Abstract
The Kruppel-like factor 5 (KLF5/IKLF) belongs to the Kruppel family of genes which bind GC-rich DNA elements and activate or repress their target genes in a promoter context and/or cellular environment-dependent manner. In the present study, we used the Gal4 fusion assay system to characterize the mechanism of transactivation by KLF5. We demonstrated that the transactivation function of KLF5 was enhanced by CREB-binding protein (CBP) and blocked by wild-type but not mutant E1A. Over expression of CBP reversed the inhibition effect of E1A. With various lengths of KLF5 fusion protein, the transactivation functions were localized to 156 amino acid residues at the N-terminal region and 133 amino acid residues adjacent to the Zn finger motif. We mapped the CBP and KLF5 interaction domain to the N-terminal region of CBP (amino acids 1–232) and the N-terminal region of KLF5 (amino acids 1–238) where one of the activation functions resides. The histone acetyltransferase (HAT) activity of CBP does not play a role in the transactivation function of KLF5 nor does it acetylate KLF5 in vitro. However, phosphorylation is important in KLF5 transactivation activity. Inhibition of protein kinase activity by H7 or calphostin C blocked both full-length and N-terminal fragment (amino acids 1–238) KLF5 activities. Mutation at a potential protein kinase C phosphorylation site within the CBP interaction domain of KLF5 reduces its transactivation function. Furthermore, using the GST pull-down approach, we showed that phosphorylation of KLF5 enhances its interaction with CBP. The results of the present study provide a mechanism for KLF5 transactivation function.
INTRODUCTION
KLF5 is the fifth member of the Kruppel gene family that was cloned and characterized (1–3). Like the other family members, KLF5 binds GC-rich sequences and plays multiple regulatory roles in development and differentiation (4–8). KLF5 is expressed in mouse embryo (9) and assumes a tissue-selective pattern of expression in the adult (2). Due to the abundance of KLF5 message present in the intestine, it was also named as intestine-enriched Kruppel-like factor (IKLF) (2,3). KLF5 has been shown to enhance the cellular proliferation of intestinal epithelium (10) and to play a role in smooth muscle cell differentiation (11) and the expression of smooth muscle cell-specific genes (12). Recently, a KLF5-null mouse was generated (8), and the homozygote knockout mice died before embryonic day 8.5, thus re-enforcing the important role of this transcription factor in development. Interestingly, heterozygous KLF5 knockout mice (Klf5+/–) showed diminished response to external stress (8). Besides functioning alone, KLF5 physically interacts with retinoic acid receptor (8) and in synergy with transcription factor NF-κB (13) in regulation of its target genes. In a transiently transfected cell system, we found that KLF5 could activate or repress reporter genes in different cell lines, suggesting that KLF5 may exert dual functions dependent on the cellular environment (2). These observations suggest that KLF5 is a pleiotropic transcription factor that participates in a wide range of biological functions and is essential for survival. Thus, it is important to identify the molecular mechanisms underlying the activation or repression function of KLF5 in order to have a better understanding of its biological roles.
Recent data indicated that numerous transcription factors mediate their effect via recruitment of CBP (14–16). CBP was discovered originally as a binding protein of phosphorylated CREB (17). The protein exerts its function through multiple mechanisms. CBP interacts with components of the basal transcription complex, and the sequence-specific transcription factor thus serves as a ‘bridging factor’ (18–21). CBP also interacts with transcription factors through specific domains, thus providing a platform for the assembly of a multiprotein complex for highly efficient transcription (22–26). The intrinsic histone acetyltransferase (HAT) activity of CBP has been a key factor in modifying chromatin structure (27,28), and hyperacetylation of histones has been associated with active chromatin (29,30). Thus, acetylation of histone loosens the chromatin structure, facilitates the interaction of transcription factors and their cognate DNA elements, and enhances the transcriptional activities of target genes. Emerging evidence demonstrated that the CBP HAT also acetylates non-histone proteins such as EKLF (31), GATA-1 (32), TFIIE and TFIIF (33) and p53 (34), and enhances their functional activity. As well as acetylation, phosphorylation of the transcription factors by kinases also promotes recruitment of the coactivator and interaction between them (35–37). In this report, we explored the potential molecular mechanisms of KLF5 in transactivation function. We used the Gal4 fusion assay system in HEC-1B cells as the model and showed that KLF5 has two activation domains, and the N-terminal activation domain teams up with the coactivator CBP to enhance the KLF5 activation function further. Our study is the first to show that phosphorylation of KLF5 promotes CBP interaction with its N-terminal region, and that this phosphorylation, through a protein kinase C (PKC) signaling pathway, plays an important role in its activation function.
MATERIALS AND METHODS
Materials
The human endometrial carcinoma cell line HEC-1B (HTB-113) was obtained from ATCC (Rockville, MD). Tissue culture components were obtained from Gibco-BRL Life Technologies, Inc. (Gaithersburg, MD). The commercial CBP antibody was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Polyclonal rabbit antiserum of GST–KLF5 has been described previously (2). Unfractionated whole histone from calf thymus (histone type II-A) was purchased from Sigma (St Louis, MO). [35S]Methionine, S-linkage glutathione–Sepharose beads, [γ-33P]ATP and [3H]acetyl CoA were purchased from Amersham Pharmacia Biotech, and [14C]chloramphenicol from NEN Life Science Products (Boston, MA). H7 (Ellagic Acid), Rp-8-Br-cAMPS (adenosine 3′,5′-cyclic monophosphorothioate, 8-bromo, Rp-isomer), calphostin C and the catalytic unit of PKC from rat brain were obtained from Calbiochem-Novabiochem Corp. (La Jolla, CA).
Plasmids
Reporter plasmid, 5×(Gal4)–CAT, was obtained from Clontech (Palo Alto, CA). Expression plasmids pBK-CMV-KLF5 and GST–KLF5 (2) were described previously. Wild-type and mutant constructs of CBP (32) were generous gifts from Gerd A. Blobel (University of Pennsylvania School of Medicine, Philadelphia, PA), E1A wild type and the mutant (38) were from E. Moran (Fels Institute for Cancer Research and Molecular Biology, Temple University, Philadelphia, PA), and Gal4–CBP fragments (21) were from D. Swope (NIEHS, Research Triangle Park, NC). For the Gal4 fusion constructs, the full-length KLF5 recovered from plasmid pBK-CMV-KLF5 by AvaI–KpnI cutting was blunted by filling-in with Klenow enzyme. The KLF5 cDNA was then inserted into the SmaI site of the Gal4 DNA-binding domain (DBD) vector (pM, Clontech) and the correct sequence with in-frame reading with Gal4 DBD was verified. The deletion of KLF5 and mutation of KLF5 S153 to A (KLF5 S153A) were carried out using the transformer site-directed mutagenesis kit (Clontech, Palo Alto, CA) according to the manufacturer’s instructions [constructs KLF5 (1–82), (239–372) and (322–457) were contributed by Il Kwon of Gene Regulation Section, LRDT, NIEHS]. The expression plasmid for PKC catalytic unit (pKC-7) was a generous gift from Eric Olson (University of Texas, M.D. Anderson Cancer Center, Houston, TX). The plasmids were purified either by CsCl centrifugation or by column chromatography (QIAfilter Maxi column, Qiagen, Santa Cruz, CA). The sequence of all plasmids was verified by automatic sequencing.
Cell culture and transient transfection assays
HEC-1B cells were maintained in Eagle’s minimum essential medium (EMEM) supplemented with 10% fetal bovine serum (FBS) (Life Technologies, Inc) under 5% CO2. Prior to transfection, cells were split into 6-well plates and cultured for 24 h. Transient transfections were performed with Effectene reagent (Qiagen) according to the manufacturer’s instructions. The cells were transfected with 1.0–1.4 µg of total DNA consisting of chloramphenicol acetyltransferase (CAT) reporter construct, the expression plasmids, the internal control (pCH110, β-galactosidase) and salmon sperm DNA or empty vectors to make up the final DNA concentration. At 24 h after transfection, cells were washed and cell extracts prepared. CAT enzyme and the β-galactosidase activities were measured as previously described (39). The CAT activities were normalized against β-galactosidase activities. All experiments were repeated at least three times with duplicate samples in each experiment, and data are presented as mean ± SD.
Western blot analysis
Various Gal4–KLF5 or Gal4–CBP (full-length or fragments) fusion constructs (0.5 µg) were transfected into HEC-1B cells. The cell lysates were prepared 24 h after transfection. About 10 µg of protein from each cell lysate was loaded and separated on the NuPAGE 4–12% Bis-Tris gel with MOPS running buffer (Invitrogen, Carlsbad, CA). The SeeBlue pre-stained standard (Invitrogen) was used as molecular weight marker. After electrophoresis, the proteins were blotted onto PVDF membrane and visualized with anti-Gal4 DBD (1–147) rabbit polyclonal IgG (0.2 µg/ml) (Santa Cruz). In the proteasome pathway inhibition study, the cells were transfected with Gal4–KLF5 (1–372) fusion construct for 24 h. Before harvesting, the cells were treated with proteasome inhibitor MG-132 (Calbiochem-Novabiochem Corp) at 10 µM for 0, 5 or 8 h. About 30 µg of protein was loaded and separated on the NuPAGE gel, and the western blot was exposed to X-ray film for 30 min.
RT–PCR
Total RNA from HEC-1B cells transfected with Gal4–KLF5 or Gal4–CBP (full-length or fragments) fusion constructs was isolated using the RNeasy Mini Kit (Qiagen). The Applied Biosystems (Foster City, CA) GeneAmp RNA PCR Kit components were used for both reverse transcription and PCRs. The primers used to encode a 101 bp Gal4 DBD are as follows: forward primer, 5′-GTGCGCCAAGTGTCTGAAGA-3′; reverse primer, 5′-GCCTTGATTCCACTTCTGTCAGAT-3′. The primers used to encode a 144 bp β-actin are: forward primer, 5′-GACAGGATGCAGAAGGAGATCAC-3′; and reverse primer, 5′-GCTGATCCACATCTGCTGGAA-3′. The RT–PCR product of β-actin served as a reference to normalize the Gal4 DBD RT–PCRs, as well as for controlling the contamination of genomic DNA.
Immunoprecipitation–histone acetyltransferase (IP-HAT) assay
An IP-HAT assay was conducted as described (27). Briefly, HEC-1B cells were grown to confluence in 100 mm culture dishes, washed twice with phosphate-buffered saline (PBS) and lysed with IPH buffer [50 mM Tris–HCl pH. 8.0, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and protease inhibitor cocktail]. The cell lysate was left on ice for 20 min to complete the reaction before clearing by centrifugation at 12 000 g for 10 min. The following steps were carried out at 4°C. Anti-CBP antibody was added to 1 ml of supernatants for 2 h before protein A–Sepharose beads were added, and the mixture was rotated gently overnight. The immune complexes formed overnight were pelleted by slow centrifugation. The precipitation was washed with IPH buffer three times. After final washing, the immune complex was suspended in 30 µl of IPH buffer and the whole histone (25 µg) or purified GST or GST–KLF5 fusion proteins (5 µg) and 1 µl of 0.25 mCi/ml [3H]acetyl CoA (5.1 Ci/mmol; Amersham; 1 Ci = 37 GBq) were added. The HAT assay was performed at 30°C for 30 min. The [3H]acetyl incorporation into histones, GST or GST fusion proteins was determined by liquid scintillation counting of reactions spotted onto p81 (Whatman) filters.
In vitro phosphorylation
GST, GST–KLF5, GST–KLF5 (1–238) and GST–KLF5 (239– 372) fusion proteins were prepared from Escherichia coli BL21 cells after isopropyl-β-d-thiogalactopyranoside (IPTG) induction of an overnight culture. The bacteria were disrupted by sonication, and fusion proteins were isolated with a 50% slurry of glutathione–Sepharose beads. Proteins bound to beads were phosphorylated by the catalytic subunits of rat brain PKC. The reaction was carried out in 24 µl of reaction buffer [50 mM MES pH 6.0, 1.25 mM EGTA, 12.5 mM MgCl2 and 5.7 µCi of [γ-33P]ATP (10 mCi/ml, 2500 Ci/mmol)] and 6 ng of PKC for 30 min at 25°C. Control samples were incubated similarly but without PKC. The phosphorylation reaction was terminated by addition of NuPAGE sample buffer followed by boiling. Both samples and molecular marker (wide range protein standard, Mark12) were separated on a NuPAGE 4–12% Bis-Tris gel, stained with Colloidal Blue Staining Kit (Novex), dried and visualized by autoradiography.
GST pull-down assay
GST–KLF5 fusion protein was phosphorylated by 125 mM ATP as described above. After the reactions, the bound and phosphorylated GST–KLF5 was washed with GST pull-down buffer (20 mM HEPES, pH 7.5, 0.1 mM EDTA, 75 mM KCl, 2.5 mM MgCl2, 0.05% NP-40) three times. The phosphorylated GST–KLF5 fusion proteins were then incubated with the in vitro translated and [35S]methionine-labeled CBP for 2 h at 4°C. After washing, the binding of 35S-labeled CBP to GST–KLF5 fusion protein was analyzed in NuPAGE and visualized by autoradiography. The developed autoradiograms were scanned with a ChemiImager™ 4000 (Alpha Inotech Corp.), and the intensity of each band was subjected to quantitative analysis with AlphaEase™ (V. 3.3) software.
RESULTS
Identified the activation domain of KLF5
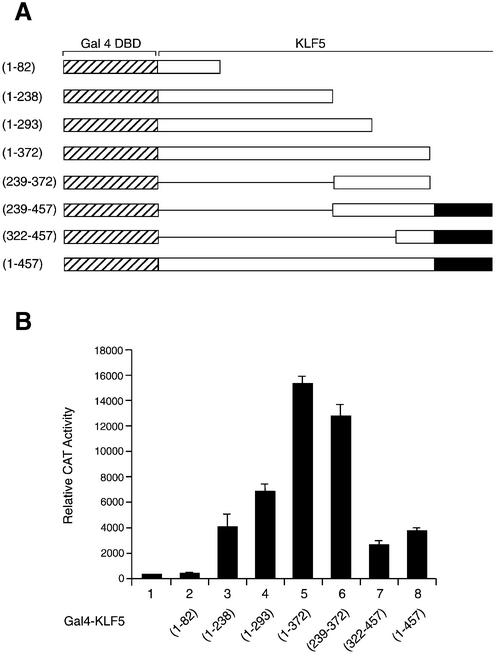
KLF5 is a pleiotropic transcription factor functioning as a positive and negative regulator (2,10,40,41). Using a series of chimeric constructs containing the Gal4 DBD and various lengths of KLF5 coding sequence (Fig. 1A), we examined the KLF5 activity in HEC-1B cells with the 5×(Gal4)–CAT reporter (Fig. 1B). The full-length KLF5 (Fig. 1B, lane 8, 1–457) activated the reporter ∼10-fold over empty vector; however, fragment 1–372 (lane 5) showed robust activity and activated the reporter 39-fold above the Gal4 vector and 4-fold above the full-length KLF5. Most of the activation activity of KLF5 resided within a 133 amino acid fragment 239–372 (lane 6). This observation is consistent with the previous report that the 239–372 fragment containing the proline-rich region of KLF5 houses the activation function (41). The present study identified an additional activation domain in the N-terminal region of KLF5 between amino acid 82 and 238 (lane 3). The KLF5 protein or the peptide fragment containing the Zn finger domain showed lower activity (lanes 7 and 8); nonetheless, all fragments demonstrated activation function except fragment 1–82 (lane 2). To ensure that the fusion constructs were being expressed in HEC-1B cells, extracts of the transfected cells were subjected to western blot analysis with antibody to Gal4 DBD. The predicated sizes of the fusion protein were observed (Fig. 1C). It was surprising to find that proteins expressed from constructs with high transcriptional activity, such as KLF5 (1–457) (lane 2), (1–372) (lane 3) and (239–372) (lane 8), were undetectable in the western blot. It is possible that these proteins become unstable and are degraded readily after expression. Recently, proteasome-mediated degradation of transcriptional activators was found to correlate with activation domain potency in vivo (42), and the ubiquitylation domain of the transcription factor is closely associated or even overlapping with the activation domain (43,44). This phenomenon has gained a lot of attention (45–47). To provide an example showing that the active fragment of KLF5 was degraded rapidly through the proteasome-mediated pathway, we treated the cells with proteasome inhibitor (MG-132) for various times after Gal4– KLF5 (1–372) was transfected into HEC-1B cells (Fig. 1D). As expected, this fusion protein was detected by western blotting after MG-132 treatment (compare lanes 1 and 3). To demonstrate that the various Gal4–KLF5 fragments indeed were transcribed in the cells after transfection, total RNA of the transfected cells was isolated and subjected to RT–PCR of the Gal4 DBD region, and β-actin was used as internal control (Fig. 1E). The results confirmed that the observed difference in transactivation activities of the KLF5 fragments was intrinsic in nature.
Figure 1.
Mapping the KLF5 transactivation domain. (A) Schematic presentation of the fusion protein containing the Gal4 DBD and the full-length or fragments of KLF5. The striped area is the Gal4 DBD; the solid box is the Zn finger region of KLF5 (2); and the open box and the numbers indicate the amino acid position of the KLF5 fragments. (B) Identification of the activation domain of KLF5 in HEC-1B cells. Gal4 DBD or Gal4–KLF5 fragment constructs (0.5 µg each) were transfected with 5×(Gal4)–CAT reporter (0.1 µg) into HEC-1B cells. The CAT activity was measured 24 h later. CAT activity was normalized with β-galatosidase activity. The experiments were repeated three times with duplicate samples, and the results are presented as means ± SD. (C) Expression of Gal4–KLF5 (full-length or fragments) fusion protein in transfected cells. Equal amounts of fusion constructs (0.5 µg) were transfected into HEC-1B cells, and cell lysates were prepared 24 h later. About 10 µg of protein from transfected cells were subjected to western blot analysis with antibody against Gal4 DBD and detected by ECL. The X-ray film was exposed for 10 min. Molecular weight markers (kDa) are shown on the left. (D) Proteasome inhibitor stabilized the KLF5 (Gal4–KLF5 1–372) fusion protein in transfected cells. Gal4–KLF5 (1–372) constructs (0.5 µg) were transfected into HEC-1B cells for 24 h. Cells were treated with MG-132 at 10 µM for 0, 5 or 8 h before preparing the cell lysate. About 30 µg of protein from transfected cells were subjected to western blot analysis with antibody against Gal4 DBD and detected by ECL. The X-ray film was exposed for 30 min. Molecular weight markers (kDa) are shown on the left. The arrow indicates the Gal4–KLF5 (1–372) fusion protein. (E) Various Gal4–KLF5 constructs were transcribed equally in HEC-1B cells. Total RNA was prepared from each transfected cell with the indicated construct. The mRNA levels for the fusion constructs and β-actin were determined by RT–PCR. M, 100 bp DNA ladder.
CBP enhances KLF5 transactivation function
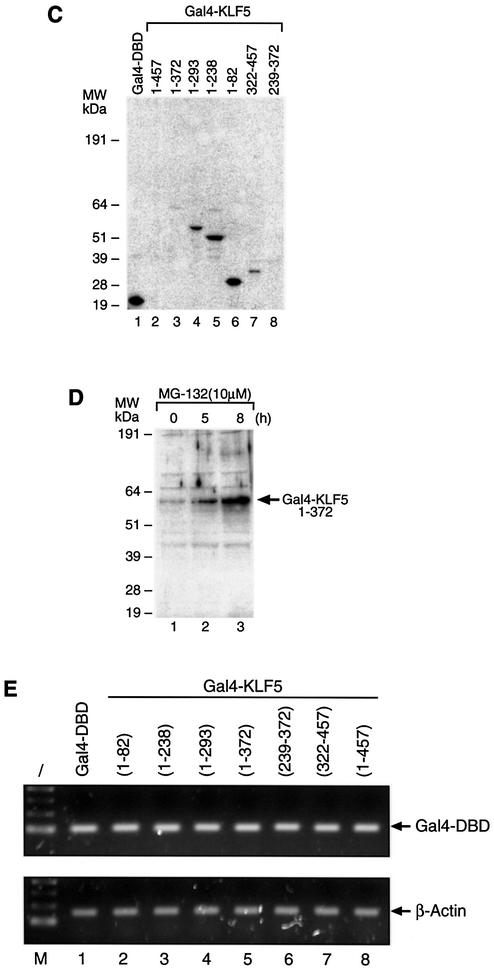
To investigate whether CBP is involved in KLF5 transactivation function, we co-transfected a constant amount of KLF5 and increasing amounts of CBP expression constructs into HEC-1B cells (Fig. 2A). The results demonstrated that CBP enhances the transactivation function of KLF5 in a dose-dependent manner. A common approach to show the relevance of CBP in the complexity of gene regulation is to examine the effect of adenovirus E1A oncoprotein on transcription (48). E1A blocks CBP coactivator function by binding to the same region that interacts with TFIIB and components of the RNA polymerase II holoenzyme complex (19,20,49). E1A also inhibits the general transcription factor (50,51) and is involved in chromatin remodeling (52). Overexpression of wild-type E1A efficiently inhibits KLF5-dependent activation, but not mutant E1A which lacks the CBP-binding site (50) (Fig. 2B). Overexpression of CBP fully reversed the E1A repression and further enhanced the KLF5 transactivation activity. This result demonstrated that CBP is an activation partner of KLF5.
Figure 2.
CBP enhances KLF5 transactivation function in HEC-1B cells. (A) Dose response. Increasing concentration of CBP expression constructs with constant amounts of KLF5 expression constructs; the reporters were transfected into HEC-1B cells. Constructs are Gal4 KLF5 expression vector and 5×(Gal4)–CAT reporter (0.1 µg). (B) Effect of E1A. KLF5 transactivation function can be blocked by wild-type but not by mutant E1A, and overexpression of CBP reverses E1A’s effect. Gal4–KLF5, wild-type or mutant E1A, CBP expression constructs (0.4 µg each) and reporter construct, 5×(Gal4)–CAT (0.1 µg), were transfected into HEC-1B cells as indicated. E1Awt, wild-type; E1Adel (2–36), mutant; Gal4, empty expression vector. The experiments were repeated three times with duplicate samples and the results are presented as means ± SD.
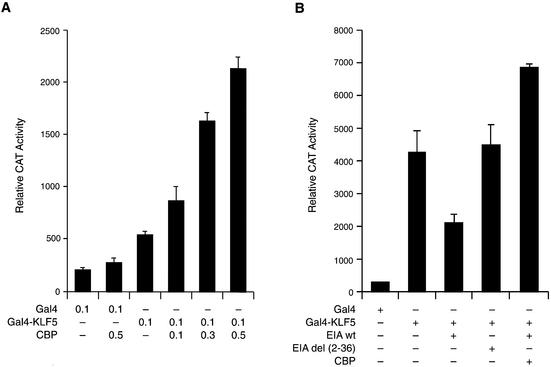
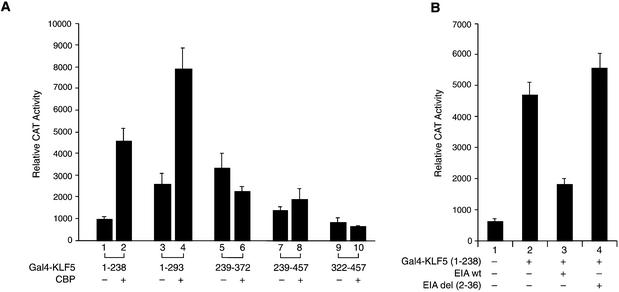
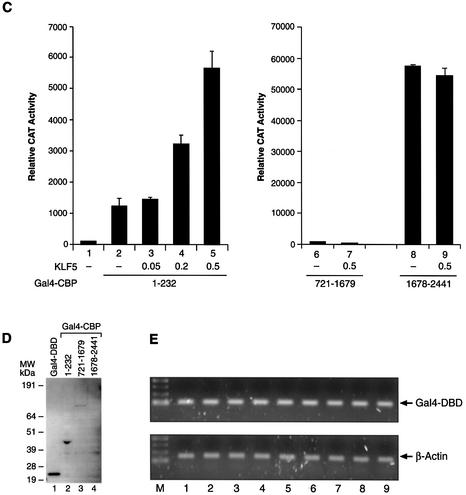
The KLF5 region that is responsible for the CBP action was analyzed with a series of chimeric constructs in HEC-1B cells (Fig. 1A). Various KLF5 fragment plasmids and the 5×(Gal4)–CAT reporter were transfected with empty vector or CBP expression constructs into HEC-1B cells, and the reporter activities were measured 24 h later (Fig. 3A). Among the five constructs analyzed, overexpression of CBP stimulated the activity of the KLF5 fragment comprising amino acids 1–238 (lanes 1–4). There was no CBP effect with KLF5 fragments at the C-terminal region (lanes 5–10), suggesting that CBP exerts its effect through the N-terminal region up to amino acid 238. This region coincides with one of the activation domains in KLF5 (Fig. 1B).
Figure 3.
(Opposite) Effect of CBP on the transactivation activity of various KLF5 fragments. (A) CBP enhances the activity of the KLF5 N-terminal region. Constant amounts of 5×(Gal4)–CAT (0.1 µg) and Gal4–KLF5 fragment constructs (0.05 µg each) were transfected with CBP expression constructs (0.5 µg) or empty vector into HEC-1B cells. The CAT activity was measured 24 h later. (B) Effect of E1A on the activation activity of the KLF5 N-terminal region. The Gal4–KLF5 (1–238) transactivation activity can be blocked by wild-type but not by mutant E1A. E1Awt, wild-type; E1Adel (2–36), mutant. (C) The N-terminal but not the C-terminal region of CBP enhances KLF5 transactivation activity. Left panel (lanes 1–5): increasing concentrations of pBK-CMV-KLF5 expression constructs and a constant amount of CBP (1–232) (0.05 µg) were transfected into HEC-1B cells. Right panel: co-expression of KLF5 expression constructs and CBP (721–1679) (lanes 6 and 7, 0.5 µg) or CBP (1678–2441) (lanes 8 and 9, 0.05 µg) in HEC-1B cells. The 5×(Gal4)–CAT reporter activities were measured 24 h later. CAT activity was normalized with β-galactosidase activity. The experiments were repeated three times with duplicate samples, and the results are presented as means ± SD. (D) Western blot analysis of Gal4–CBP (fragments) fusion protein in transfected cells. Equal amounts of fusion constructs (0.5 µg) were transfected into HEC-1B cells, and cell lysates were prepared 24 h later. About 10 µg of protein from transfected cells were subjected to western blot analysis with antibody against Gal4 DBD and detected by ECL. The X-ray film was exposed for 10 min. Molecular weight markers (kDa) are shown on the left. (E) Transcription of various Gal4–CBP constructs in HEC-1B cells. Total RNA was isolated from cells transfected with the indicated fusion construct (same as C). The mRNA levels of Gal4–CBP and β-actin were determined by RT–PCR. M, 100 bp DNA ladder.
Additional support for the involvement of CBP in KLF5 activity through the N-terminal region came from E1A blocking experiments (Fig. 3B). Overexpression of wild-type, but not the mutant E1A, effectively inhibited the transactivation activity of KLF5 (1–238) (lanes 3 and 4). We also located the CBP region that participates in KLF5 transactivation function. A constant amount of Gal4–CBP (1–232) constructs was co-transfected with an increasing concentration of KLF5 expression plasmids, and the 5×(Gal4)–CAT activities were measured (Fig. 3C, left panel). The reporter activity was dramatically increased by increasing the expression of KLF5 in HEC-1B cells, highlighting the importance of the N-terminal region of both proteins in the activation function. The C-terminal regions of CBP were also tested; however, there were no significant effects on the reporter activities even when both CBP and KLF5 were present (Fig. 3C, right panel). The expression of each CBP fusion construct was examined by western blot analysis (Fig. 3D). Expression of CBP (1–232) (lane 2) and (721–1679) (lane 3) was found; however, the CBP (721–1679) was less stable. The active fragment CBP (1678–2441) was unstable in HEC-1B cells and not detectable by western blot (lane 4). Regardless of the intrinsic activities of the different regions of CBP, the Gal4–CBP fusion constructs were transcribed equally in the transfected cells as monitored by RT–PCR of the Gal4-DBD (Fig. 3E).
Phosphorylation but not acetylation plays a role in KLF5 transactivation function
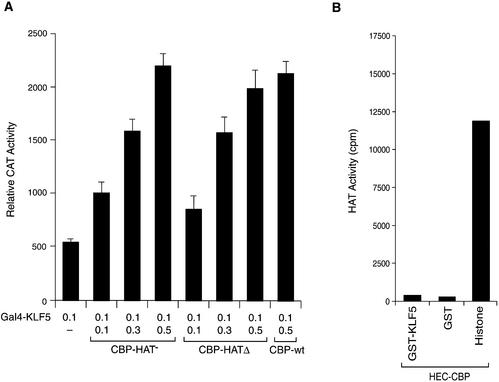
CBP/p300 contains a HAT domain and has strong HAT activity that plays a role in disrupting the repressive chromatin structure (28,52,53). It has also been shown that CBP acetylates EKLF, a member of the Kruppel family, and enhances its ability to transctivate the β-globin promoter in erythroid cells (31). Whether HAT activity of the CBP is required for the enhancement of the KLF5 transcriptional activity was examined. It was found that the mutant CBP constructs whose HAT activity was destroyed by point mutation (CBP-HAT–) (54) or by deletion (CBP-HATΔ) (55) enhanced KLF5 activation function as the wild-type CBP (Fig. 4A). In addition, CBP did not acetylate KLF5 in vitro either, as the CBP immunoprecipitated from HEC-1B cells did not acetylate KLF5 protein, whereas the control histone mixture was heavily acetylated (Fig. 4B). Thus, the acetylation function of CBP does not play a role in KLF5-dependent transactivation in the transiently transfected cell system, and KLF5 is not a substrate for CBP HAT in in vitro conditions.
Figure 4.
CBP HAT activity does not affect KLF5 transactivation function. (A) Effect of mutation or deletion of CBP HAT. Increasing concentrations of mutant CBP expression constructs whose HAT activity was destroyed by mutation or deletion and 0.1 µg of KLF5 expression constructs were transfected into HEC-1B cells. Gal4 KLF5 and 5×(Gal4)–CAT (0.1 µg) were used. CBP-HAT–, mutation at HAT; CBP-HATΔ, HAT deleted. The experiments were repeated three times with duplicate samples, and the results are presented as means ± SD. (B) CBP HAT does not acetylate KLF5 in vitro. CBP from HEC-1B cells was immunoprecipitated with CBP antibody. In vitro acetylation of GST, GST–KLF5 and histone mixture was conducted as described. The results were the average of two experiments.
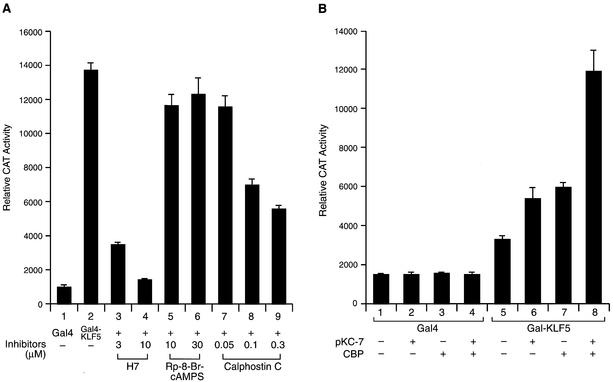
Protein phosphorylation is another mechanism that plays a major role in modulating the activity of transcription factors (36). To address whether phosphorylation is involved in KLF5 transactivation function, we transfected HEC-1B cells with Gal4–KLF5 expression constructs and 5×(Gal4)–CAT reporters, followed by a treatment with various protein kinase inhibitors (Fig. 5A). The H7 serine/threonine kinase inhibitor (56) blocked >70% of the KLF5 transactivation activity at 3 µM and inhibited most of the activity at 10 µM (lanes 3 and 4). Calphostin C, a PKC-specific inhibitor (57), also effectively blocked the KLF5 transactivation function (lanes 7–9), whereas the PKA-specific inhibitor, Rp-8-Br-cAMPS (58), did not (lanes 5 and 6). The result demonstrated that phosphorylation through the PKC pathway has a positive influence on the activity of KLF5s. The relationship between KLF5 phosphorylation and CBP was explored by co-expression of CBP and pKC-7 in HEC-1B cells (Fig. 5B). Under such conditions, both pKC-7 and CBP increase KLF5’s activity by themselves (lanes 6 and 7, respectively); however, when co-expressed (lane 8), the KLF5 transactivation activity was greatly elevated (compare lanes 5 and 8). As a control, neither pKC-7 nor CBP activates the Gal4 construct without the presence of KLF5 (lanes 1–4). These experiments suggested that CBP stimulates KLF5 activity more efficiently with the phosphorylated form. In addition, the phosphorylation rather than acetylation of KLF5 is important for its activation function.
Figure 5.
Phosphorylation is important in KLF5 transactivation function. (A) Protein kinase inhibitors block KLF5 transactivation activity. Constant amounts of 5×(Gal4)–CAT (0.1 µg) and Gal4–KLF5 (0.5 µg) were transfected into HEC-1B cells. The transfected cells were treated with various concentrations of protein kinase inhibitors, H7, Rp-8-Br-cAMPS or calphostin C, for 24 h before the CAT activities were measured. (B) Synergistic effect of CBP and PKC on KLF5 transactivation function. Overexpression of the PKC catalytic unit, pKC-7 and CBP enhances KLF5 transactivation activity. Reporters, Gal4 and Gal4–KLF5 (0.1 µg) together with pKC-7 (0.1 µg) and CBP (0.2 µg) expression vector alone or together were co-transfected into HEC cells as indicated. The experiments were repeated twice with duplicate samples. The results are presented as means ± SD.
Phosphorylation of KLF5 at the N-terminal region promotes CBP interaction
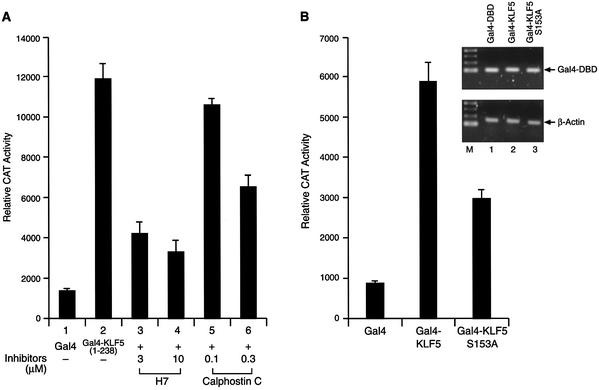
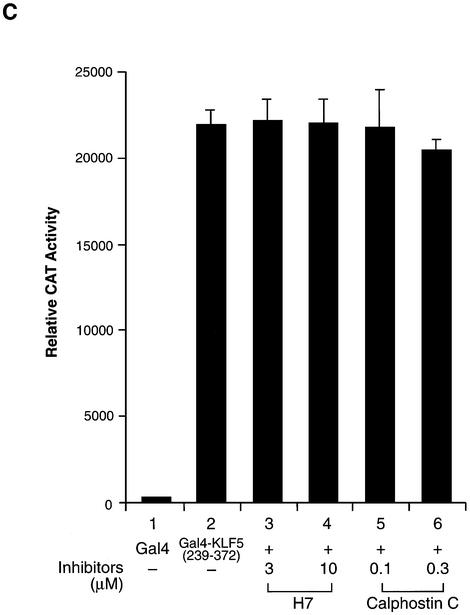
Since CBP enhances KLF5 transactivation function through the N-terminal region, we examined the effect of kinase inhibitor on the KLF5 (1–238). As expected, H7 and calphostin C efficiently suppressed the activity of KLF5 (1–238) (Fig. 6A). Within the CBP interaction domain of KLF5, there are a number of potential phosphorylation sites, and one of them is a potential PKC phosphorylation site (2). The significance of this PKC site for KLF5 function was examined. We found that KLF5’s activity was reduced to half when the serine in the potential PKC phosphorylation site was mutated to alanine (Fig. 6B, S153A) Nonetheless, the S153A mutant still retains significant transactivation activity. Both wild-type and mutant KLF 5 were unstable and undetectable by western blotting (data not shown) even though their mRNAs were transcribed and measured equally in the transfected cells (Fig. 6B, inset). There are a number of potential casein kinase II phosphorylation sites and a PKC site located in the KLF5 that may also be involved in modulating the activity of KLF5 transactivation function. This could explain the retention of significant transactivation activity by the S153A KLF5 mutant. KLF5 (239–372) housed an activation function (Fig. 1B). Whether or not phosphorylation plays a role in transactivation in this region of the protein was investigated. Interestingly, both H7 and calphostin C have no effect on the activity of Gal4–KLF5 (239–372) in HEC-1B cells (Fig. 6C). These results demonstrated that phosphorylation mainly affects the activity of the KLF5 N-terminal domain.
Figure 6.
Phosphorylation of the N-terminal region of KLF5 is important in transactivation function. (A) Protein kinase inhibitors block the transactivation function of the KLF5 N-terminus. Gal4–KLF5 (1–238) plasmids were transfected with the reporter into HEC-1B cells. Various concentrations of protein kinase inhibitors, H7 and calphostin C, were added to the culture for 24 h before CAT measurement. (B) Effect of mutation at a potential PKC phosphorylation site within the KLF5 N-terminus. Reporter (0.1 µg) and Gal4, Gal4–KLF5 or Gal4–KLF5 S153A (0.5 µg) were co-transfected into HEC-1B cells and the reporter activities measured 24 h after transfection. The experiments were repeated three times with duplicate samples. The results are presented as means ± SD. Inset: transcription of Gal4–KLF5 and Gal4–KLF5 S153A fusion constructs. The mRNA levels for the fusion constructs and β-actin were determined by RT–PCR of total RNA of transfected cells. M, 100 bp DNA ladder. (C) Protein kinase inhibitors did not block the transactivation function of the KLF5 C-terminal activation domain. Identical experiments to those in (A), except that Gal4–KLF5 (239–372) was transfected with the reporter into HEC-1B cells.
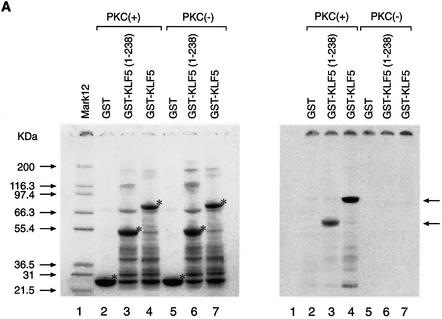
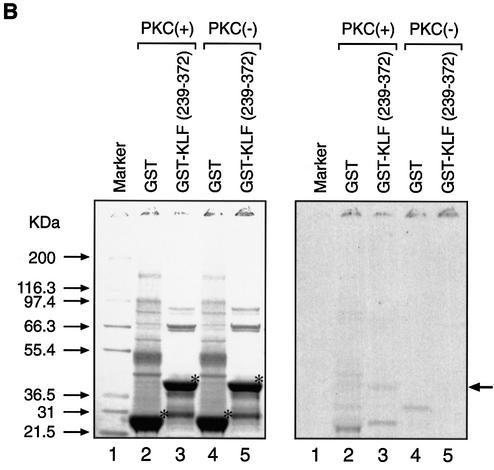
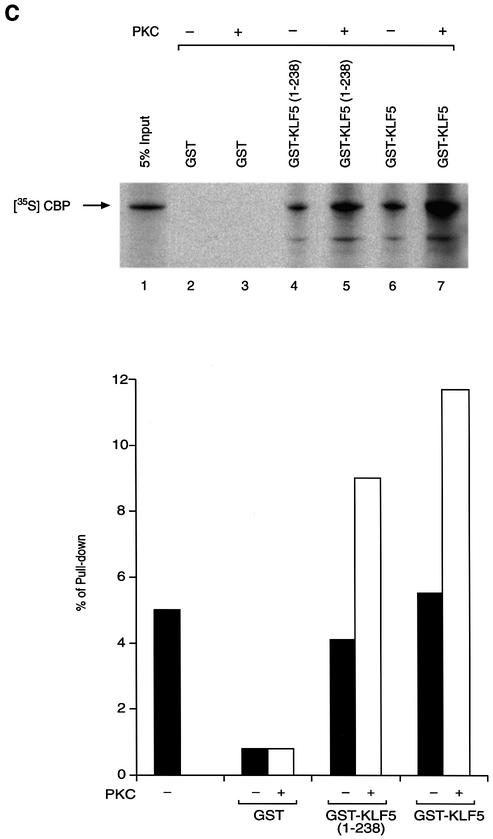
Since phosphorylation at the N-terminal region of KLF5 where CBP exerts its effect contributed to the KLF5 activation function, we explored the relationship of protein–protein interaction between CBP and phosphorylated KLF5. We first established the phosphorylation conditions and demonstrated that KLF5 can be phosphorylated in vitro, and then we performed GST pull-down experiments (Fig. 7). Equal amounts of GST, GST–KLF5 and GST–KLF5 (1–238) fusion proteins were phosphorylated with or without rat brain PKC catalytic subunit and radiolabeled ATP in vitro (Fig. 7A, left panel, compare lanes 2 and 5, 3 and 6, and 4 and 7). The same gel was then exposed to X-ray film and the autoradiogram verified the successful phosphorylation of GST–KLF5 (1–238) and GST–KLF5 (right panel, lanes 3 and 4, arrows) by PKC. As expected, GST–KLF5 (239–372) was not phosphorylated by PKC (Fig. 7B, right panel, lanes 3 and 5). The same in vitro phosphorylation conditions were used to phosphorylate KLF5 with cold ATP, and we performed the GST pull-down experiments with the in vitro 35S-labeled CBP (Fig. 7C, upper panel). GST did not interact with CBP (lanes 2 and 3), while both the N-terminal fragment KLF5 (1–238) and the full-length KLF5 protein interacted with CBP, regardless of the phosphorylation status (lanes 4–7). Nonetheless, in vitro phosphorylation of KLF5 significantly enhanced the interaction between KLF5 and CBP (compare lanes 4 with 5, and 6 with 7). The data were analyzed further by the ChemiImager system and the intensity of the bands was quantified (Fig. 7C, bottom panel). Phosphorylation of KLF5 in vitro increases its interaction with CBP by 2-fold.
Figure 7.
(Previous page) Effect of KLF5 phosphorylation and CBP interaction. (A) In vitro phosphorylation of KLF5 by PKC. Left panel: colloidal blue stain. The expressed GST, GST–KLF5 and GST–KLF5 (1–238) proteins are indicated by an asterisk. The molecular weight markers are indicated. Right panel: autoradiogram of the [γ-33P]ATP-labeled GST–KLF5 and GST–KLF5 (1–238) (indicated by arrows). The exposure time was 12 h. (B) KLF5 (239–372) was not phosphorylated by PKC in vitro. Identical experiment to that in (A), except that GST–KLF5 (239–372) was used. Left panel: colloidal blue stain. The GST and the fusion protein are marked by an asterisk. Right panel: autoradiogram. GST–KLF5 (239–372) was not phosphorylated by PKC even after long exposure (24 h) of the X-ray film (arrow). (C) KLF5 phosphorylation enhances CBP interaction. Top panel: autoradiogram of the GST pull-down assay. The arrow indicates the labeled CBP that was pulled-down by the GST–KLF5 or GST–KLF5 (1–238) fusion proteins. Bottom panel: quantitation of the [35S]CBP intensity by imaging analysis. The integrated density value is presented as a percentage, with the input intensity as 5%.
DISCUSSION
KLF5 plays an important role in development and differentiation (7,8). It regulates genes both positively and negatively; however, the molecular mechanism of KLF5 action currently is unknown. The present study demonstrated that the coactivator CBP contributes significantly to the KLF5 transactivation function by interaction with the N-terminal activation domain of KLF5. Functional studies together with in vitro protein interaction analyses showed that the CBP acetyltransferase activity is not involved in the KLF5 transactivation function but phosphorylation of KLF5 is. Our study is the first to show that phosphorylation through a PKC signaling pathway at the CBP interaction domain of KLF5 promotes CBP interaction and enhances KLF5 functional activity.
Previously we have demonstrated that KLF5 binds GCII of the lactoferrin promoter as a homodimer in vitro (2), and recently the homodimerization domain was mapped to amino acids 239–353 (13). Whether KLF5 could heterodimerize with other Kruppel family members or bind GC regions in a cooperative fashion is not clear. The recent discoveries that KLF5 interacts and/or cooperates with other transcription factors to activate or repress its target gene have widened the potential roles of KLF5 in gene regulation and increased the complexity of molecular mechanisms of KLF5 activation function (8,13). CBP interacts with numerous transcription factors and provides a platform for the assembly of enhanceosomes (23,59) or acts as a bridging factor to facilitate the formation of a pre-initiation complex (19,20,49). We showed, by GST pull-down and one-hybrid interaction experiments, that CBP physically interacts with the KLF5 N-terminal activation domain and enhances its transactivation function. The N-terminal activation domain of KLF5 does not have any notable feature to suggest its activation function. The second activation domain juxtaposed to the Zn finger domains between amino acid 239 and 372 contains a proline-rich sequence that has been identified as the KLF5 activation domain (41). Nonetheless, this region did not interact with CBP (Fig. 3A) nor with either the initiation factors or TBP (41). Additionally, it was not the target site for PKC phosphorylation (Figs 6C and 7B). It is possible that another coactivator(s) is involved with this region and plays a role in KLF5 transactivation function. Post-translational modification of the activation domain that increases the KLF5 activation function autonomously is another likely mechanism. The two activation domains identified in KLF5 may synergize with each other to bring out its maximum effect in certain cellular environments and in response to different signals. The well-characterized family members EKLF/KLF1, LKLF/KLF2 and GKLF/KLF4 also mapped their activation domain to the N-terminus at amino acids 1–125 (60,61), and CBP also played a major role in the activation function; however, no interaction domain was identified (31,61). A second centrally located transactivation subdomain was identified recently in EKLF (62) in addition to the previously mapped subdomains which carry the activation and inhibition function (31). It is probable that multiple regions of the KLF protein can interact with the coactivator or corepressor depending on the functional state of the KLFs, the context of the environment and the promoter of their target genes.
It is interesting to note that the KLF5 interaction domain on CBP is at the region mainly involved in nuclear receptor interaction, and the majority of transcription factors interact with CBP via the three Zn fingers and CREB-binding domain (15). Since CBP possesses acetyltransferase activity (63), it not only acetylates histone and remodels the structure of chromatin, but also acetylates non-histone transcription factors, including the KLF member (31). Surprisingly, we found that the HAT region of CBP was not needed to enhance KLF5 transactivation function because deletion or mutation of the HAT region had no effect on its ability to augment the transactivation of KLF5. Likewise, CBP immunoprecipitated from the cell did not acetylate KLF5 in vitro (Fig. 4). Although these data indicated that KLF5 is not a substrate for CBP HAT and the HAT activity is not required in transiently introduced templates, it did not preclude the need for HAT activity at the chromatin level where the natural gene is regulated by KLF5.
Protein phosphorylation has been an important mechanism by which a transcription activator recruits coactivators such as phosphorylation of CREB at Ser133 leading to the recruitment of CBP (17). This discovery was followed by genetic, biochemical and molecular modeling studies supporting the importance of phosphorylation of CREB and interaction with CBP (19,64). Surprisingly, other transcription factors can interact with the CBP at the CREB-binding site in a phosphorylation-independent manner, presumably through α-helical structure (65,66). KLF5 contains a number of potential casein kinase II, PKC and cAMP phosphorylation sites (2). Two of the signaling pathways, PKC and cAMP, were investigated, and we found that the PKC pathway affects KLF5’s transactivation and interactive functions (Figs 5 and 6). Although phosphorylation of KLF5 is not an obligatory requirement for its function and CBP interaction, phosphorylation at the PKC target site within the CBP-binding region substantially strengthened the interaction between KLF5 and CBP, thus enhancing the KLF5 activation activity. It has been shown that EKLF transcriptional activity was modulated by phosphorylation of a protein kinase casein kinase II site in the transactivation region (67). Like the EKLF, several potential casein kinase II phosphorylation sites are present within the CBP interaction domain of KLF5. It will be interesting to determine whether the casein kinase II pathway is also involved in KLF5 transactivation function and CBP interaction. In conclusion, KLF5 possesses two activation domains, and the N-terminal one collaborated with CBP to exert maximum transactivation function.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Masahiko Negishi, Deborah Swope and Trevor Archer for critically reading the manuscript and supplying useful comments. We thank Wesley Gladwell for technical support, and the NIEHS sequencing core laboratory for the automatic sequencing. We appreciate the reagents provided by Drs Il Kown, D. Swope, G. A. Blobel, E. Moran and E. Olson. The paper was edited by L. Moore.
REFERENCES
- 1.Sogawa K., Imataka,H., Yamasaki,Y., Kusume,H., Abe,H. and Fujii-Kuriyama,Y. (1993) cDNA cloning and transcriptional properties of a novel GC box-binding protein, BTEB2. Nucleic Acids Res., 21, 1527–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi H., Zhang,Z., Wang,X., Liu,S. and Teng,C.T. (1999) Isolation and characterization of a gene encoding human Kruppel-like factor 5 (IKLF): binding to the CAAT/GT box of the mouse lactoferrin gene promoter. Nucleic Acids Res., 27, 4807–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conkright M.D., Wani,M.A., Anderson,K.P. and Lingrel,J.B. (1999) A gene encoding an intestinal-enriched member of the Kruppel-like factor family expressed in intestinal epithelial cells. Nucleic Acids Res., 27, 1263–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turner J. and Crossley,M. (1999) Mammalian Kruppel-like transcription factors: more than just a pretty finger. Trends Biochem. Sci., 24, 236–240. [DOI] [PubMed] [Google Scholar]
- 5.Philipsen S. and Suske,G. (1999) A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res., 27, 2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang D.T., Pevsner,J. and Yang,V.W. (2000) The biology of the mammalian Kruppel-like family of transcription factors. Int. J. Biochem. Cell Biol., 32, 1103–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bieker J.J. (2001) Kruppel-like factors: three fingers in many pies. J. Biol. Chem., 276, 34355–34358. [DOI] [PubMed] [Google Scholar]
- 8.Shindo T., Manabe,I., Fukushima,Y., Tobe,K., Aizawa,K., Miyamoto,S., Kawai-Kowase,K., Moriyama,N., Imai,Y., Kawakami,H. et al. (2002) Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nature Med., 8, 856–863. [DOI] [PubMed] [Google Scholar]
- 9.Ohnishi S., Laub,F., Matsumoto,N., Asaka,M., Ramirez,F., Yoshida,T. and Terada,M. (2000) Developmental expression of the mouse gene coding for the Kruppel-like transcription factor KLF5. Dev. Dyn., 217, 421–429. [DOI] [PubMed] [Google Scholar]
- 10.Sun R., Chen,X. and Yang,V.W. (2001) Intestinal-enriched Kruppel-like factor (Kruppel-like factor 5) is a positive regulator of cellular proliferation. J. Biol. Chem., 276, 6897–6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adam P.J., Regan,C.P., Hautmann,M.B. and Owens,G.K. (2000) Positive- and negative-acting Kruppel-like transcription factors bind a transforming growth factor beta control element required for expression of the smooth muscle cell differentiation marker SM22alpha in vivo. J. Biol. Chem., 275, 37798–37806. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe N., Kurabayashi,M., Shimomura,Y., Kawai-Kowase,K., Hoshino,Y., Manabe,I., Watanabe,M., Aikawa,M., Kuro-o,M., Suzuki,T. et al. (1999) BTEB2, a Kruppel-like transcription factor, regulates expression of the SMemb/Nonmuscle myosin heavy chain B (SMemb/NMHC-B) gene. Circ. Res., 85, 182–191. [DOI] [PubMed] [Google Scholar]
- 13.Sur I., Unden,A.B. and Toftgard,R. (2002) Human Kruppel-like factor5/KLF5: synergy with NF-kappaB/Rel factors and expression in human skin and hair follicles. Eur. J. Cell Biol., 81, 323–334. [DOI] [PubMed] [Google Scholar]
- 14.Giles R.H. (1998) Update CBP/p300 transgenic mice. Trends Genet., 14, 214. [DOI] [PubMed] [Google Scholar]
- 15.Goodman R.H. and Smolik,S. (2000) CBP/p300 in cell growth, transformation, and development. Genes Dev., 14, 1553–1577. [PubMed] [Google Scholar]
- 16.Janknecht R. and Nordheim,A. (1996) Regulation of the c-fos promoter by the ternary complex factor Sap-1a and its coactivator CBP. Oncogene, 12, 1961–1969. [PubMed] [Google Scholar]
- 17.Chrivia J.C., Kwok,R.P., Lamb,N., Hagiwara,M., Montminy,M.R. and Goodman,R.H. (1993) Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature, 365, 855–859. [DOI] [PubMed] [Google Scholar]
- 18.Abraham S.E., Lobo,S., Yaciuk,P., Wang,H.G. and Moran,E. (1993) p300, and p300-associated proteins, are components of TATA-binding protein (TBP) complexes. Oncogene, 8, 1639–1647. [PubMed] [Google Scholar]
- 19.Kwok R.P., Lundblad,J.R., Chrivia,J.C., Richards,J.P., Bachinger,H.P., Brennan,R.G., Roberts,S.G., Green,M.R. and Goodman,R.H. (1994) Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature, 370, 223–226. [DOI] [PubMed] [Google Scholar]
- 20.Nakajima T., Uchida,C., Anderson,S.F., Lee,C.G., Hurwitz,J., Parvin,J.D. and Montminy,M. (1997) RNA helicase A mediates association of CBP with RNA polymerase II. Cell, 90, 1107–1112. [DOI] [PubMed] [Google Scholar]
- 21.Swope D.L., Mueller,C.L. and Chrivia,J.C. (1996) CREB-binding protein activates transcription through multiple domains. J. Biol. Chem., 271, 28138–28145. [DOI] [PubMed] [Google Scholar]
- 22.Chen H., Lin,R.J., Schiltz,R.L., Chakravarti,D., Nash,A., Nagy,L., Privalsky,M.L., Nakatani,Y. and Evans,R.M. (1997) Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell, 90, 569–580. [DOI] [PubMed] [Google Scholar]
- 23.Kamei Y., Xu,L., Heinzel,T., Torchia,J., Kurokawa,R., Gloss,B., Lin,S.C., Heyman,R.A., Rose,D.W., Glass,C.K. et al. (1996) A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell, 85, 403–414. [DOI] [PubMed] [Google Scholar]
- 24.Sheppard K.A., Rose,D.W., Haque,Z.K., Kurokawa,R., McInerney,E., Westin,S., Thanos,D., Rosenfeld,M.G., Glass,C.K. and Collins,T. (1999) Transcriptional activation by NF-kappaB requires multiple coactivators. Mol. Cell. Biol., 19, 6367–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith C.L., Onate,S.A., Tsai,M.J. and O’Malley,B.W. (1996) CREB binding protein acts synergistically with steroid receptor coactivator-1 to enhance steroid receptor-dependent transcription. Proc. Natl Acad. Sci. USA, 93, 8884–8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X.J., Ogryzko,V.V., Nishikawa,J., Howard,B.H. and Nakatani,Y. (1996) A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature, 382, 319–324. [DOI] [PubMed] [Google Scholar]
- 27.Bannister A.J. and Kouzarides,T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. [DOI] [PubMed] [Google Scholar]
- 28.Ogryzko V.V., Schiltz,R.L., Russanova,V., Howard,B.H. and Nakatani,Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 87, 953–959. [DOI] [PubMed] [Google Scholar]
- 29.Allfrey V., Faulkner,R.M. and Mirsky,A.E. (1964) Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl Acad. Sci. USA, 51, 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner B.M. (1998) Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell Mol. Life Sci., 54, 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W. and Bieker,J.J. (1998) Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc. Natl Acad. Sci. USA, 95, 9855–9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung H.L., Lau,J., Kim,A.Y., Weiss,M.J. and Blobel,G.A. (1999) CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol. Cell. Biol., 19, 3496–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imhof A., Yang,X.J., Ogryzko,V.V., Nakatani,Y., Wolffe,A.P. and Ge,H. (1997) Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol., 7, 689–692. [DOI] [PubMed] [Google Scholar]
- 34.Gu W. and Roeder,R.G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell, 90, 595–606. [DOI] [PubMed] [Google Scholar]
- 35.Du K., Asahara,H., Jhala,U.S., Wagner,B.L. and Montminy,M. (2000) Characterization of a CREB gain-of-function mutant with constitutive transcriptional activity in vivo. Mol. Cell. Biol., 20, 4320–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hunter T. and Karin,M. (1992) The regulation of transcription by phosphorylation. Cell, 70, 375–387. [DOI] [PubMed] [Google Scholar]
- 37.Lambert P.F., Kashanchi,F., Radonovich,M.F., Shiekhattar,R. and Brady,J.N. (1998) Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem., 273, 33048–33053. [DOI] [PubMed] [Google Scholar]
- 38.Wang H.G., Rikitake,Y., Carter,M.C., Yaciuk,P., Abraham,S.E., Zerler,B. and Moran,E. (1993) Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to the control of cell growth. J. Virol., 67, 476–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi H. and Teng,C. (1996) Promoter-specific activation of mouse lactoferrin gene by epidermal growth factor involves two adjacent regulatory elements. Mol. Endocrinol., 10, 732–741. [DOI] [PubMed] [Google Scholar]
- 40.Dang D.T., Zhao,W., Mahatan,C.S., Geiman,D.E. and Yang,V.W. (2002) Opposing effects of Kruppel-like factor 4 (gut-enriched Kruppel-like factor) and Kruppel-like factor 5 (intestinal-enriched Kruppel-like factor) on the promoter of the Kruppel-like factor 4 gene. Nucleic Acids Res., 30, 2736–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kojima S., Kobayashi,A., Gotoh,O., Ohkuma,Y., Fujii-Kuriyama,Y. and Sogawa,K. (1997) Transcriptional activation domain of human BTEB2, a GC box-binding factor. J. Biochem., 121, 389–396. [DOI] [PubMed] [Google Scholar]
- 42.Molinari E., Gilman,M. and Natesan,S. (1999) Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. EMBO J., 18, 6439–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salghetti S.E., Muratani,M., Wijnen,H., Futcher,B. and Tansey,W.P. (2000) Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc. Natl Acad. Sci. USA, 97, 3118–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salghetti S.E., Caudy,A.A., Chenoweth,J.G. and Tansey,W.P. (2001) Regulation of transcriptional activation domain function by ubiquitin. Science, 293, 1651–1653. [DOI] [PubMed] [Google Scholar]
- 45.Tansey W.P. (2001) Transcriptional activation: risky business. Genes Dev., 15, 1045–1050. [DOI] [PubMed] [Google Scholar]
- 46.Natesan S., Molinari,E., Rivera,V.M., Rickles,R.J. and Gilman,M. (1999) A general strategy to enhance the potency of chimeric transcriptional activators. Proc. Natl Acad. Sci. USA, 96, 13898–13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ottosen S., Herrera,F.J. and Triezenberg,S.J. (2002) Transcription. Proteasome parts at gene promoters. Science, 296, 479–481. [DOI] [PubMed] [Google Scholar]
- 48.Bannister A.J. and Kouzarides,T. (1995) CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J., 14, 4758–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim T.K., Kim,T.H. and Maniatis,T. (1998) Efficient recruitment of TFIIB and CBP-RNA polymerase II holoenzyme by an interferon-beta enhanceosome in vitro. Proc. Natl Acad. Sci. USA, 95, 12191–12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kraus V.B., Moran,E. and Nevins,J.R. (1992) Promoter-specific trans-activation by the adenovirus E1A12S product involves separate E1A domains. Mol. Cell. Biol., 12, 4391–4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song C.Z., Loewenstein,P.M., Toth,K. and Green,M. (1995) Transcription factor TFIID is a direct functional target of the adenovirus E1A transcription-repression domain. Proc. Natl Acad. Sci. USA, 92, 10330–10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Q., Imhof,A., Collingwood,T.N., Urnov,F.D. and Wolffe,A.P. (1999) p300 stimulates transcription instigated by ligand-bound thyroid hormone receptor at a step subsequent to chromatin disruption. EMBO J., 18, 5634–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolffe A.P. and Hayes,J.J. (1999) Chromatin disruption and modification. Nucleic Acids Res., 27, 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Korzus E., Torchia,J., Rose,D.W., Xu,L., Kurokawa,R., McInerney,E.M., Mullen,T.M., Glass,C.K. and Rosenfeld,M.G. (1998) Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science, 279, 703–707. [DOI] [PubMed] [Google Scholar]
- 55.Martinez-Balbas M.A., Bannister,A.J., Martin,K., Haus-Seuffert,P., Meisterernst,M. and Kouzarides,T. (1998) The acetyltransferase activity of CBP stimulates transcription. EMBO J., 17, 2886–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jarvis W.D., Turner,A.J., Povirk,L.F., Traylor,R.S. and Grant,S. (1994) Induction of apoptotic DNA fragmentation and cell death in HL-60 human promyelocytic leukemia cells by pharmacological inhibitors of protein kinase C. Cancer Res., 54, 1707–1714. [PubMed] [Google Scholar]
- 57.Svetlov S. and Nigam,S. (1993) Calphostin C, a specific protein kinase C inhibitor, activates human neutrophils: effect on phospholipase A2 and aggregation. Biochim. Biophys. Acta, 1177, 75–78. [DOI] [PubMed] [Google Scholar]
- 58.Gjertsen B.T., Mellgren,G., Otten,A., Maronde,E., Genieser,H.G., Jastorff,B., Vintermyr,O.K., McKnight,G.S. and Doskeland,S.O. (1995) Novel (Rp)-cAMPS analogs as tools for inhibition of cAMP-kinase in cell culture. Basal cAMP-kinase activity modulates interleukin-1 beta action. J. Biol. Chem., 270, 20599–20607. [DOI] [PubMed] [Google Scholar]
- 59.Wadgaonkar R., Phelps,K.M., Haque,Z., Williams,A.J., Silverman,E.S. and Collins,T. (1999) CREB-binding protein is a nuclear integrator of nuclear factor-kappaB and p53 signaling. J. Biol. Chem., 274, 1879–1882. [DOI] [PubMed] [Google Scholar]
- 60.Chen X. and Bieker,J.J. (1996) Erythroid Kruppel-like factor (EKLF) contains a multifunctional transcriptional activation domain important for inter- and intramolecular interactions. EMBO J., 15, 5888–5896. [PMC free article] [PubMed] [Google Scholar]
- 61.Geiman D.E., Ton-That,H., Johnson,J.M. and Yang,V.W. (2000) Transactivation and growth suppression by the gut-enriched Kruppel-like factor (Kruppel-like factor 4) are dependent on acidic amino acid residues and protein–protein interaction. Nucleic Acids Res., 28, 1106–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pandya K., Donze,D. and Townes,T.M. (2001) Novel transactivation domain in erythroid Kruppel-like factor (EKLF). J. Biol. Chem., 276, 8239–8243. [DOI] [PubMed] [Google Scholar]
- 63.Blobel G.A. (2000) CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood, 95, 745–755. [PubMed] [Google Scholar]
- 64.Parker D., Ferreri,K., Nakajima,T., LaMorte,V.J., Evans,R., Koerber,S.C., Hoeger,C. and Montminy,M.R. (1996) Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol. Cell. Biol., 16, 694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cardinaux J.R., Notis,J.C., Zhang,Q., Vo,N., Craig,J.C., Fass,D.M., Brennan,R.G. and Goodman,R.H. (2000) Recruitment of CREB binding protein is sufficient for CREB-mediated gene activation. Mol. Cell. Biol., 20, 1546–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oliner J.D., Andresen,J.M., Hansen,S.K., Zhou,S. and Tjian,R. (1996) SREBP transcriptional activity is mediated through an interaction with the CREB-binding protein. Genes Dev., 10, 2903–2911. [DOI] [PubMed] [Google Scholar]
- 67.Ouyang L., Chen,X. and Bieker,J.J. (1998) Regulation of erythroid Kruppel-like factor (EKLF) transcriptional activity by phosphorylation of a protein kinase casein kinase II site within its interaction domain. J. Biol. Chem., 273, 23019–23025. [DOI] [PubMed] [Google Scholar]