interactions (1): (i) charge mediated binding could take place between the mono-cationic H-258 and charged backbone phosphate. This will occur with a stoichiometric ratio of [H-258]/[DNA] (here [DNA] is expressed in terms of phosphates) = 1:1; (ii) a free H-258 molecule can bind to another H-258 molecule already bound to the DNA in the minor groove or to a backbone phosphate; and (iii) electric linear dichroism (ELD) studies demonstrated that H-258 can bind unspecifically as an intercalator to G:C-rich sequences in the major groove (37).
The proposition of unspecific binding of H-258 with DNA has further been supported by titration rotational viscometric studies using CT ds DNA (4). However, these types of unspecific binding were not observed at isolated sites, e.g. at AATT or AAATTT. Here no aggregation, ligand superposition or ‘tilting’ would be possible that might be the consequence of some kind of co-operativity on an alternating polymer like poly[d(A:T)] (38). Although the affinity constant of H-258 for poly[d(A:T)2] has been reported to be 60 times stronger than for poly[d(G:C)2] (39), the binding of H-258 to A:T-rich sequences is not as straightforward as it was originally concluded. A continuous variation method for the H-258–poly[d(A:T)2] complex revealed distinct stoichiometries for one, two, three, four and six H-258 molecules per five A:T base pairs (1).
In this communication, we report about absorption, emission and time-resolved fluorescence measurements to elucidate the different binding modes of H-258 with CT ds DNA. Fluorescence quantum yield measurements at various [H-258]/[DNA bp] ratios show that the extent of unspecific binding increases with the [H-258]/[DNA bp] ratio. Furthermore, time-resolved fluorescence data indicate that non-specifically bound H-258 molecules are more exposed to the surrounding water than specifically bound molecules. Fluorescence studies on single H-258–CT ds DNA complexes adsorbed on amino-modified cover slides suggest that non-specifically bound H-258 molecules are more exposed to the aqueous surround accompanied by a shorter fluorescence lifetime and lower photostability. Monitoring of the fluorescence lifetime of single H-258–CT ds DNA complexes with millisecond time resolution reveals that the fraction of molecules which shows a shorter fluorescence lifetime exhibits a lower photostability, i.e. the fluorescence lifetime increases with time due to selective photobleaching of non-specifically bound H-258 molecules.
MATERIALS AND METHODS
Materials
H-258 (Sigma), CT ds DNA (Fluka, batch no. 89370), Na2HPO4·2H2O and NaH2PO4·H2O (Fluka) were used as received. According to the manufacturer the CT ds DNA was already sonicated to avoid strand–strand interactions. The purity of H-258 was confirmed by HPLC using a RP18 column and a gradient of 0–75% acetonitrile in 0.1 M aqueous triethylammonium acetate (pH 7.0). DNA solutions (typically 2 × 10–4 M in terms of base pairs, i.e. 4 × 10–4 M in terms of nucleotides) were prepared in 5 × 10–2 M phosphate buffer (pH 7.0). All solutions were prepared and diluted using Millipore (Milli Q) water. The different [H-258]/[DNA bp] ratios of 0.05, 0.15 and 0.20 were prepared by addition of H-258 to DNA solutions yielding final H-258 concentrations of 10, 30 or 40 µM, respectively.
Determination of the concentrations of H-258 in the bound state
It was observed that H-258 is quite effective in causing precipitation of double-stranded DNA and during this precipitation, a considerable amount of H-258 is also lost (40). To avoid precipitation, doubly concentrated H-258 solutions were allowed to mix dropwise in the doubly concentrated DNA solutions under constant stirring. H-258 concentrations in the bound state were determined following the method of Comings (40) using a Cary 500 UV-VIS-NIR spectrophotometer (Varian, Darmstadt, Germany) with 1 cm standard quartz cuvettes. In accordance with the literature (17), the UV-Vis data show that no free H-258 was present in the solution.
Fluorescence intensity and lifetime measurements
Steady state fluorescence and excitation spectra were measured using a LS100 spectrometer (PTI, Wedel, Germany). Fluorescence quantum yields were determined using quinine sulfate in 0.1 N H2SO4 (fluorescence quantum yield, Φf,standard = 0.70) (41) as standard and the following relation (equation 1):
Φf,sample = Φf,standard × (Fsample / Fstandard) × (Astandard at 360nm / Aunknown at 360 nm)1
Here Φf,sample represents the unknown fluorescence quantum yield of the fluorophore, and F is the integrated fluorescence intensity (in this case between 380 and 600 nm after excitation at 360 nm). A is the absorbance at 360 nm in 1 cm cuvettes. The fluorescence lifetimes were measured using the time correlated single photon counting (TCSPC) technique with an instrument from IBH (model 500MC; Glasgow, UK). The solutions were filtered through a membrane filter with 0.45 µm pore width (Millex-H SLHA 025 BS; Millipore, France) before lifetime measurements. This procedure removes dust particles and air bubbles which otherwise interfere with the measurements. For efficient excitation a frequency doubled TiSa laser (Mira 900; Coherent, Santa Clara, CA) providing ∼300 fs (full-width half-maximum) laser pulses at 360 nm, was coupled into the IBH spectrometer. The repetition rate of the laser was reduced to 3.8 MHz using a pulse picker (Pulse Select; APE, Berlin, Germany). A reference signal of the titanium sapphire laser was used as start signal for the time-to-amplitude (TAC) converter. To exclude polarization effects, fluorescence was observed under the magic angle (54.7°). Typically, 5000 photon counts were collected in the maximum channel using 2056 channels. The decay parameters were determined by least-squares deconvolution, and their quality was judged by the reduced χ2 values and the randomness of the weighted residuals. In the case that a mono-exponential model was not adequate to describe the measured decay, a multi-exponential model was used to fit the data (equation 2):
I(t) = I(0)Σaiτi2
Here ai are the pre-exponential factors describing the fraction of the excited species (Σai =1) and τi denote their lifetimes.
Fluorescence imaging of single DNA–H-258 complexes
For imaging, H-258–CT ds DNA complexes were adsorbed on standard cover slides with a thickness of 170 µm and dried under nitrogen. Prior to use the cover slides were washed with 3.5% HF followed by treatment with 1% (3-aminopropyl) trimethoxysilane (Fluka) for 10 min and a final washing step with water. A treatment with ∼10–10 M solutions of the H-258–DNA complexes yielded an areal density of <1 DNA molecule/µm2. A titanium sapphire laser (360 nm, 3.8 MHz, pulse width ∼300 fs, average excitation power 1 kW/cm2) and an avalanche photodiode were used in a sample scanning confocal microscope to image and monitor the fluorescence from single complexes immobilized on glass cover slips. The collimated laser beam was sent through a quarter-wave retarder oriented to ensure circularly polarized light, and then directed into the back port of an inverted microscope. Within the microscope the laser beam was reflected by a dichroic mirror (385 DLRP; Omega Optics, Brattleborro, VT) and focused to a tight spot by a water-immersion objective (63×/1.2 W; Zeiss, Germany). Fluorescence collected by the same objective was spatially and spectrally filtered using a 100 µm pinhole and a bandpass filter (500DF40; Omega Optics), and focused onto the active area of a single-photon counting avalanche photodiode (SPAD). The sample x and y positions were controlled using a closed-loop piezo stage with a parallel PC interface for imaging the sample and positioning individual fluorescing spots in the focused excitation beam. An automated search routine operates as follows. The sample is scanned across a 20 µm line (the x-axis). In the event that a signal above a threshold count rate of 5 kHz is observed, the stage returns to the line position of maximum intensity and records the fluorescence continuously until the molecule is photobleached. The y-axis position is stepped by 300 nm and the process is repeated. The signal of the SPAD was fed into a TCSPC PC interface board (SPC-630; Becker & Hickl, Berlin, Germany) and the laser sync signal (3.8 MHz) was connected to the board’s sync input. Time-resolved data were acquired using the FIFO-mode in which the microscopic arrival time and the macroscopic arrival time are registered for each fluorescence photon detected. For efficient determination of fluorescence lifetimes of single H-258–DNA complexes, a maximum likelihood estimator (MLE) algorithm was applied (42).
RESULTS AND DISCUSSION
Ensemble absorption and emission characteristics of free and bound H-258
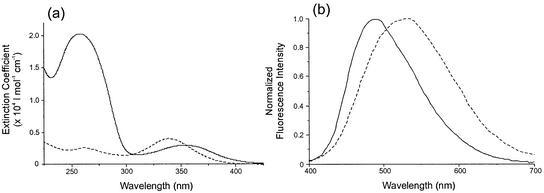
Table 1 gives the spectroscopic properties (λabs, λem, ε, Φf,rel, τi, ai) of free H-258 and its DNA complexes at [H-258]/[DNA bp] ratios of 0.05, 0.15 and 0.20 in 50 mM phosphate buffer, pH 7.0. To test for aggregation or dimer formation, a 6 × 10–6 M aqueous solution of H-258 (50 mM phosphate buffer, pH 7.0) was heated to 95°C. No significant changes in the absorption spectra of free H-258 that would point to an aggregation were observed (14,36,43). However, downward-curved deviations from linearity were observed in the 250– 380 nm range of the absorption spectrum at bis-benzimidazole concentrations above 40 µM in aqueous solution (17). This is in favor of a dimer contribution or weak association with otherwise unchanged properties as the monomer. Therefore, H-258 concentrations of ≤40 µM were used in our fluorescence experiments. Upon binding of H-258 to CT ds DNA the absorption maximum shows a bathochromic shift from 340 to 355 nm. In accordance with previous measurements, the absorption spectrum of H-258 is significantly broadened when bound to DNA (Fig. 2a) (17,35,40,44,45). The bathochromic shift in the absorption spectra upon binding indicates a more hydrophobic environment as a result of reduced solvation (14,46). At higher [H-258]/[DNA bp] ratios the red-shift in the absorption spectrum is less pronounced (Table 1). With increasing [H-258]/[DNA bp] ratio, the absorption spectrum shifts towards shorter wavelengths from 355 to 347 nm. The observed changes in the position and shape of the absorption spectra of H-258–DNA complexes have been attributed to additional binding of H-258 molecules at G:C sites with increasing [H-258]/[DNA bp] ratio (35,47).
Table 1. Spectroscopic characteristics [absorption maximum (λabs), emission maximum (λem), extinction coefficient (ε), relative fluorescence quantum yield (Φf,rel), fluorescence lifetimes (τi) with corresponding amplitudes (ai), of free H-258 and H-258 (10 µM) bound to CT ds DNA ([H-258]/[DNA bp] ratio of 0.05)] in phosphate buffer, pH 7.0.
| λabs (nm) | λem (nm) | ε (l mol–1 cm–1) | Φf,rel | τ1 (ns)/a1 | τ2 (ns)/a2 | τ3 (ns)/a3 | |
|---|---|---|---|---|---|---|---|
| H-258 | 340 | 525 | 4.2 × 104 | 0.02 | 0.34/0.78 | 3.50/0.22 | –/– |
| [H-258]/[DNA bp] = 0.05 | 355 | 485 | 3.0 × 104 | 0.58 | 0.63/0.02 | 3.61/0.98 | –/– |
| [H-258]/[DNA bp] = 0.15 | 349 | 485 | 3.0 × 104 | 0.28 | 0.45/0.08 | 1.73/0.26 | 4.05/0.66 |
| [H-258]/[DNA bp] = 0.20 | 347 | 485 | 3.0 × 104 | 0.19 | 0.52/0.11 | 1.82/0.29 | 4.05/0.60 |
The fluorescence decays were measured at the emission maxima, respectively.
Figure 2.
Relative absorption spectra (a) and normalized emission spectra (b) of 10 µM solutions of H-258 bound to DNA (—) ([H-258]/[DNA bp] = 0.05) and free H-258 (- - -) in 50 mM phosphate buffer, pH 7.0.
In contrast to the absorption spectra, the fluorescence spectra of H-258 exhibit a significant blue-shift upon binding to DNA in 50 mM phosphate buffer, pH 7.0 (Fig. 2b). The emission maximum of free H-258 (λem = 525 nm) is shifted ∼40 nm towards the blue when bound to CT ds DNA (λem = 485 nm). The extent of this hypsochromic shift does not depend on the [H-258]/[DNA bp] ratio. It has to be pointed out that the absolute values of published fluorescence emission maxima of the free and DNA-bound H-258 differ slightly while the relative shifts in absorption and emission maxima are similar (17,35,36,40,44,45,48). However, any deviation in the emission maximum may well be caused by the use of different dye concentrations, buffers and ionic strength. The solvatochromic shifts observed in absorption (red-shift) and fluorescence (blue-shift) upon DNA binding, i.e. the decrease of the Stokes shift with decreasing polarity of the surrounding medium, is frequently the result of changes in the dipole moment upon excitation or decay to the ground state. Similar shifts have been found for other fluorescent DNA dyes (17,49). The magnitude of this shift is dependent not only on the size of the change in dipole moment but also on the location of the dye in the DNA structure. For example, properly groove-bound dyes are more protected from the polar (aqueous) environment than non-specifically bound or adsorbed dye molecules. This is directly reflected in the decrease of the red-shift of the absorption maximum with increasing [H-258]/[DNA bp] ratio (Table 1).
Fluorescence quantum yield of free and bound H-258
The fluorescence quantum yield of H-258 increases significantly upon binding to DNA (Table 1). At pH 7.0, H-258 exhibits a fluorescence quantum yield, Φf, of 0.02, which increases to Φf = 0.58 for H-258 bound to CT ds DNA at a [H-258]/[DNA bp] ratio of 0.05 in 50 mM phosphate buffer, pH 7.0. At higher [H-258]/[DNA bp] ratios of 0.15 and 0.20, Φf decreases from 0.58 to 0.28, and 0.19, respectively. The measured decrease in Φf of H-258 at [H-258]/[DNA bp] ratios ≥0.1 confirms that at higher [H-258]/[DNA bp] ratios unspecific binding of H-258 to CT ds DNA occurs (1,36, 47,49). The observed shifts in the absorption spectra and changes in fluorescence quantum yield of H-258 with increasing [H-258]/[DNA bp] ratios evidence that unspecific bound H-258 molecules are less tightly bound to the DNA. Consequently, the absorption maximum shifts towards the value of free H-258 accompanied by a reduced fluorescence quantum yield. Since free H-258 exhibits a very small fluorescence quantum yield, the emission spectrum remains almost unaffected. These findings imply that unspecific bound H-258 molecules are more exposed to the surrounding water than specifically bound molecules.
Time-resolved fluorescence characteristics
Cosa et al. reported that the fluorescence decay of free H-258 ([H-258] ∼ 10–4 M) in Tris-EDTA buffer (pH 7.4) recorded with a streak camera can be described by a biexponential model with lifetimes of τ1 = 3.33 ns (17%) and τ2 = 0.18 ns (83%) (49). Using TCSPC, the fluorescence decay of 1 × 10–6 M solutions of free H-258 in Tris–EDTA buffer pH 7.0 were fitted successfully by three components likewise with a predominant contribution (>80%) from a short lifetime component of ∼100 ps (50). As shown in Table 1, in our study, the fluorescence decays measured from 10–5 M solutions of free H-258 in phosphate buffer (pH 7.0) were fitted best using a biexponential model with a short lifetime of 0.34 ns (78%), and a longer lifetime of 3.5 ns (22%). Independent of the spectroscopic method used, the fluorescence decay of H-258 in aqueous buffer pH ∼7.0 is dominated by a short component (∼80%) of 100–340 ps. Measurements at higher concentrations (10–4 M) yield similar results. The major competing decay channels of the first excited state of H-258 in aqueous solution are internal conversion and fluorescence as major and minor processes, respectively. As known from transient absorption spectroscopic measurements, intersystem crossing into the triplet states plays no role (17). The properties of the excited state of bis-benzimide dyes in aqueous solvents are strongly controlled by the proton concentration. The ground-state of H-258 has pKa values at 3.5, 5.5, 8.5 and 9.8 (16). They refer to protonation at the benzimidazole units and deprotonation at the phenolic hydroxyl and the end-standing nitrogen of the piperazoline unit (15–17). For example, the fluorescence quantum yield of H-258 increases dramatically upon protonation of the benzimidazole units at pH ∼4.0 (17). The reason for the low quantum yield in neutral water, as compared with pH 4, is not fully understood (16,17).
Kalninsh et al. (16) have shown that one benzimidazole substituted with both the phenol and piperazine units does not exhibit low fluorescence in neutral aqueous solutions as occurs with the bis-benzimidazole dyes. This indicates that two benzimidazole units, one with the piperazine as a substituent and the other one with the phenol or its alkylated form, are required for efficient fluorescence quenching in neutral water. At low pH values the piperazine unit is protonated with both nitrogen atoms being involved in the hydrogen bonding. Hence, a certain degree of charge transfer from the protonated piperazine-bis-benzimide subunit to the phenol-bis-benzimide is feasible resulting in a stronger planar bond between the two piperazines. Previous calculations show that this charge transfer does occur and the interconnecting bond between the two benzimidazole units becomes planar and stronger in the excited state (16). NMR studies support the idea that H-258 binds to the minor groove of double-stranded DNA resulting in a planar conformation involving the phenol and the two benzimidazole units (29). At higher pH values the piperazine unit is mostly non-protonated. Consequently charge transfer in the first excited singlet state is prevented and internal conversion via intramolecular rotation represents the dominant deactivation pathway. That is, the fluorescence decay is dominated by a short lifetime component (Table 1).
Upon binding to CT ds DNA the fluorescence quantum yield and the contribution of the long lifetime component increases (Table 1). Our data show that at a [H-258]/[DNA bp] ratio of 0.05, the fluorescence decay is dominated (>90%) by a component with a lifetime >3.5 ns. When bound in the minor groove, the contours of the groove will constrain the bond torsion angles between the rings of the H-258 molecule (18). Theoretical calculations have revealed that H-258 molecule is less twisted when bound to the DNA (47) than in solution (18). It seems reasonable therefore to assume that the longer lifetime component of ∼3.5 ns arises from the conformer with both benzimidazole units in a planar conformation. This is the predominant conformation in double-stranded DNA due to tight binding in the minor groove and its contribution decreases in single-stranded DNA where the binding is more flexible (49). In aqueous H-258 solutions at pH ∼7, the planar conformation enabling charge transfer is less pronounced and most of the excited state energy is released via rotation along the benzimide axis. The fluorescence decay exhibits only ∼20% of the long lifetime component (∼3.5 ns). The high amplitude of >90% for the long lifetime component indicates that at low [H-258]/[DNA bp] ratios, H-258 binds almost quantitatively in the minor groove of DNA. As also shown in Table 1, with increasing [H-258]/[DNA bp] ratio the fluorescence decays have to be described using at least three exponents with a short lifetime τ1 of ∼500 ps, a medium lifetime τ2 of ∼1.8 ns and a long lifetime τ3 of ∼4 ns.
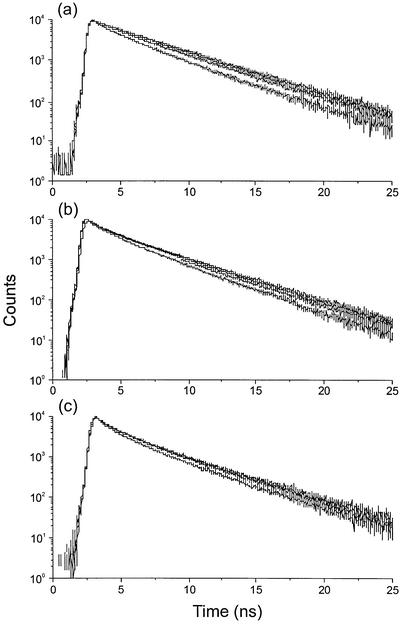
Figure 3 shows the decay profiles measured from DNA-bound H-258 at different [H-258]/[DNA bp] ratios and different detection wavelengths. Here clear distinctive features among the [H-258]/[DNA bp] ratios as well as among the different wavelengths for a particular [H-258]/[DNA bp] ratio can be observed. The data in Table 2 clearly demonstrate that at low [H-258]/[DNA bp] ratios, the influence of the shorter decay component with a lifetime <1 ns is almost negligible especially at longer detection wavelengths, i.e. at a [H-258]/[DNA bp] ratio of 0.05, predominantly sequence-specific minor groove binding occurs and the contribution from unspecific binding is very low. Time-resolved fluorescence spectra show that the fluorescence emission of H-258 in aqueous buffer at pH 7.4 shifts towards the red with time (49). Together with our results obtained from fluorescence lifetime measurements at different detection wavelengths, the data demonstrate that the planar conformer with the higher dipole moment (resulting from charge transfer interactions) exhibits a long fluorescence lifetime of 3.5–4 ns.
Figure 3.
Fluorescence decays recorded from H-258 solutions in 50 mM phosphate buffer (pH 7.0) bound to DNA at different [H-258]/[DNA bp] ratios of 0.05 (10 µM) (a), 0.15 (30 µM) (b) and 0.20 (40 µM) (c) and different detection wavelengths of 440, 460, 480 and 500 nm. With increasing detection wavelength the fluorescence lifetime increases. In the case of a [H-258]/[DNA bp] ratio of 0.05 the fluorescence decay recorded at 500 nm can be fitted almost mono-exponentially.
Table 2. Fluorescence lifetimes of H-258 bound to CT ds DNA at different [H-258]/[DNA bp] ratios of 0.05, 0.15 and 0.20 corresponding to H-258 concentrations of 10, 30 and 40 µM, respectively, in phosphate buffer (50 mM), pH 7.0.
| Detection wavelength (nm) | τ1 (ns)/a1 | τ2 (ns)/a2 | τ3 (ns)/a3 | χ2 |
|---|---|---|---|---|
| [H-258]/[DNA bp] = 0.05 | ||||
| 440 | 0.67/0.08 | 3.07/0.91 | –/– | 1.220 |
| 460 | 0.71/0.04 | 3.49/0.96 | –/– | 1.261 |
| 480 | 0.63/0.02 | 3.61/0.98 | –/– | 1.189 |
| 500 | 0.49/0.02 | 3.78/0.98 | –/– | 1.143 |
| 520 | 0.58/0.01 | 3.84/0.99 | –/– | 1.023 |
| 540 | 0.58/0.01 | 3.84/0.99 | –/– | 1.116 |
| [H-258]/[DNA bp] = 0.15 | ||||
| 440 | 0.40/0.10 | 1.58/0.37 | 3.73/0.53 | 1.214 |
| 460 | 0.46/0.08 | 1.75/0.31 | 4.03/0.61 | 1.280 |
| 480 | 0.45/0.08 | 1.73/0.26 | 4.05/0.66 | 1.275 |
| 500 | 0.48/0.09 | 1.85/0.25 | 4.13/0.66 | 1.286 |
| 520 | 0.55/0.10 | 1.98/0.24 | 4.19/0.66 | 1.274 |
| 540 | 0.51/0.11 | 2.14/0.29 | 4.33/0.60 | 1.112 |
| [H-258]/[DNA bp] = 0.20 | ||||
| 440 | 0.41/0.12 | 1.56/0.33 | 3.81/0.55 | 1.201 |
| 460 | 0.54/0.12 | 1.91/0.30 | 4.09/0.58 | 1.465 |
| 480 | 0.47/0.10 | 1.74/0.27 | 4.02/0.63 | 1.348 |
| 500 | 0.49/0.11 | 1.82/0.26 | 4.09/0.63 | 1.414 |
| 520 | 0.50/0.12 | 1.79/0.23 | 4.06/0.64 | 1.222 |
| 540 | 0.52/0.10 | 1.77/0.20 | 4.15/0.70 | 1.188 |
Excitation was performed at 360 nm.
At higher [H-258]/[DNA bp] ratios the amplitude of the short lifetime component shows a considerable increase independent of the detection wavelength (Table 2). In addition, a lifetime component of 1.5–2.0 ns with an amplitude of 20–40% appears at [H-258]/[DNA bp] ratios of 0.15 and 0.20. Simultaneously, the quality of the fits decreases as reflected by the higher χ2 values. This points towards a lifetime distribution rather than a three-exponential fit, i.e. the occurrence of multiple binding modes of H-258 at higher [H-258]/[DNA bp] ratios. It is intuitively easy to understand that weaker binding sites are filled-up at higher [H-258]/[DNA bp] ratios. These weaker bound H-258 molecules are more exposed to the aqueous environment. Since, the fluorescence quantum yield of this state is very low, the influence on the emission maximum is negligible. On the other hand, the amplitudes of the shorter fluorescence lifetime components are larger at shorter detection wavelengths (compare amplitudes at 440 nm with amplitudes at 500 nm in Table 2). This gives further evidence that the blue-shifted species with short fluorescence lifetimes represent weakly bound H-258 molecules.
Confocal fluorescence lifetime imaging microscopy (CFLIM)
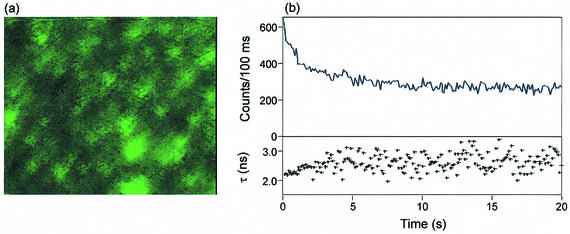
To elucidate whether different binding modes of H-258 molecules induce different photostabilities, we performed single-molecule measurements using CFLIM (51). Due to increased photobleaching rates, fluorescence detection of single molecules using UV excitation is very demanding (52). Efforts to detect single freely diffusing H-258 in phosphate buffer failed. Therefore, we restricted our study to detect individual CT ds DNA molecules in 50 mM phosphate buffer (pH 7.0) bound with H-258 at [H-258]/[DNA bp] ratios >0.05. Assuming a molecular weight of CT ds DNA of ∼5 × 106 Da, approximately 3000 H-258 molecules can bind to a single CT ds DNA. Hence, the fluorescence of individual DNA molecules should be detectable, even at high photobleaching rates. To induce binding of the negatively charged DNA on the glass surface, cover slides were silanized with 3-aminopropyltrimethylsilane. As a result of surface modification with positive groups, individual H-258 labeled CT ds DNA molecules bind at the surface and can be detected (Fig. 4a). To record single-molecule transients, individual fluorescing spots were positioned in the focused excitation beam and the fluorescence was recorded continuously for 60 s. As expected for a multichromophoric system, the count rate decreases continuously with time due to subsequent photobleaching of individual H-258 molecules. The fluorescence lifetimes calculated in 100 ms steps using a mono-exponential MLE algorithm show that the fluorescence lifetime increases simultaneously with continuous photobleaching (Fig. 4b). This indicates that those H-258 molecules that bind at higher [H-258]/[DNA bp] ratios not only exhibit a shorter fluorescence lifetime but also a reduced photostability. It can be concluded that those H-258 molecules that are more exposed to the surrounding water, i.e. the weakly bound fraction, bleach first. The observed dependence of the photostability on the binding mode of H-258 on DNA might be explained by the increased oxygen accessibility of weakly bound H-258 molecules.
Figure 4.
(a) Single molecule fluorescence scanning confocal image (7.5 × 10.0 µm) of CT ds DNA labeled with H-258 (40 µM) at a [H-258]/[DNA bp] ratio of 0.2 in 50 mM phosphate buffer, pH 7.0. DNA molecules bind to the amino-modified glass cover slide due to electrostatic interactions. Imaging was performed at room temperature with a resolution of 50 nm/pixel, λexc = 360 nm, average excitation power ∼1 kW/cm2 and 6 ms integration time per pixel (intensity: 0–50 counts/6 ms). (b) Fluorescence intensity and lifetime transient of a H-258 labeled DNA molecule ([H-258]/[DNA bp] = 0.2) with a time resolution of 100 ms. The transients demonstrate that weaker bound H-258 molecules show shorter fluorescence lifetimes and lower photostability.
Conformations of specific and unspecific bound H-258
NMR studies show that binding of H-258 molecules to the minor groove of DNA results in a planar conformation of H-258 involving the phenol and the two benzimidazole units (29). Therefore, a certain degree of charge transfer from the protonated piperazine-bis-benzimidazole unit to the phenol bis-benzimidazole unit might occur upon excitation. As a result, the interconnecting bond between the two benzimidazole units becomes planar and stronger in the excited state (16). This implies that the longer component of the fluorescence decay of minor groove bound H-258 arises from the conformer having both benzimidazole units in a planar conformation. Specific binding of this conformer in the more hydrophobic environment of the minor groove lowers its internal conversion rate. Thus, both the decrease of internal conversion and the increase in hydrophobicity due to specific minor groove binding contribute to the origin of the longer component of the fluorescence decay.
From the ELD data available in literature (37,39), it can be envisaged that H-258 molecules partially intercalate one of their benzimidazole units and the attached phenol ring between two G:C pairs at higher H-258 concentrations thereby placing the second benzimidazole into the major groove with the positively charged N-terminal piperazine ring protruding outside the DNA helix. Furthermore, it is known that the fluorescence quantum yield of related heterocompounds, such as DAPI, is more strongly enhanced upon addition of poly[d(A:T)] than poly[d(G:C)] (53). However, why is the fluorescence quantum yield and lifetime of these H-258 molecules reduced. The answer could be that a high loading of the dye molecules in DNA leads to steric hindrance in such a way that either partial intercalation between G:C pairs occurs and/or a twisted conformation is favored (27,29,47). These H-258 molecules are more exposed to the surrounding water. Furthermore, non-radiative deactivation via intramolecular rotation is possible. Therefore, fluorescence quantum yield, lifetime and photostability decrease slightly. Our data suggest that the lifetime component of 1.5–2.0 ns with an amplitude of 20–40% appearing at [H-258]/[DNA bp] ratios of 0.15 and 0.20 is due to partial intercalation. Since unwinding of the helix takes place during intercalation (54), partial intercalation of H-258 molecules in the G:C sequences also explains the unwinding of the DNA helix occurring at [H-258]/[DNA bp] >0.05 as observed by atomic force microscopy (55).
The data in Table 2 also show that with increasing [H-258]/[DNA bp] ratios a shorter fluorescence decay component (∼500 ps) occurs (11% at [H-258]/[DNA bp] = 0.2). From the lifetimes of the free H-258 molecules, we envisage that this component arises from unspecific stacking or dynamic adsorption of H-258 molecules onto DNA. These molecules are strongly exposed to water comparable to free H-258 with very low fluorescence quantum yield, short lifetime and very low photostability.
CONCLUSIONS
The spectroscopic parameters of H-258–CT ds DNA complexes at various [H-258]/[DNA bp] ratios have been characterized using steady-state absorption and fluorescence as well as time-resolved fluorescence spectroscopy. Overall, the data obtained are consistent with previous reports but the additional information gained, especially at higher [H-258]/[DNA bp] ratios, clearly demonstrates that H-258 exhibits different binding modes. Up to [H-258]/[DNA bp] ratios of 0.05, exclusively specific minor groove binding occurs and the fluorescence decay can be described almost by a mono-exponential model with a long fluorescence lifetime of ∼3.5 ns. Additional binding at higher [H-258]/[DNA bp] ratios results in blue-shifted absorption maxima, lower fluorescence quantum yield and shortened fluorescence lifetimes. This indicates that unspecific binding of H-258 with DNA occurs by (i) partial intercalation at G:C sequences reflected by a lifetime component of 1.5–2.0 ns (20–40%) and (ii) stacking or adsorption on DNA where H-258 molecules exhibit a lifetime of ∼500 ps and are more exposed to the aqueous environment. Single-molecule transients of fluorescence intensity together with lifetime demonstrate that the water exposure is accompanied not only by a reduced fluorescence intensity and lifetime but by a strong reduction in photostability. Our data demonstrate that H-258 cannot be used as an efficient molecular probe at [H-258]/[DNA bp] ratios >0.05.
Acknowledgments
ACKNOWLEDGEMENTS
A.A. is grateful to the DAAD for the financial support and to the authorities of the Rajdhani College and University of Delhi for giving the permission to work in the group of Dr M. Sauer. The authors also thank Professor Dr J. Wolfrum for his keen interest and support in this work.
REFERENCES
- 1.Loontiens F., Regenfuss,P., Zechel,A., Dumortier,L. and Clegg,R.M. (1990) Binding characteristics of Hoechst 33258 with calf thymus DNA, poly[d(A-T)] and d(CCGGAATTCCGG): multiple stoichiometries and determination of tight binding with a wide spectrum of site affinities. Biochemistry, 29, 9029–9039. [DOI] [PubMed] [Google Scholar]
- 2.Hilwig I. and Gropp,A. (1972) Studies of the constitutive heterochromatin in mammalian chromosomes with a new fluorochrome. Exp. Cell Res., 75, 122–123. [DOI] [PubMed] [Google Scholar]
- 3.Latt S.A. (1973) Microfluorometric detection of deoxyribonucleic acid replication in human metaphase chromosomes. Proc. Natl Acad. Sci. USA, 70, 3395–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinmetzer K. and Reinert,K.-E. (1998) Multimode interaction of Hoechst 33258 with eukaryotic DNA; quantitative analysis of the DNA conformational changes. J. Biomol. Struct. Dyn., 15, 779–791. [DOI] [PubMed] [Google Scholar]
- 5.Bailly C. (2000) Topoisomerase I poisons and suppressors as anticancer drugs. Curr. Med. Chem., 7, 39–58. [DOI] [PubMed] [Google Scholar]
- 6.Soderlind K.-J., Gorodetsky,B., Singh,A.K., Bachur,N.B., Miller,G.G. and Lown,J.W. (1999) Bis-benzimidazole anticancer agents: targeting human tumor helicases. Anti-Cancer Drug Design, 14, 19–36. [PubMed] [Google Scholar]
- 7.Denison L., Haigh,A., D’Cunha,G. and Martin,R.F. (1992) DNA ligands as radioprotectors: molecular studies with Hoechst 33342 and Hoechst 33258. Int. J. Radiat. Biol., 61, 69–81. [DOI] [PubMed] [Google Scholar]
- 8.Adhikary A., Bothe,E., Jain,V. and von Sonntag,C. (1997) Inhibition of radiation-induced DNA strand breaks by Hoechst 33258: OH-radical scavenging and DNA radical quenching. Radioprotection, 32, C1-89–C1-90. [Google Scholar]
- 9.von Sonntag C. (1987) Techniques for measuring radiation-induced changes in DNA. In The Chemical Basis of Radiation Biology. Taylor and Francis, London, pp. 194–197.
- 10.Lyubimova N.V., Coultas,P.G., Yuen,K. and Martin R.F. (2001) In vivo radioprotection of mouse brain endothelial cells by Hoechst 33342. Brit. J. Radiol., 74, 77–82. [DOI] [PubMed] [Google Scholar]
- 11.von Sonntag C. (1991) The chemistry of free-radical-mediated DNA damage. In Glass,W.A. and Verma,M.N. (eds), Physical and Chemical Mechanisms in Molecular Radiation Biology. Plenum Press, New York, NY.
- 12.Adhikary A., Bothe,E., von Sonntag,C. and Jain,V. (1997) DNA radioprotection by bisbenzimidazol derivative Hoechst 33258: model studies on the nucleotide level. Radiat. Res., 148, 493–494. [Google Scholar]
- 13.von Sonntag C., Bothe,E., Ulanski,P. and Adhikary,A. (1999) Radical transfer reactions in polymers. Radiat. Phys. Chem., 55, 599–603. [Google Scholar]
- 14.Adhikary A., Bothe,E., Jain,V. and von Sonntag,C. (2000) Pulse radiolysis of the DNA-binding ligands Hoechst 33258 and 33342 in aqueous solution. Int. J. Radiat. Biol., 76, 1157–1166. [DOI] [PubMed] [Google Scholar]
- 15.Umetskaya V.N. and Rozanov,Y.M. (1990) Mechanism of the interaction of DNA with the fluorescent dye Hoechst 33258. Biophysika, 35, 399–401. [PubMed] [Google Scholar]
- 16.Kalninsh K.K., Pestov,D.V. and Roshchina,Y.K. (1994) Absorption and fluorescence spectra of the probe Hoechst 33258. J. Photochem. Photobiol. A: Chem., 83, 39–47. [Google Scholar]
- 17.Görner H. (2001) Direct and sensitized photoprocesses of bis-benzimidazole dyes and the effects of surfactants and DNA. Photochem. Photobiol., 73, 339–348. [DOI] [PubMed] [Google Scholar]
- 18.Alemán C., Adhikary,A., Zanuy,D. and Casanovas,J. (2002) On the protonation equilibrium for the benzimidazole derivative Hoechst 33258: an electronic molecular orbital study. J. Biomol. Struct. Dyn., 20, 301–310. [DOI] [PubMed] [Google Scholar]
- 19.Pullman A. and Pullman,B. (1981) Molecular electrostatic potential of the nucleic acids. Q. Rev. Biophys., 14, 289–380. [DOI] [PubMed] [Google Scholar]
- 20.Squire C.J., Baker,L.J., Clark,G.R., Martin,R.F. and White,J. (2000) Structures of m-iodo Hoechst–DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design. Nucleic Acids Res., 28, 1252–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson H., Gao,Y.G., Bauer,C., Roberts,C., Switzer,C. and Wang,A.H.-J. (1998) 2′-Deoxyguanosine adopts more than one tautomer to form base pairs with thymidine observed by high-resolution crystal structure analysis. Biochemistry, 37, 10897–1095. [DOI] [PubMed] [Google Scholar]
- 22.Spink N., Brown,D.G., Skelly,J.V. and Neidle,S. (1994) Sequence-dependent effects in drug–DNA interaction: the crystal structure of Hoechst 33258 bound to the d(CGCAAATTTGCG)2 duplex. Nucleic Acids Res., 22, 1607–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vega M.C., Garcia-Saez,I., Aymami,J., Eritja,T., van der Marel,G.A., van Boom,J.H., Rich,A. and Coll,M. (1994) Three dimensional crystal structure of the A-tract dodecamer d(CGCAAATTTCGCG)2 complexed with the minor groove-binding drug Hoechst 33258. Eur. J. Biochem., 222, 721–726. [DOI] [PubMed] [Google Scholar]
- 24.Quintana J.R., Lipanov,A.A. and Dickerson,R.E. (1991) Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer CGCGAATTCGCG. Biochemistry, 30, 10294–10306. [DOI] [PubMed] [Google Scholar]
- 25.de Carrondo M.A.A.F.C.T., Coll,M., Aymami,J., Wang,A.H.-J., van der Marel,G.A., van Boom,J.H. and Rich,A. (1989) Binding of Hoechst dye to d(CGCGATATCGCG) and its influence on the conformation of the DNA fragment. Biochemistry, 28, 7849–7859. [DOI] [PubMed] [Google Scholar]
- 26.Teng M.-K., Usman,N., Frederick,C.A. and Wang,A.H.-J. (1988) The molecular structure of the complex of Hoechst 33258 and the DNA dodecamer d(CGCGAATTCGCG). Nucleic Acids Res., 16, 2671–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pjura P.E., Grzeskowiak,K. and Dickerson,R.E. (1987) Binding of Hoechst 33258 to the minor groove of B-DNA. J. Mol. Biol., 197, 257–271. [DOI] [PubMed] [Google Scholar]
- 28.Parkinson J.A., Barber,J., Buckingham,B.A. and Doughlas,K.T. (1992) Hoechst 33258 and its complex with the oligonucleotide d(CGCGAATTCGCG)2: 1H NMR assignments and dynamics. Magn. Reson. Chem., 30, 1064–1069. [Google Scholar]
- 29.Fede A., Labhardt,A., Bannwarth,W. and Leupin,W. (1991) Dynamics and binding mode of Hoechst 33258 to (GTGGAATTCCAC)2 in the 1:1 solution complex as determined by two dimensional 1H NMR. Biochemistry, 30, 11377–11388. [DOI] [PubMed] [Google Scholar]
- 30.Searle M.S. and Embrey,K.J. (1990) Sequence-specific interaction of Hoechst 33258 with the minor groove of an adenine-tract complex studied by 1H NMR spectroscopy. Nucleic Acids Res., 18, 3753–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parkinson J.A., Barber,J., Douglas,K.T., Rosamond,J. and Sharples,D. (1990) Minor groove recognition of the self-complementary duplex d(CGCGAATTCGCG)2 by Hoechst 33258: a high-field NMR study. Biochemistry, 29, 10181–10190. [DOI] [PubMed] [Google Scholar]
- 32.Loontiens F.G., McLaughlin,L.W., Diekmann,S. and Clegg,R.M. (1991) Binding of Hoechst 33258, 4′6-diamidino-2-phenylindole to self-complementary decadeoxynucletides with modified exocyclic base substituents. Biochemistry, 30, 182–189. [DOI] [PubMed] [Google Scholar]
- 33.Abu-Daya A., Brown,P.M. and Fox,K.R. (1995) DNA sequence preferences of several AT-selective minor groove binding ligands. Nucleic Acids Res., 23, 3385–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steiner R.F. and Sternberg,H. (1979) The interaction of Hoechst 33258 with natural and biosynthetic nucleic acids. Arch. Biochem. Biophys., 197, 580–588. [DOI] [PubMed] [Google Scholar]
- 35.Mikhailov M.V., Zasedatelev,A.S., Krylov,A.S. and Gurskii,G.V. (1981) Mechanism of AT base pairs recognition by molecules of dye ‘Hoechst 33258’. Mol. Biol. (Mosk.), 15, 690–705. [PubMed] [Google Scholar]
- 36.Stokke T. and Steen,H.B. (1985) Multiple binding modes for Hoechst 33258 to DNA. J. Histochem. Cytochem., 33, 333–338. [DOI] [PubMed] [Google Scholar]
- 37.Colson P., Bailly,C. and Houssier,C. (1996) Electric linear dichroism as a new tool to study sequence preference in drug binding to DNA. Biophys. Chem., 58, 125–140. [DOI] [PubMed] [Google Scholar]
- 38.Breusegem S.Y., Clegg,R.M. and Loontiens,F.G. (2002) Base sequence specificity of Hoechst 33258 and DAPI binding to five (A/T)4 DNA sites with kinetic evidence for more than one high-affinity Hoechst 33258–AATT complex. J. Mol. Biol., 315, 1049–1061. [DOI] [PubMed] [Google Scholar]
- 39.Bailly C., Colson,P., Hénichart,J.-P. and Houssier,C. (1993) The different binding modes of Hoechst 33258 to DNA studied by electric linear dichroism. Nucleic Acids Res., 21, 3705–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Comings D.E. (1975) Mechanisms of chromosome banding VIII. Hoechst 33258–DNA interactions. Chromosoma (Berl.), 52, 229–243. [DOI] [PubMed] [Google Scholar]
- 41.Scott T.G., Spencer,R.D., Leonard,N.J. and Weber,G.J. (1970) Synthetic spectroscopic models related to coenzymes and base pairs. V. Emission properties of NADH. Studies of fluorescence lifetimes and quantum efficiencies of NADH, AcPyADH, [reduced acetylpyridineadenine dinucleotide] and simplified synthetic models. J. Am. Chem. Soc., 92, 687–695. [Google Scholar]
- 42.Tellinghuisen J., Goodwin,P.M., Ambrose,W.P., Martin,J.C. and Keller,R.A. (1994) Analysis of fluorescence lifetime data for single molecules in flowing sample streams. Anal. Chem., 66, 64. [Google Scholar]
- 43.Haq I., Ladbury,J.E., Chowdhry,B.Z., Jenkins,T.C. and Chaires,J.B. (1997) Specific binding of Hoechst 33258 to the d(CGCAAATTTGCG)2 duplex: calorimetric and spectroscopic studies. J. Mol. Biol., 271, 244–257. [DOI] [PubMed] [Google Scholar]
- 44.Bontemps J., Houssier,C. and Fredericq,E. (1975) Physico-chemical study of the complexes of ‘33258 Hoechst’ with DNA and nucleohistone. Nucleic Acids Res., 2, 971–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Latt S.A. and Wohlleb,J.C. (1975) Optical studies of the interaction of 33258 Hoechst with DNA, chromatin and metaphase chromosomes. Chromosoma (Berl.), 52, 297–316. [DOI] [PubMed] [Google Scholar]
- 46.Jin R. and Breslauer,K.J. (1988) Characterization of the minor groove environment in a drug–DNA complex: bisbenzimide bound to the poly[d(AT)].poly[d(AT)] duplex. Proc. Natl Acad. Sci. USA, 85, 8939–8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moon J.-H., Kim,S.K., Sehlstedt,U., Rodger,A. and Nordén,B. (1996) DNA structural features responsible for sequence-dependent binding geometries of Hoechst 33258. Biopolymers, 38, 593–606. [DOI] [PubMed] [Google Scholar]
- 48.Sailer B.L., Nastasi,A.J., Valdez,J.G., Steinkamp,J.A. and Crissman,H.A. (1997) Differential effects of deuterium oxide on the fluorescence lifetimes and intensities of dyes with different modes of binding to DNA. J. Histochem. Cytochem., 45, 165–175. [DOI] [PubMed] [Google Scholar]
- 49.Cosa G., Focsaneanu,K.S., McLean,J.R., McNamee,J.P. and Scaiano,J.C. (2001) Photophysical properties of fluorescent DNA-dyes bound to single- and double-stranded DNA in aqueous buffered solution. Photochem. Photobiol., 73, 585–599. [DOI] [PubMed] [Google Scholar]
- 50.Härd T., Fan,P. and Kearns,D.R. (1990) A fluorescence study of the binding of Hoechst 33258 and DAPI to the halogenated DNAs. Photochem. Photobiol., 51, 77–86. [DOI] [PubMed] [Google Scholar]
- 51.Tinnefeld V., Buschmann,V., Herten,D.P., Han,K.T. and Sauer,M. (2000) Confocal fluorescence lifetime imaging microscopy (FLIM) at the single molecule level. Single Mol., 1, 215–223. [Google Scholar]
- 52.Eggeling C., Brand,L. and Seidel,C.A.M. (1997) Laser-induced fluorescence of coumarin derivatives in aqueous solution: photochemical aspects for single molecule detection. Bioimaging, 5, 105–115. [Google Scholar]
- 53.Szabo A.G., Krajcarski,D.T., Cavatorta,P., Casotti,L. and Barcellona,L. (1986) Excited state pKa behavior of DAPI. A rationalization of the fluorescence enhancement of DAPI in DAPI–nucleic acid complexes. Photochem. Photobiol., 44, 143–150. [Google Scholar]
- 54.Bloomfeild V.A., Crothers,D.M. and Tinoco,I.,Jr (1974) Physical Chemistry of Nucleic Acids. Harper and Row, New York, NY.
- 55.Utsuno K., Tsuboi,M., Katsumata,S. and Iwamoto,T. (2002) Visualization of complexes of Hoechst 33258 and DNA duplexes in solution by atomic force microscopy. Chem. Pharm. Bull., 50, 216–219. [DOI] [PubMed] [Google Scholar]