Abstract
Eukaryotic DNA polymerase (pol) α is a complex of four subunits. The subunits in the yeast Saccharomyces cerevisiae are: 167, 79, 62 and 48 kDa polypeptides. The p79 subunit has no known enzymatic functions, but it is essential for growth and chromosomal DNA replication. We have analyzed the interaction between the subunits of yeast pol α, particularly p167 and p79, using a yeast two-hybrid screen and deletion analysis. We have identified the interaction sites in each of these two subunits leading to p167·p79 complex formation, and correlated our results with the available genetic data. A detailed two-hybrid analysis, using the p79 gene as the bait and a yeast genomic DNA library, identified two independent groups of positive clones. One group that displayed strong positive interaction (δ1) with p79 represented a fusion of the p167 open reading frame at 3502 bp (Ile1168), and the second group, displaying weak positive interaction (δ2) with p79, had a fusion at 3697 bp (Asn1233) with the DNA-binding domain of the yeast Gal4 transcription factor. A detailed deletion analysis of the downstream region indicated the existence of two subdomains that interact with p79. Subdomain I encompasses a 65 amino acid segment between Ile1168 and Phe1232, and subdomain II is a 25 amino acid segment between Glu1259 and Leu1283. Deletion and two-hybrid interaction analysis of the p79 subunit of pol α revealed a complementary region with two subdomains: a 67 amino acid segment between Asn189 and Gln255 (I) and a 68 amino acid segment between Glu256 and Met323 (II). The p79 subdomains I and II appeared to interact with the p167 subdomains I and II, respectively. Analysis of interaction between p62 and various deletion clones of p167 did not result in an unambiguous and stable positive interaction in the two-hybrid screen between these two subunits. A strong interaction between p167 and p62 would probably require the presence of either p79 or p48 in the complex.
INTRODUCTION
Three nuclear DNA polymerases (pols) that may be involved in chromosomal DNA replication in eukaryotic cells are: pol α, pol δ and pol ε (1–3). In addition, a number of other pols have been discovered recently that are involved in DNA repair (1–7). Pols α, δ and ε have been implicated primarily through genetic and biochemical studies of chromosomal DNA replication in yeast and other eukaryotes (1–6). Pol α and δ are clearly involved in nuclear DNA replication, whereas the role of pol ε in DNA synthesis remains somewhat unclear (5–7). Pol ε appears to be involved in DNA repair as well as in intra-S phase checkpoint control (8,9). Among these three replicative enzymes, pol α has the unique property of exhibiting both primase and polymerase activities (5,6). Thus, it is of central importance in DNA replication, since it initiates DNA synthesis de novo by synthesizing short RNA/DNA primers.
Pol α is a hetero-tetrameric protein consisting of four subunits of 167, 79, 62 and 48 kDa. The largest subunit, p167, is the catalytic subunit. The p62 and p48 subunits are the primase subunits. Previous studies have demonstrated the existence of a large number of domains in the p167 subunit (10,11). Wang et al. (10) have defined six regions based on the evolutionary conservation of residues among the eukaryotic homologs of p167 (I–VI). Miyazawa et al. have defined an additional five (A–E) conserved domains (11). The most conserved is domain I, located between the residues Asp998 and Gly1005 (12). In addition, two Zn finger motifs (-C-X2-4-C-Xn-C-X2-4-C) are also present in the C-terminus, with unknown functional roles.
Unlike p167, p79 is unique in that there are no known motifs in this polypeptide that can be attributed to an enzymatic function. Despite the fact that p79 is essential for the function of pol α, the role(s) of p79 during pol α priming and DNA replication remains unclear (13). In human pol α, it has been demonstrated that p79 binds to the SV40 large T antigen, which is a DNA helicase, and is involved in SV40 viral DNA replication (14). Moreover, it has been shown to play an essential role in the early stages of the replication process and always exists as a tight complex with p167 (15). In addition, Brooke and Dumas (16) have demonstrated that p79 modulates the rate and extent of complex formation of p167 with p49 and p62.
The nature of pol α assembly and the functions of its various subunits, particularly p79, remain unclear. To elucidate the specific role of each pol α subunit, it is first essential to characterize their interactions with each other. Earlier studies by Mizuno et al. (17) indicated that in the case of mouse pol α, the corresponding mouse subunits interact with the large subunit at its C-terminus. However, the precise locations of the subunit interaction sites in each of these two polypeptides remain unknown. In this study, we have used a budding yeast two-hybrid screen to delineate the specific sites essential for the interaction of p167 and p79. The results have allowed us to determine the precise locations of the interaction sites in the structural assembly of pol α.
MATERIALS AND METHODS
Nucleic acids, enzymes and other reagents
Oligonucleotides were purchased from Sigma-Genosys and Fisher Chemicals Inc. (Pittsburgh, PA). All other reagents used in this study were ACS reagent or spectroscopy grade and obtained from Aldrich Chemical Co. (Milwaukee, WI). Vectors and reagents for the two-hybrid methods were from Clontech Laboratories, Palo Alto, CA. The Saccharomyces cerevisiae genomic DNA library in pACT vector was obtained as a gift from Dr S. Elledge of Baylor College of Medicine (18). The library was amplified in Escherichia coli and used for screening in two-hybrid protocols. Monoclonal antibodies against the Gal4 activation and binding domains were obtained from Clontech Laboratories.
Plasmids and DNA
The full-length p79 gene [POL12 open reading frame (ORF)] was PCR amplified using oligonucleotide primers containing an NcoI site in the 5′ end and a XhoI site in the 3′ end. The PCR-amplified DNA was cloned into pAS2-1 vector (Clontech Laboratories) at NcoI and SalI sites, and the resultant plasmid pAS2p79 was transformed in the budding yeast strain, Y190 (in GAL4 2H-2; MATa, ura3-52, his3-200, ade2-101, lys2-801, trp1-901, leu2-3, 112, gal4Δ, gal80Δ, cyhr2, LYS2::GAL1UAS-HIS3TATA-HIS3, MEL1 URA3:: GAL1UAS-GAL1TATA-lacZ). Expression of the fusion protein was verified by western blot analysis using mouse monoclonal antibody against the Gal4 DNA-binding domain (DBD). The p79 and p167 deletion fragments were created by PCR amplification of the p79 and p167 genes using the oligonucleotides shown in Figure 1A. The p79 deletion strategy is described in Figure 1B. These fragments were inserted into the NdeI and SalI sites in pAS2-1 and fused in-frame with the yeast Gal4 DBD.
Figure 1.
Oligonucleotide primers and deletion strategy. (A) Oligonucleotides used for the construction of plasmid pAS2-p79, plasmids with p79 deletions (pAS2-p79Δ0, pAS2-p79Δ1, pAS2-p79Δ2, pAS2-p79Δ3, pAS2-p79Δ4, pAS2-p79Δ5 and pAS2-p79Δ6) and plasmids containing p167 deletions (pACT-p167δ3 and pACT-p167δ4). (B) Schematic representation of p79 deletion constructs. The deletion fragments were generated by PCR amplifications as described in Materials and Methods.
All PCR amplifications were carried out using the hot start method with high-fidelity Pfu DNA polymerase (Stratagene Inc., San Diego, CA), 20 µM each of four dNTPs, 2 ng/µl primers, 10–20 ng of template DNA, and 25 cycles.
Transformation and library screening
Yeast plasmid or genomic DNA library transformation was carried out by electro-transformation using a BioRad Gene Pulser with an efficiency of ∼105 colony-forming units/µg. The yeast strain, Y190, containing pAS2p79 bait plasmid, was transformed with the pACT-yeast genomic DNA library and plated onto minimal SD plates lacking leucine (Leu–), tryptophan (Trp–) and histidine (His–) and containing 25 mM 3-amino triazole (3-AT) (SD-Trp–/Leu–/His–+3AT) (18). Putative positive colonies were replica plated at 48 and 96 h, respectively.
Filter-lift β-galactosidase assay
The putative positive colonies were screened for β-galactosidase activity by the filter-lift assay using VWR 75 mm filters (19). Whatman filters (75 mm in diameter) were pre-soaked in buffer Z (60 mM Na2HPO4, 60 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 38 mM β-mercaptoethanol pH 7.0) containing X-gal. Yeast colonies were streaked onto filters using a sterile toothpick. The streaked filters were frozen in an aluminum boat floating on liquid N2. Frozen filters were thawed by incubation at room temperature for 5–15 min. The freeze– thaw cycle was repeated three times. Filters were then placed on pre-soaked filters and incubated at 30°C for 1–2 h. Positive clones generally produced blue to bluish-green color in ∼1 h.
Quantitative β-galactosidase assay
The β-galactosidase activity of all positive interactors, as determined by the filter-lift assays, subsequently was quantified using the liquid o-nitrophenyl-β-d-galactopyranoside (ONPG) spectroscopic assay (20). Briefly, yeast were grown on SD-Trp–/Leu–/His–+3AT plates for 3–5 days at 30°C. Several colonies from each plate were inoculated into 5 ml of liquid SD-Trp–/Leu–/His–+3AT medium and grown overnight at 30°C with shaking at 250 r.p.m. From the liquid culture, 1 ml was removed, diluted into 5 ml of SD-Trp–/Leu–/His–+3AT medium and grown at 30°C to mid-log phase (A600 of 0.5–0.7). Optical density at 600 nm was used to normalize β-galactosidase activity to yeast cell density. A 1 ml aliquot of each yeast culture was centrifuged briefly to pellet the cells and resuspended in 1 ml of buffer Z. After the addition of 50 µl of chloroform and 50 µl of 0.1% SDS, the cell suspension was vortexed vigorously for 10 s. Samples were pre-incubated for 5 min at 30°C, and β-galactosidase assays were initiated by addition of 200 µl of 4 mg/ml ONPG. Reactions were terminated by addition of 400 µl of 1 M Na2CO3, and reaction mixtures were centrifuged for 10 min at 13 000 g. Absorption at 420 nm (A420) was measured for each sample and converted into β-galactosidase units (21). One unit of β-galactosidase is defined as the amount which hydrolyzes 1 µmol of ONPG to o-nitrophenol and d-galactose/min/cell (21).
DNA sequence analysis
DNA sequences of positive clones were determined by manual Sanger sequencing using a Sequanase 2.0 kit from US Biochemicals Inc. (Cleveland, OH) or by automated DNA sequencing at the Kimmel Cancer Center, Thomas Jefferson University, Philadelphia, PA.
RESULTS
Identification of gene products that interact with the p79 subunit of pol α
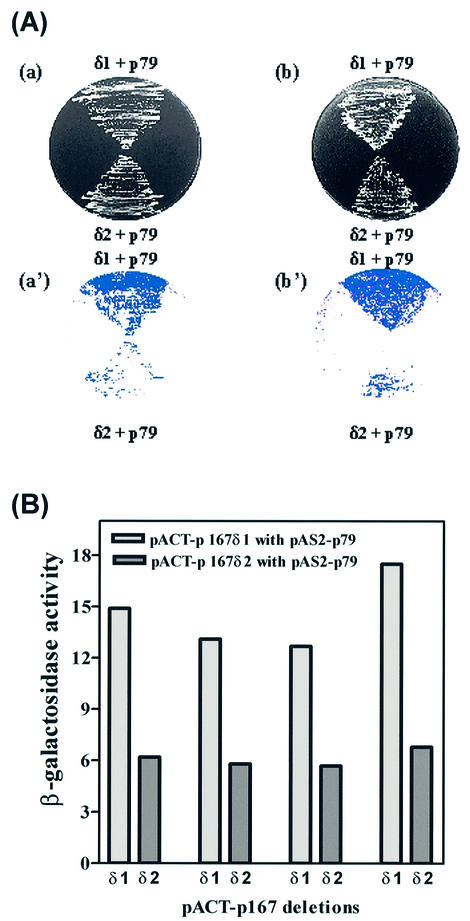
We sought to identify proteins that interact with p79 of pol α using the complete coding sequence of its gene, POL12. We have cloned the POL12 ORF into a binding domain fusion vector, pAS2-1, and the ORF was fused in-frame with the Gal4 DBD (18). The resulting plasmid pAS2-p79 was introduced into the yeast reporter strain, Y190. A fusion protein of the expected size was expressed in this strain and verified by western blotting using monoclonal antibody against the Gal4 DBD. Plasmid DNA from a pACT2-based yeast genomic DNA library containing ∼1 kb random fragments of yeast genomic DNA (obtained from Dr Steve Elledge of Baylor College of Medicine, TX) was transformed into the above strain containing pAS2-p79 (18). Approxim ately 6 × 106 transformants were plated on minimal SD-Trp–/Leu–/His–+3AT plates. We have isolated the transformants that grew on these selective SD plates after 48 and 96 h in order to distinguish between strong positive and weak positive clones. These clones were examined further for false positives. Only 48 clones were capable of growth in SD-Trp–/Leu–/His–+3AT plates and were truly positive when assayed for β-galactosidase activity using a filter-lift assay (Fig. 2A). The positive colonies were grown to mid-log phase in liquid culture, and the β-galactosidase activities were measured by spectrophotometric assay (Fig. 2B). These clones could be divided into two classes: strongly positive clones, which were isolated after 48 h, and weakly positive clones, which were isolated after 96 h. The former account for 21 of the 48 total clones, the latter for 23, with the remaining four clones uncertain. The results of the spectrophotometric assay mirrored the growth characteristics of these clones in the selective medium. The average β-galactosidase activity of the strongly positive group was ∼14 U, while that of the weakly positive group was ∼6 U.
Figure 2.
Interactions between p167 and p79 subunits. (A) The Y190 yeast strain was co-transformed with p167 deletions δ1/δ2 with pAS2-p79, where δ1 and δ2 refer to plasmids pACT-p167δ1 and pACT-p167δ2, respectively. The colonies were picked and re-streaked on SD- SD-Trp–/Leu– (a) and SD-Trp–/Leu–/His–+3AT plates (b) and incubated for 3 and 4 days, respectively. (a′) and (b′) represent the results from the X-gal colony filter-lift assay for the respective plates. (B) β-Galactosidase activity was determined quantitatively using the liquid culture assay as described in Materials and Methods for the interaction between pACT-p167δ1/δ2 and pAS2-p79.
Plasmids of the positive yeast colonies were transferred directly into E.coli strain KC8 (Leu– strain) by electroporation and selected for Leu+ so that only the activation domain or pACT recombinant plasmids could be selected. Plasmid DNA from each clone was purified and the insert DNA was sequenced in order to determine the identity of the selected clones. Analysis of the DNA sequences of these plasmids indicated that the majority of these 48 clones contained portions of p167 coding sequence. One group (pACTp167δ1) contained a fusion of the Gal4 DBD with the p167 sequence starting at 3502 bp (ATG as +1) or the Ile1168 codon, and the other group (pACTp167δ2) contained a fusion of the Gal4 DBD with p167 starting at 3697 bp or the Asn1233 codon (Table 1). The clones that were isolated as strongly β-galactosidase positive by their rapid growth (48 h) had a fusion at Ile1168, and the clones that were weakly positive grew only slowly (96 h) and contained a fusion at Asn1233.
Table 1. Plasmids used in this study.
| Plasmid | Description |
|---|---|
| pAS2-1-p79 | GAL4(1–147) TRP1-p79 (1–706) |
| pAS2-1-p79(Δ0) | GAL4(1–147) TRP1-p79 (189–706) |
| pAS2-1-p79(Δ1) | GAL4(1–147) TRP1-p79 (255–706) |
| pAS2-1-p79(Δ2) | GAL4(1–147) TRP1-p79 (323–706) |
| pAS2-1-p79(Δ3) | GAL4(1–147) TRP1-p79 (439–706) |
| pAS2-1-p79(Δ4) | GAL4(1–147) TRP1-p79 (492–706) |
| pAS2-1-p79(Δ5) | GAL4(1–147) TRP1-p79 (495–706) |
| PAS2-1-p79(Δ6) | GAL4(1–147) TRP1-p79 (547–706) |
| PACT2-p167δ1 | GAL4(768–881) LEU2-p167(1167–1468) |
| PACT2-p167δ2 | GAL4(768–881) LEU2-p167(1233–1468) |
| PGADT7-p167δ3 | GAL4(768–881) LEU2-p167(1259–1468) |
| PGADT7-p167δ4 | GAL4(768–881) LEU2-p167(1284–1468) |
| PGBKT7-p62 | GAL4(1–147) TRP1-p62 |
| PGADT7-p62 | GAL4(768–881) LEU2-p62 |
| PGBKT7-p48 | GAL4(1–147) TRP1-p48 |
| PGADT7-p48 | GAL4(768–881) LEU2-p48 |
In order to define the p79-binding site in p167, we compared the amino acid sequence of this region with those of other eukaryotic p167 homologs (Fig. 3). Alignment of the region between Val1161 and Cys1290 indicated >50% homology in this domain between the seven p167 polypeptides. The conserved domain E, as described by Miyazawa et al. (11), is a prominent part. Thr1169 corresponds to Ser1180 in mouse pol α. Izumi et al. (22) have shown that the mutation in Ser1180 in this subunit of mouse pol α leads to a temperature-sensitive phenotype as well as a general defect in the catalytic activity of mouse pol α.
Figure 3.
Sequence alignment and conservation of the binding domain sequence of the p167 subunit. The alignment was carried out using the SEQWEB 3.0 PRETTY analysis program (GCG Corporation, Madison, WI). The location of domain E, as described previously (11), is underlined.
Identification of protein interaction site(s) in the p79 subunit of pol α
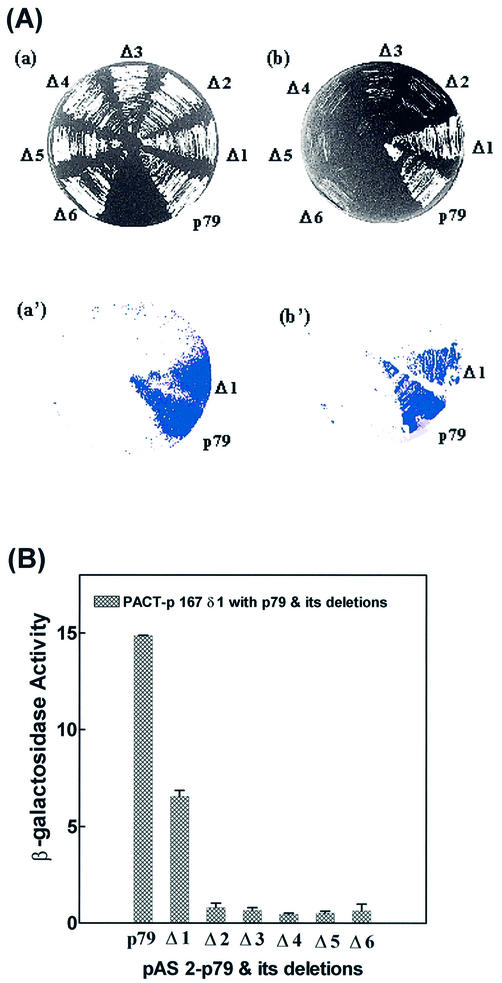
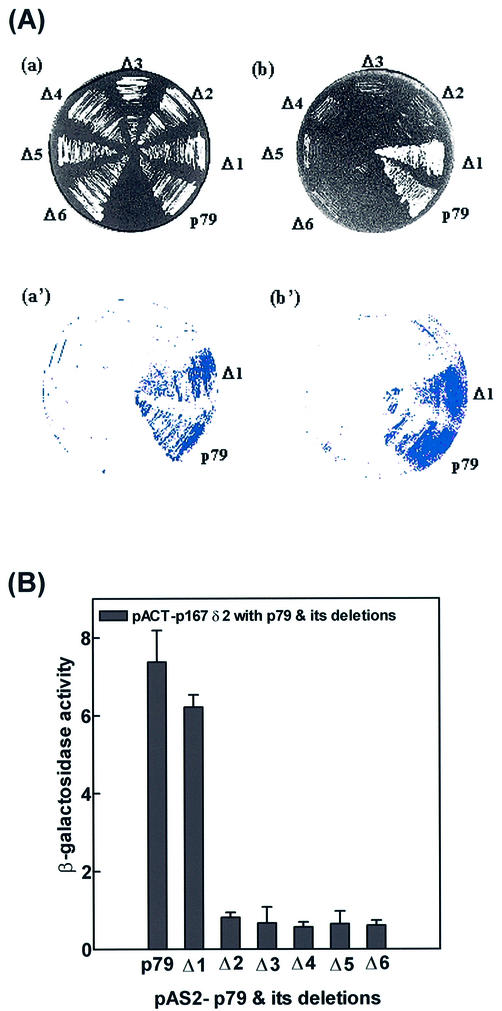
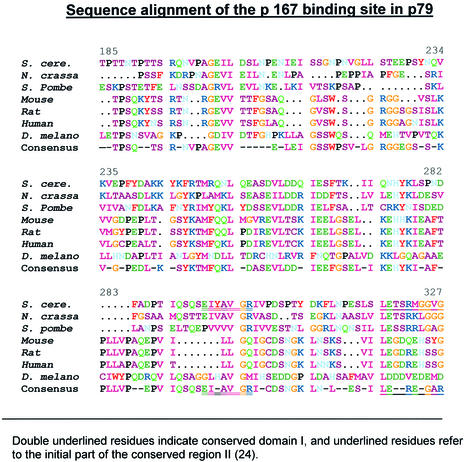
We have carried out a deletional two-hybrid analysis to identify the p167-binding site in p79. The construction of deletions in the p79 gene is shown in Figure 1B. For each deletion, the PCR-amplified truncated gene fragment was inserted in the pAS2-1 vector with in-frame fusion to the Gal4 DBD (Table 1). Yeast strain Y190 was co-transformed with p79 deletion clones in pAS2-1 vector and either pACTp167δ1 or pACTp167δ2 to examine interaction of p79 deletions. The N-terminal ends of the deletion clones were as follows: p79Δ1, Glu256; p79Δ2, Gly324; p79Δ3, His440; p79Δ4, Ser493; p79Δ5, Asn498; p79Δ6, Gly548 (Fig. 1, Table 1). These six deletion clones and wild-type p79 were analyzed for positive interactions with pactp167δ1 (strong interactor) and pactp167δ2 (weak interactor) in a two-hybrid screen. All of the clones were found to be negative except for the wild type and p79Δ1. These two clones grew almost equally well in SD-Trp–/Leu– and SD-Trp–/Leu–/His–+3AT plates with both p167 deletions (Figs 4A and 5A, Table 2). However, in a β-galactosidase filter-lift assay, the wild-type p79 clone was consistently stronger than the p79Δ1 clone. Further quantitation of β-galactosidase activities of these clones indicated that the β-galactosidase activity of pACTp79Δ1 was ∼50% of that observed with the wild type when the activation domain interactor was pACTp167δ1 (Fig. 4B). However, with pACTp167δ2 as the activation domain interactor, the wild-type p79 and p79Δ1 were indistinguishable in both the β-galactosidase assay and growth in SD-Trp–/Leu–/His–+3AT medium (Fig. 5A and B, Table 2). Comparison of their interactions with pACTp167δ1 and pACTp167δ2 indicated that a sequence immediately upstream of Glu256 in p79 is important in the overall interaction of the two subunits. However, this sequence is not required for interaction with pACTp167δ2 that also lacks an N-terminal sequence that is necessary for optimal interaction of p167 with p79. Consequently, it is possible that the region between Ile1168 and Asn1233 interacts with the N-terminal sequence immediately adjacent to Glu256, and then binding sites in the two subunits orient parallel in N→C directions. The sequence alignment of p79 in different species is given in Figure 6.
Figure 4.
Interactions between p167δ1 and p79 deletions. (A) The Y190 yeast strain was co-transformed with pACT-p167δ1 and pAS2-p79 and its deletions Δ1–Δ6. All p79 deletions were constructed in pAS2-1 vector as described in Materials and Methods. Colonies were picked and replica- plated on SD-Trp–/Leu– (a) and SD-Trp–/Leu–/His–+3AT plates (b), and incubated for 3 and 4 days, respectively. (a′) and (b′) represent results from the X-gal colony filter-lift assay for the respective plates. (B) β-Galactosidase activity was determined quantitatively using the liquid culture assay as described in Materials and Methods for the interaction between pACT-p167δ1 and pAS2-p79 and its deletions.
Figure 5.
Interactions between p167δ2 and p79 deletions. (A) The Y190 yeast strain was co-transformed with pACT-p167 δ2, pAS2-p79 and its deletions Δ1–Δ6. All p79 deletions were constructed in pAS2 vector as described in Materials and Methods. Colonies were picked and replica- plated on SD-Trp–/Leu– (a) and SD-Trp–/Leu–/His–+3AT plates (b), and incubated for 3 and 4 days, respectively. (a′) and (b′) represent the results from the X-gal colony filter-lift assay for the respective plates. (B) β-Galactosidase activity was determined quantitatively using the liquid culture assay as described in Materials and Methods for the interaction between pACT-p167 δ2 and pAS2-p79 deletions.
Table 2. Phenotypes of various two-hybrid interactions: growth on SD-Trp–/Leu– and SD-Trp–/Leu–/His– +3AT plates.
| Plasmid | SD-Trp–/Leu– | SD-Trp–/Leu–/His– +3AT |
|---|---|---|
| p79+ p167δ1 | + | + |
| p79(Δ0) + p167δ1 | + | + |
| p79(Δ1) + p167δ1 | + | + |
| p79(Δ2) + p167δ1 | + | – |
| p79(Δ3) + p167δ1 | + | – |
| p79(Δ4) + p167δ1 | + | – |
| p79(Δ5) + p167δ1 | + | – |
| p79(Δ6) + p167δ1 | + | – |
| p79 + p167δ2 | + | + |
| p79(Δ0) + p167δ2 | + | + |
| p79(Δ1) + p167δ2 | + | + |
| p79(Δ2) + p167δ2 | + | – |
| p79 + p167δ3 | + | + |
| p79(Δ0) + p167δ3 | + | + |
| p79(Δ1) + p167δ3 | + | + |
| p79 + p167δ4 | + | – |
| p79(Δ0) + p167δ4 | + | – |
| p79(Δ1)+ p167δ4 | + | – |
| p62 + p167δ1 | + | – |
| p62 + p167δ2 | + | – |
| p62 + p167δ3 | + | – |
| p62 + p167δ4 | + | – |
| p62 + p79 | + | – |
| p62 + p79(Δ0) | + | – |
| p62 + p79(Δ1) | + | – |
| p48 + p167δ1 | + | – |
| p48 + p167δ2 | + | – |
| p48 + p167δ3 | + | – |
| p48 + p167δ4 | + | – |
| p48 + p79 | + | – |
| p48 + p79(Δ0) | + | – |
| p48 + p79(Δ1) | + | – |
| p62 + p48 | + | + |
Figure 6.
Sequence alignment and conservation of the binding domain sequence in the p79 subunit. Alignment was carried out as described in the legend of Figure 3. The locations of previously defined (24) regions I and II are underlined.
Detailed mapping of the interaction sites in p167 and p79
In order to define the binding sites further, we have created two deletion clones p167δ3 and p167δ4 in the region downstream of Asn1233 (Table 1). Deletion clone p167δ3 contained a deletion of amino acids 1–1258 and started from Glu1259 (pACTp167δ3), and deletion clone p167δ4 contained a deletion of residues 1–1283 and started from Glu1284 (pACTp167δ4). We have examined the interactions of these two deletions with p79 deletions: pAS2-p79, pAS2-p79Δ1 and pAS2-p79Δ2 by growth on SD-Trp–/Leu–/His–+3AT plates, the colony filter-lift β-galactosidase assay and a semi- quantitative liquid culture β-galactosidase assay.
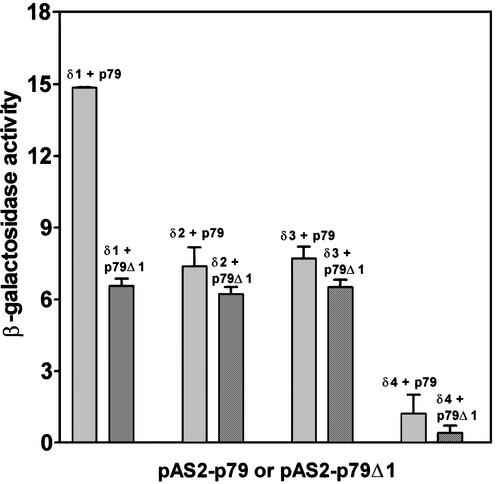
The yeast strain Y190 was co-transformed with each of these p167 deletions and pAS2-p79, pAS2-p79Δ1 or pAS2-p79Δ2 plasmids. These six sets of transformants were plated on SD-Trp–/Leu– and SD-Trp–/Leu–/His–+3AT plates and incubated for 3 and 4 days, respectively, and the results of subsequent analyses are presented in Figure 7 and Table 2. The pACTp167δ3 deletion was found to be positive upon interaction with both pAS2-p79 and pAS2-p79Δ1 (Fig. 7). The semi-quantitative β-galactosidase assay using mid-log phase yeast cells indicated that the pACTp167δ3–pAS2-p79 interaction had ∼50% activity compared to that observed with the pACTp167δ1–pAS2-p79 interaction. However, the pACTp167δ3–pAS2-p79Δ1 interaction was found to be comparable with p167δ3–pAS2-p79. In addition, interactions of both pACTp167δ3 and pACTp167δ2 with either pAS2-p79 or pAS2-p79Δ1 were indistinguishable in the semi-quantitative liquid culture β-galactosidase assay. These interactions had ∼50% in activity compared with that observed with p167δ1–pAS2-p79. These results have profound implications for our understanding of the organization of the interaction domain of p167. In particular, it appears that two subdomains of this region make significant contributions to the interaction between the p167 and p79 subunits: a 65 amino acid sequence between Ile1168 and Phe1232 and a 25 amino acid sequence between Glu1259 and Leu1283. In contrast, the region between Asn1233 and Asn1258 did not appear to contribute to the interaction, and it most probably acts as a spacer region between the two interacting subdomains (Fig. 7). Deletions downstream of Glu1284 (p167δ4) did not appear to show any significant level of interaction. Therefore, we concluded that the downstream region may not make any significant contribution to this interaction.
Figure 7.
Interaction of pACT-p167 deletions with p79. β-Galactosidase activities were determined quantitatively using the liquid culture assay as described in Materials and Methods for the interaction of pACT-p167 deletions δ1/δ2/δ3/δ4 with pAS2-p79 as well as with its deletion pAS2-p79 Δ1.
None of the p167 deletions, as described above, showed any positive interaction with p79Δ2. Therefore, deletion of the N-terminal residues 1–323 of p79 eliminated any interaction with p167, which is comparable with that observed earlier with p167δ1 and p167δ2 (Figs 4 and 5, Table 1). However, deletion of residues 1–254 in p79 (p79Δ1) resulted in attenuation of its interaction with p167 (Figs 4 and 5). Consequently, we have created a deletion in the region upstream of residue 254. Deletion clone p79Δ0, in which residues 1–188 were deleted, had β-galactosidase activity comparable with that of p79 itself and it appeared to maintain the same levels of interaction with the various p167 deletions as wild-type p79. These results indicated that at least the major part of the interaction domain in p79 is confined in a 133 amino acid segment between residues Asn189 and Met323. Next, we have carefully examined the organization of this region in terms of its interactions with the two subdomains of p167. The results, presented in Figure 7, demonstrate that subdomain I in p167δ1 appears to interact with the subdomain between Asn189 and Gln255. Interestingly, these subdomains in both p167 and p79 are similar in size, which could be of some significance in their interactions. The other subdomain, consisting of the region between Glu256 and Met323, appeared to interact with subdomain II of p167. Unlike subdomain I, these two subdomains are not comparable in size.
Interactions between the primase subunit p62 and p167
Earlier studies demonstrated that p62 also interacts with p167 in the C-terminal domain (17). Therefore, we have examined the interactions of C-terminal deletions of p167, as described above, with the p62 ORF fused to the Gal4 DBD (Table 1). We have also examined the interactions between p62 and p167 in a number of different ways using the two-hybrid screen (Table 2). In these two-hybrid screens, we did not detect any significant interaction between these two subunits. In analyses involving growth in SD-Trp–/Leu–/His–+3AT plates as well as colony filter-lift assays, we did not find any definitive and measurable interactions between these subunits. We have also examined the interaction between these two subunits in the opposite orientation by fusing p62 with the Gal4 activation domain and p167δ1 with the Gal DBD (data not shown). In combination with studies reported earlier, it appears that a stable interaction between these subunits may require the presence of p79 or p48 (12,17). It could be due to induction of appropriate conformational changes associated with p167 and p79/p48 interaction or a direct physical interaction of p62 with p167, and either p79 or p48. Considering the fact that these subunits are large, which may allow combined multiple interactions with p62, the second possibility appears quite reasonable. In addition, we did not find any interaction between p48 and p167 or p79, as expected (Table 2). However, we observed positive interactions between p48 and p62, as anticipated (Table 2).
DISCUSSION
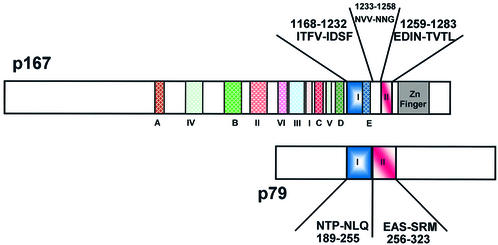
Subunit interaction domain(s) in the p167 subunit
Results of two-hybrid analysis revealed that two short polypeptide subdomains in the C-terminus of p167 (subunit A) are responsible for interaction with p79 (subunit B). The sequence between these two subdomains, Asn1233–Gly1258, did not appear to be involved in the p167–p79 interaction. Among the 12 known conserved domains of p167, domain E is present in subdomain I, between Leu1206 and Arg1235 (11). Therefore, domain E may have a function in mediating the protein–protein interaction between p167 and p79.
The p167 subdomain I appeared to exhibit a high degree (42%) of evolutionary conservation. The observed sequence variability could account for the species specificity. Izumi et al. (22) have shown that a mutation of Ser1180 in this region of the mouse subunit, which is equivalent to Thr1169 in S.cerevisiae, resulted in a temperature-sensitive phenotype in mouse pol α. They have demonstrated that this mutation leads to severe enzymatic defects in DNA synthesis at the permissive as well as the non-permissive temperatures (22). In conjunction with the results presented here, it appears that this mutation in p79 impairs its interaction with p167. An impaired interaction between these two subunits could explain the observed temperature-sensitive phenotype of the mouse pol α mutant.
Earlier studies with in vivo deletion analysis and nuclear transport of the mouse subunit B and pol α subassemblies pointed to the involvement of nearly the entire C-terminal domain of p167 (17). Here, we demonstrate that in budding yeast, the Zn finger domain is not likely to be involved in the p167–p79 interaction, unlike that observed with mouse pol α (17). These results correlate well with the recent report by Smith and Nasheuer (23) on the involvement of the C-terminus of p167 in the assembly of human pol α.
Precise localization of the subunit interaction sites in p79
An important function of the N-terminal region in human subunit B has been elucidated by Collins et al. (14) using an in vitro SV40 viral DNA replication system. A part of this region appears to interact with the T-antigen of SV40 virus, although the exact location of the binding site remains unknown. Foiani et al. have demonstrated that in yeast p79, insertion of two amino acids between Leu261 and Asp262 leads to a complete loss of in vivo function of pol α, i.e. cell growth, whereas similar insertions at other locations did not have such drastic effects (15). Sequence analyses of subunit B of human pol α have indicated that the two conserved sequences, as proposed by Makiniemi et al. (24), are present in subdomain II in the yeast subunit B (Fig. 6).
The p167-binding site in p79 is located between Asn189 and Met323. The results of insertion and deletion analysis of p79 by Foiani et al. (15) indicate that Asn189–Ser225 have little impact on pol α function. Taken together, these results indicate that the interaction of subdomain I is probaby localized to the region between Thr226 and Gln255, although we cannot rule out the possibility that the residues between Asn189 and Ser225 may make a minor contribution to the overall interaction. Subdomain II of p79 is located between Glu256 and Met323. Two-hybrid analysis indicates that p79 subdomain II interacts with p167 subdomain II. Similar to the binding site in p167, the binding site in p79 may also have a spacer region that we have not detected in our deletion and two-hybrid analysis.
The two-hybrid analysis of the interaction of p79 deletions with p167δ1 and p167δ2 revealed that a major interaction site of p167 is present in the region between these two sequences. Further analysis of p167 deletions, p167δ3 and p167δ4, has enabled us to postulate a model that displays the precise location of the interacting binding sites of p167 and p79 (Fig. 8). Subdomain I of p167 interacts specifically with subdomain I of p79, and the same is true for subdomains II. The gap between the two subdomains of p167 did not appear to participate in the interaction with p79, but it may provide a site for interaction of other pol α subunits, such as p62. For yeast pol α, our results indicated that the Zn finger domain is not essential for p162 and p79 interaction. These results are consistent with those of Smith and Nasheuer (23) demonstrating that the Zn finger domain is not essential for interaction between these two subunits in human pol α.
Figure 8.
Schematic representation of the domains involved during the interaction of p167 with p79. Regions I–IV as defined by Wang et al. (10) and regions A–E as mentioned by Miyazawa et al. (11) are indicated with different colors in the p167 polypeptide. The amino acid sequences of subdomains I (blue) and II (red) are as shown in Figures 3 and 6.
Interaction between p62 and p167
In our detailed two-hybrid analysis, we did not detect any measurable interaction of p62 with any of the clones including the wild-type p79. The possibility that the two-hybrid screen employed here is simply not sensitive enough to detect this interaction cannot be ruled out. Brooke et al. (16) have demonstrated that p167 can form a complex with the primase hetero-dimer (p62 + p48) in yeast, which is supported by the results of Smith and Nasheuer (23). The rate and extent of formation of the p167·p62·p48 ternary complex were significantly enhanced by p79 (16). However, these studies did not demonstrate a direct interaction of mouse subunit C with mouse subunit A in the absence of subunit B. Taken together, these results suggest that in yeast, either p79 or p48 is required for a stable p62–p167 interaction. A second possibility is that in the absence of p48, p62 interacts with both p167 and p79 in order to form a stable complex, and such a complex is not formed with either one of them. It is clear from both of these studies that p167 and p79 form a strong and stable complex. Therefore, the interaction of p62 with both of these subunits (p167 and p62) in a three-dimensional geometry is a likely possibility and may have profound consequences for the assembly and function of pol α. Further structure–function analyses including structural determination of these polypeptides are essential for understanding the assembly and mechanism of action of this unique enzyme.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Drs Robert Nagele and Michael Henry of this department, and Dr Randall Hammond of Johnson and Johnson Inc. for critical review of this manuscript. This work was supported by grant (GM-36002) from the National Institute of General Medical Sciences of the National Institutes of Health of the US Public Health Service.
REFERENCES
- 1.DePamphilis M.L. (1993) Eukaryotic DNA replication: anatomy of an origin. Annu. Rev. Biochem., 62, 29–63. [DOI] [PubMed] [Google Scholar]
- 2.Linn S. (1991) How many pols does it take to replicate nuclear DNA? Cell, 66, 185–187. [DOI] [PubMed] [Google Scholar]
- 3.Wang T.S. (1996) DNA polymerases. In DePamphillis,M.L. (ed.), DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 461–493.
- 4.Haracska L., Prakash,L. and Prakash,S. (2002) Role of human DNA polymerase kappa as an extender in translesion synthesis. Proc. Natl Acad. Sci. USA, 99, 16000–16005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang T.S. (1991) Eukaryotic DNA polymerases. Annu. Rev. Biochem., 60, 513–552. [DOI] [PubMed] [Google Scholar]
- 6.Waga S. and Stillman,B. (1994) Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature, 369, 207–212. [DOI] [PubMed] [Google Scholar]
- 7.Zhou J.Q., He,H., Tan,C.K., Downey,K.M. and So,A.G. (1997) The small subunit is required for functional interaction of DNA polymerase delta with the proliferating cell nuclear antigen. Nucleic Acids Res., 25, 1094–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vlatkovic N., Guerrera,S., Li,Y., Linn,S., Haines,D.S. and Boyd,M.T. (2000) MDM2 interacts with the C-terminus of the catalytic subunit of DNA polymerase epsilon. Nucleic Acids Res., 28, 3581–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishida C., Reinhard,P. and Linn,S. (1988) DNA repair synthesis in human fibroblasts requires DNA polymerase delta. J. Biol. Chem., 263, 501–510. [PubMed] [Google Scholar]
- 10.Wang T.S., Wong,S.W. and Korn,D. (1989) Human DNA polymerase alpha: predicted functional domains and relationships with viral DNA polymerases. FASEB J., 3, 14–21. [DOI] [PubMed] [Google Scholar]
- 11.Miyazawa H., Izumi,M., Tada,S., Takada,R., Masutani,M., Ui,M. and Hanaoka,F. (1993) Molecular cloning of the cDNAs for the four subunits of mouse DNA polymerase alpha–primase complex and their gene expression during cell proliferation and the cell cycle. J. Biol. Chem., 268, 8111–8122. [PubMed] [Google Scholar]
- 12.Copeland W.C. and Wang,T.S. (1993) Mutational analysis of the human DNA polymerase alpha. The most conserved region in alpha-like DNA polymerases is involved in metal-specific catalysis. J. Biol. Chem., 268, 11028–11040. [PubMed] [Google Scholar]
- 13.Nasheuer H.P., Moore,A., Wahl,A.F. and Wang,T.S. (1991) Cell cycle-dependent phosphorylation of human DNA polymerase alpha. J. Biol. Chem., 266, 7893–7903. [PubMed] [Google Scholar]
- 14.Collins K.L., Russo,A.A., Tseng,B.Y. and Kelly,T.J. (1993) The role of the 70 kDa subunit of human DNA polymerase α in DNA replication. EMBO J., 12, 4555–4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foiani M., Marini,F., Gamba,D., Lucchini,G. and Plevani,P. (1994) The B subunit of the DNA polymerase alpha–primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol., 14, 923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brooke R.G. and Dumas,L.B. (1991) Reconstitution of the Saccharomyces cerevisiae DNA primase–DNA polymerase protein complex in vitro. The 86-kDa subunit facilitates but is not required for complex formation. J. Biol. Chem., 266, 10093–10098. [PubMed] [Google Scholar]
- 17.Mizuno T., Yamagishi,K., Miyazawa,H. and Hanaoka,F. (1999) Molecular architecture of the mouse DNA polymerase alpha–primase complex. Mol. Cell. Biol., 19, 7886–7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durfee T., Becherer,K., Chen,P., Yeh,S.H., Yang,Y., Kilburn,A., Lee,W. and Elledge,S. (1993) The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev., 7, 555–569. [DOI] [PubMed] [Google Scholar]
- 19.Breeden L. and Nasmyth,K. (1985) Regulation of yeast Ho gene. Cold Spring Harbor Symp. Quant. Biol., 50, 643–650. [DOI] [PubMed] [Google Scholar]
- 20.Flick J.J. and Johnston,M. (1990) Two systems of glucose repression of the GAL1 promoter in Saccharamyces cerevisiae. Mol. Cell. Biol., 10, 4757–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller J. (1992) A Short Course in Bacterial Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. p 74–90.
- 22.Izumi M., Miyazawa,H., Harakawa,S., Yatagai,F. and Hanaoka,F. (1994) Identification of a point mutation in the cDNA of the catalytic subunit of DNA polymerase alpha from a temperature-sensitive mouse FM3A cell line. J. Biol. Chem., 269, 7639–7644. [PubMed] [Google Scholar]
- 23.Smith R.W. and Nasheuer,H.P. (2002) Control of complex formation of DNA polymerase alpha–primase and cell-free DNA replication by the C-terminal amino acids of the largest subunit p180. FEBS Lett., 527, 143–146. [DOI] [PubMed] [Google Scholar]
- 24.Makiniemi M., Pospjech,H., Kilpelainen,S., Jokela,M., Vihinen,M. and Syvaojaj,E. (1999) A novel family of DNA polymerase associated B subunits. Trends Biochem. Sci., 24, 14–16. [DOI] [PubMed] [Google Scholar]