Abstract
N1-methyladenosine (m1A) is found at position 58 in the T-loop of many tRNAs. In yeast, the formation of this modified nucleoside is catalyzed by the essential tRNA (m1A58) methyltransferase, a tetrameric enzyme that is composed of two types of subunits (Gcd14p and Gcd10p). In this report we describe the cloning, expression and characterization of a Gcd14p homolog from the hyperthermophilic bacterium Thermus thermophilus. The purified recombinant enzyme behaves as a homotetramer of ∼150 kDa by gel filtration and catalyzes the site- specific formation of m1A at position 58 of the T-loop of tRNA in the absence of any other complementary protein. S-adenosylmethionine is used as donor of the methyl group. Thus, we propose to name the bacterial enzyme TrmI and accordingly its structural gene trmI. These results provide a key evolutionary link between the functionally characterized two-component eukaryotic enzyme and the recently described crystal structure of an uncharacterized, putative homotetrameric methyltransferase Rv2118c from Mycobacterium tuberculosis. Interest ingly, inactivation of the T.thermophilus trmI gene results in a thermosensitive phenotype (growth defect at 80°C), which suggests a role of the N1-methylation of tRNA adenosine-58 in adaptation of life to extreme temperatures.
INTRODUCTION
One of the characteristics of functional mature tRNA is the presence of a variety of chemically altered nucleosides that are formed by enzymatic modification of the primary transcript during the sequential process of tRNA maturation. To date, more than 80 different modifications have been identified in tRNA from various organisms, the type and location of each of these modified nucleosides being dependent on the tRNA species as well as the organism from which they originate (1,2).
Among these modified nucleosides, N1-methyladenosine (m1A) is found in position 58 in the T-loop of many tRNAs from the three domains of life. The enzyme responsible for this modification, S-adenosyl-l-methionine (AdoMet)-dependent tRNA (m1A58) methyltransferase (MTase) (EC 2.1.1.36) has been studied for a long time using cell extracts as well as purified (or partially purified) enzymes from several organisms: mammals, Tetrahymena pyriformis, Dictyostelium discoideum, Thermus flavus and Thermus thermophilus (3–9; reviewed in 10). However, the genes corresponding to these tRNA (m1A58) MTases had yet to be identified.
Recently, mutants of the yeast Saccharomyces cerevisiae affected in GCN4 translational regulation allowed to identify two essential genes (GCD10 and GCD14), encoding two non-identical subunits of a newly characterized nuclear tRNA (m1A58) MTase (54 and 44 kDa, respectively) (11). Detailed analysis of the corresponding recombinant proteins, both in vitro and in vivo, demonstrated first that the purified recombinant Gcd10p/Gcd14p enzyme behaves as a tetramer upon gel filtration with an apparent molecular weight of 200 kDa and second that Gcd14p is responsible for AdoMet binding and presumably catalysis of the methyltransfer reaction, while Gcd10p appears to be essential for tRNA binding (12). These results are in perfect agreement with computational sequence analysis (13), which suggested that only Gcd14p, and not Gcd10p, possesses a set of conserved motifs typical for the Rossmann fold MTase superfamily (reviewed in 14). Recently, Gcd14p and its orthologs from other Eukaryota were found to be closely related to a group of prokaryotic open reading frames (ORFs), which were previously annotated as hypothetical protein isoaspartate MTases in sequence databases. These prokaryotic proteins share not only the AdoMet-binding site, but also other strongly conserved motifs and invariant residues with the Gcd14p family. Hence, it has been hypothesized that they all correspond to true tRNA (m1A58) MTases (15). On the other hand, orthologs of the Gcd10p protein were found only in Eukaryota. Interestingly, protein fold-recognition analysis (a procedure of assessment of whether a given protein sequence is compatible with the structure of any other protein in the database) revealed that, despite the absence of the characteristic MTase motifs, Gcd10p is structurally and evolutionarily related to Gcd14p (15). Both protein families share the same MTase domain, but it seems that Gcd10p has lost the catalytic and cofactor-binding residues in the course of divergent evolution, and presumably has been adapted to fulfill the RNA-binding and/or structural role (15). These observations suggest that tRNA (m1A58) MTases from yeast and possibly all eukaryotic species evolved by gene duplication and subfunctionalization to produce a heteromultimeric protein complex, while their prokaryotic orthologs remained homomultimers encoded by a single gene (15).
More recently, the crystal structure of a bacterial Gcd14p ortholog, the Rv2118c protein from Mycobacterium tuberculosis, has been determined at 1.98 Å resolution (16). The crystallographic analysis demonstrated that Rv2118c exhibits a ‘classical’ MTase fold, that it binds AdoMet and forms a homotetramer with two pairs of tight homodimers stabilized by a relatively small number of contacts. Unfortunately, the MTase activity of Rv2118c has not been demonstrated explicitly, neither in vivo nor in vitro. Hence, it remains to be confirmed if the prokaryotic orthologs of Gcd14p share the RNA (m1A) specificity with the yeast enzyme. Indeed, while the orthologous relationship of proteins from distinct domains of life detected by sequence and/or structural analysis generally allows prediction of a common type of enzymatic reaction, it does not necessarily imply an identical physiological function. For instance, it has been demonstrated that RrmJ and Trm7p, true orthologs from Bacteria and Eukaryota, share the same 2′-O-ribose MTase activity, but modify distinct targets: rRNA and tRNA, respectively (17,18). Likewise, one of the S.cerevisiae orthologs of E.coli tRNA-pseudouridine synthase TruA, acting at positions 38, 39 and 40 of the anticodon arm of E.coli tRNAs, is Pus1p catalyzing the formation of pseudouridine at eight positions in S.cerevisiae tRNAs (positions 26–28, 34–36, 65 and 67) but not at positions 38–40 (19).
In this report, we demonstrate that in T.thermophilus, a newly identified very close homolog of Rv2118c from M.tuberculosis has tRNA (m1A58) MTase activity in vivo and in vitro. This result provides evidence for not only an evolutionary, but also a functional relationship between the bacterial and eukaryotic MTases, related to Rv2118c and Gcd14p, respectively. In view of its newly established function, the bacterial tRNA (m1A58) MTase is named TrmI and its coding gene trmI. Interestingly, inactivation of the T.thermophilus trmI gene results in a thermosensitive phenotype (growth defect at 80°C), which suggests a role of the N1-methylation of tRNA adenosine-58 in adaptation of life to extreme temperatures.
MATERIALS AND METHODS
Sequence information for the T.thermophilus trmI gene has been deposited in the EMBL Nucleotide Sequence Database (accession no. AJ516007).
Strains, media, growth conditions and general procedures
Thermus thermophilus (HB27) was grown at 70°C on a rotary shaker platform in complex medium (20). Growth conditions for E.coli were described previously (21). Ampicillin was used at a concentration of 50 µg/ml and kanamycin at 30 µg/ml, except where otherwise indicated. Restriction endonucleases and T4 DNA ligase were purchased from Roche Diagnostics (Basel, Switzerland), except where otherwise indicated. Oligonucleotides were synthesized by Invitrogen (Carlsbad, CA). Sequences and purposes of oligonucleotides used in this study are given in Table 1. Thermus thermophilus genomic DNA was isolated according to Ramseier et al. (22). Escherichia coli (strain MC1061) and T.thermophilus total (bulk) tRNA were prepared as described (23). Protein concentrations were measured using the Bio-Rad (Hercules, CA) protein assay, using bovine serum albumin as a standard.
Table 1. Oligonucleotides used in this study.
| Oligonucleotides | Sequencea,b | Purpose |
|---|---|---|
| ML-1 | TATTAATACGACTCACTATAGGCCCCGTGGTGTAGTTGGTTAAC | Amplification of T.thermophilus tRNAAsp gene |
| ML-2 | TATCCTGGCGGCCCCGACGGGACTCGAACCC | Amplification of T.thermophilus tRNAAsp gene |
| ML-13 | CGTCCTAAGGTCAAAATGGTATGCGTTTTGACAC | Amplification of knt (Kmr) gene |
| ML-55 | CGTCCTAAGGTAAACGCGTGGAGGTGAAGCATGAATGGACCAATA | Amplification of knt (Kmr) gene |
| ML-56 | CGTACGCGTGTGGAGCGCTTCATTGAGGAGATCC | Amplification of T.thermophilus 16S rRNA promoter |
| ML-57 | CGTACGCGTCTGCTTGGGCTTTCGCCGCGCAGAA | Amplification of T.thermophilus 16S rRNA promoter |
| ML-83 | TATCATATGGCGTGGCCGGGACCGCTACTCCTCA | Amplification of T.thermophilus trmI gene |
| ML-84 | TATAGATCTTAGGAGCCCTTCCATCGCCTAAGGGCCAC | Amplification of T.thermophilus trmI gene |
| PGCD-1 | CGGAATTCGTGCAGAGCCTTCAGGACTG | Amplification of a 2.9 kb T.thermophilus genomic DNA fragment bearing the trmI gene |
| PGCD-2 | CGGAATTCCCTGGTAGTCGCCAAGCCTC | Amplification of a 2.9 kb T.thermophilus genomic DNA fragment bearing the trmI gene |
| PGCD-3 | CTTTGACCTGGAGCGGTACC | Checking for knt (Kmr) insertion within the T.thermophilus trmI gene |
| PGCD-4 | GCTCCCGCAAGACGGCGTGG | Checking for knt (Kmr) insertion within the T.thermophilus trmI gene |
aT7 RNA polymerase promoter is in italic.
bRestriction enzyme sites (MvaI, Bsu36I, MluI, NdeI, BglII, EcoRI) are underlined.
Cloning and T7 in vitro transcription of the T.thermophilus tRNAAsp gene
The general procedure for generating an in vitro transcript of T.thermophilus tRNAAsp is based on the method described previously (24,25). The sequence coding for T.thermophilus tRNAAsp was amplified by PCR using the ML-1 and ML-2 oligonucleotides and Pwo DNA polymerase (Roche Diagnostics). The PCR product was cloned into the SmaI site of the pUC18 vector, giving plasmid pML1 in which the tRNA sequence is flanked by a 5′ T7 promoter and a 3′ MvaI restriction site. Radioactive (32P) in vitro transcripts were obtained using MvaI-digested pML1 plasmid as a template. [α-32P]ATP and [α-32P]GTP were purchased from ICN Biomedicals (Costa Mesa, CA) and T7 RNA polymerase was purchased from Roche Diagnostics.
Cloning of the T.thermophilus trmI gene
The T.thermophilus trmI gene was amplified by PCR from T.thermophilus HB27 DNA using the ML-83 and ML-84 oligonucleotides and the GC-rich PCR System (Roche Diagnostics). The 782 bp amplified product was cloned into the pCR2.1 vector using the TOPO TA cloning kit (Invitrogen), giving the pML19 plasmid. The 769 bp NdeI–BglII fragment of pML19 was then cloned between the NdeI and BamHI restriction sites of the pET15b expression vector (Novagen, Madison, WI) resulting in the pML24 plasmid. This plasmid allowed T7 expression in E.coli of the T.thermophilus TrmI protein bearing an N-terminal His-tag.
Expression and purification of the recombinant T.thermophilus TrmI protein
The His-tagged T.thermophilus TrmI protein was expressed in E.coli strain Rosetta (DE3) (Novagen). Transformed cells were grown at 37°C in 2 l of Luria broth supplemented with ampicillin to an optical density at 660 nm (OD660) of 0.5. At this stage, isopropylthiogalactopyranoside (IPTG) (Roche Diagnostics) was added to a final concentration of 1 mM to induce recombinant protein expression. Cells were harvested after 3 h incubation at 37°C and resuspended in 100 ml of buffer A (50 mM Tris–HCl pH 8.5, 500 mM KCl) and lyzed by a 30 min sonication at 4°C using a Vibracell 75041 sonicator (40% amplitude). The lysate was cleared by centrifugation (20 000 g for 30 min) and was applied to a column of Chelating Sepharose Fast Flow (1 × 30 cm; Amersham Biosciences, Little Chalfont, UK) charged with Ni2+. The column was washed with buffer A and the adsorbed material was eluted with a linear gradient (750 ml, from 0 to 1.0 M) of imidazole in buffer A. Eluted fractions were analyzed by SDS–PAGE. The fractions containing the recombinant protein were pooled and dialyzed against buffer A supplemented with 200 mM imidazole. These high KCl and imidazole concentrations were needed to keep the protein soluble at high concentration (5 mg/ml). In the absence of imidazole, the highest concentration obtained for soluble enzyme was 0.5 mg protein/ml. The final yield was estimated to be 25 mg purified protein/l culture. Aliquots (200 µl) of the resulting preparation (5 mg protein/ml) were flash-frozen in liquid nitrogen and stored at –80°C.
tRNA MTase assays
Two types of tRNA MTase assays were used. The first method consisted of measuring the amount of 14C transferred to tRNA using [methyl-14C]AdoMet as methyl donor. The reaction mixture (200 µl) consisted of 50 mM Tris–HCl pH 8, 10 mM MgCl2, 50 µg of total (bulk) E.coli tRNA, 25 nCi [methyl-14C]AdoMet (50 mCi/mmol; Amersham Biosciences) and 5 µg of purified enzyme. After 30 min incubation at 60°C, the reaction was stopped by phenol extraction and trichloroacetic acid precipitation. The precipitate was filtered (Whatman GF/C) and the radioactivity on the filter was measured by scintillation counting. For experiments requiring the identification of the 14C-methylated nucleotide, the nucleic acids obtained after the phenol extraction were ethanol precipitated and digested overnight by 5 µg of nuclease P1 (Sigma Chemical Co., St Louis, MO). The nucleotides were separated by 2-dimensional thin layer chromatography (TLC) on 20 × 20 cm cellulose plates (Merck, Whitehouse Station, NJ) using the solvent system described previously (26): first dimension developed with isobutyric acid/conc. NH4OH/water (66:1:33 v/w/v); second dimension developed with 0.1 M sodium phosphate pH 6.8/(NH4)2SO4/n-propanol (100:60:2 v/w/v). The radioactive compounds were detected by autoradiography. The nucleotides were identified using a reference map (27).
The second type of tRNA MTase assay, involving in vitro transcribed, 32P-labeled T.thermophilus tRNAAsp as substrate, was based on the procedure previously described (28). The reaction mixture (300 µl) consisted of 50 mM Tris–HCl pH 8, 10 mM MgCl2, 106 c.p.m. of the radioactive transcript, 500 µM AdoMet and 5 µg of purified enzyme or 100 µg protein of a crude T.thermophilus extract. After the incubation period at 60°C, identification of the methylated nucleotides formed in the tRNA transcript was performed as described above. The reaction was stopped by phenol extraction and the tRNA was ethanol precipitated. The recovered radioactive tRNA was then completely digested by either nuclease P1 (1 µg) or RNase T2 (0.1 U), both from Sigma Chemical Co., in the presence of 5 µg total yeast tRNA (Roche Diagnostics) as carrier, except when otherwise stated. The nucleotides were separated by 2-dimensional TLC on 10 × 10 cm cellulose plates (see above). The radioactive compounds were detected by autoradiography. Quantification of each identified nucleotide was performed after scratching the corresponding spots off the plates and counting the radioactivity by liquid scintillation.
Plasmid constructions for trmI gene inactivation in T.thermophilus HB27
Oligonucleotides ML-56 and ML-57, bearing MluI sites, were used to amplify the 16S rRNA promoter (29) using Pwo DNA polymerase. The resulting 106 bp fragment was cloned in the SmaI site of pUC18 resulting in plasmid pML10.
The thermoresistant knt gene derivative (Kmr) encoding kanamycin nucleotidyltransferase (30) was PCR-amplified from the Thermus plasmid pYK189 (a gift of Y. Koyama) using Pwo DNA polymerase and the oligonucleotides ML-13 and ML-55. These oligonucleotides contain Bsu36I/MluI and Bsu36I restriction sites, respectively.
A 2.9 kb T.thermophilus genomic fragment bearing the trmI gene was amplified from T.thermophilus HB27 genomic DNA using the GC-rich PCR kit (Roche Diagnostics) and the oligonucleotides PGCD-1 and PGCD-2 with EcoRI restriction sites to facilitate the cloning into the cognate site of pUC18. The resulting plasmid was named pTT1.
The 100 bp MluI fragment of pML10 was ligated in its cognate site of plasmid pML11a. In the resulting construct, pML11b, the knt (Kmr) gene is transcribed from the 16S rRNA promoter. Plasmid pTT1 was digested with Bsu36I (New England Biolabs, Beverly, MA). The 509 bp Bsu36I fragment, in the coding region of trmI, was replaced by the 838 bp Bsu36I fragment of pML11b to create the ligation product pML51 used for homologous recombination. Transformation of the T.thermophilus HB27 strain was performed as described by Koyama et al. (31).
RESULTS
Cloning and expression of an ORF (trmI) of the T.thermophilus HB27 genome encoding a putative RNA (m1A) MTase
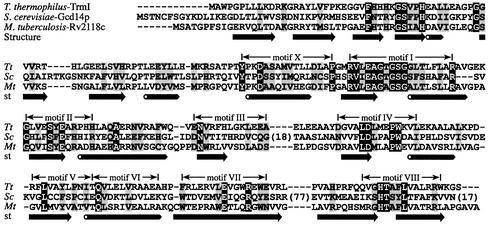
The amino acid sequence of the putative RNA (m1A) MTase Rv2118c from M.tuberculosis was used as a query in a tBLASTn search (32) of the T.thermophilus HB27 genome database at the Göttingen Genomics Laboratory (http://www.g2l.bio.uni-goettingen.de/). This search revealed a highly similar sequence (expectation value = 7 × 10–35), corresponding to a 255 amino acid ORF. PSI-BLAST search (32) of the non-redundant database at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) revealed a further 23 bacterial and archaeal homologs of Rv2118c, which are all predicted (i.e. not characterized experimentally) MTases, as well as the genuine eukaryotic tRNA (m1A58) MTase from S.cerevisiae (Gcd14p) and its eukaryotic homologs. As reported previously (15), no Gcd14p/Rv2118c orthologs could be found in E.coli and related Proteobacteria. Figure 1 shows the sequence alignment of Gcd14p, Rv2118c and their ortholog from T.thermophilus [for more extensive sequence alignment of prokaryotic and eukaryotic Gcd14p/Gcd10p family members see Bujnicki (15)]. The newly identified T.thermophilus ORF (called trmI) encodes a polypeptide which shares 39% identical plus 13% similar residues with Rv2118c and 22% identical plus 15% similar residues with ScGcd14p. This conservation includes all sequence motifs identified previously in this protein family (15).
Figure 1.
Sequence alignment of S.cerevisiae Gcd14p, M.tuberculosis Rv2118c and their T.thermophilus ortholog (TrmI). The size of insertions in Gcd14p omitted for clarity is indicated in parentheses. Highly conserved residues are shown on a black background and residues with a similar physico-chemical character are on a gray background. The experimentally determined secondary structure of M.tuberculosis Rv2118c is shown below its sequence as arrows (strands) and tubes (helices). Within the catalytic domain (central and C-terminal parts of the protein), the nine sequence motifs typical for the ‘classical’ AdoMet-dependent MTase family (14) are indicated.
The T.thermophilus trmI gene was PCR-amplified and cloned into the pET15b expression vector, allowing the production of an N-terminal His-tagged protein in E.coli (see Materials and Methods). The expression of the recombinant protein was induced with 1 mM IPTG in the E.coli strain Rosetta (DE3) carrying extra copies of tRNA genes (argU, argW, ileX, glyT, leuW, proL, metT, thrT, tyrU and thrU) specific for rare E.coli codons, to aid this expression. The His-tagged protein was purified by immobilized metal ion affinity chromatography and analyzed for its homogeneity by denaturing gel electrophoresis. Figure 2A shows that the recombinant protein is at least 90% pure, the apparent molecular weight (32 kDa) being close to the expected estimated value from the amino acid sequence (30.7 kDa).
Figure 2.
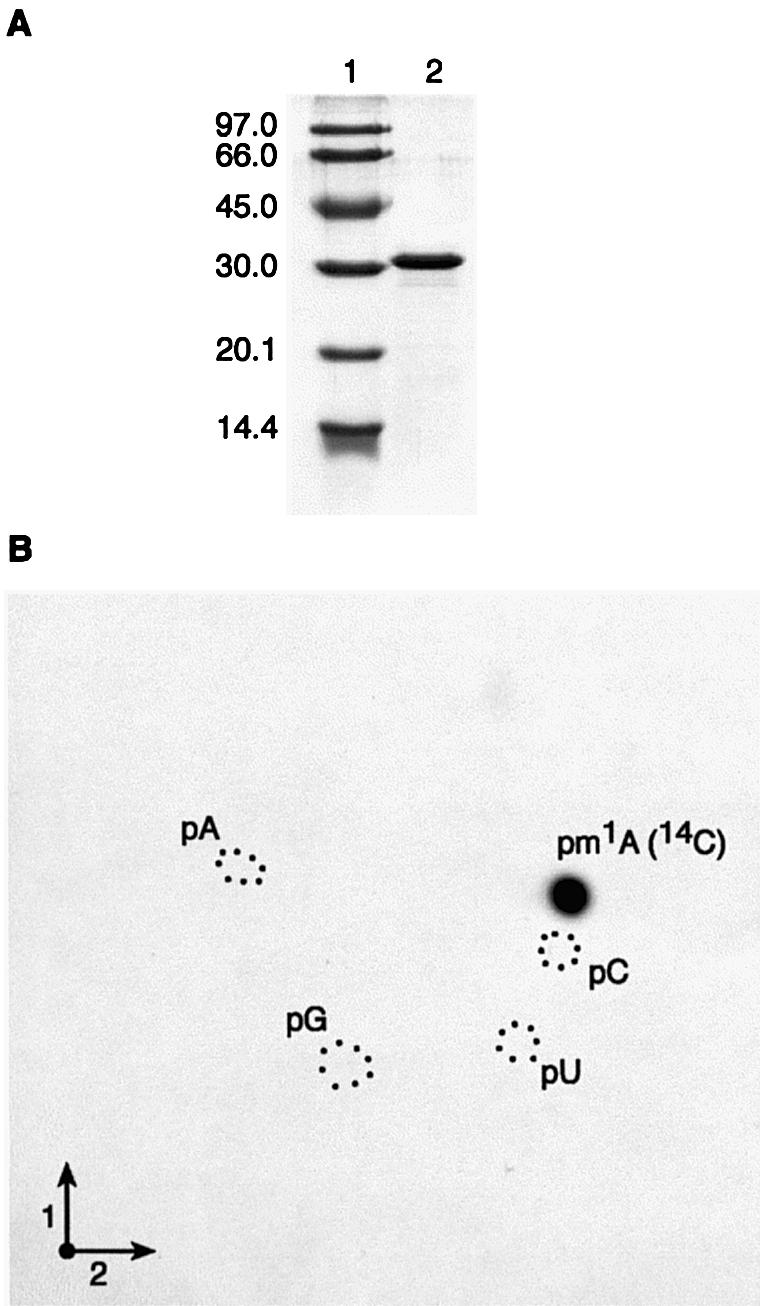
Affinity-purified T.thermophilus TrmI catalyzes the formation of m1A in E.coli tRNA in vitro. (A) SDS–PAGE analysis of the purified TrmI protein. Lane 1, molecular weight marker (Pharmacia-Biotech); lane 2, purified protein. (B) Autoradiography of a 2-dimensional chromatogram of 5′-phosphate nucleosides on a thin layer cellulose plate. Total (bulk) E.coli tRNA (50 µg) was incubated in the presence of [methyl-14C]AdoMet and 5 µg of the purified TrmI protein as described in Materials and Methods. After 30 min incubation at 60°C, the tRNA was recovered, digested by nuclease P1 and the resulting nucleotides were analyzed by 2-dimensional TLC as described in Materials and Methods. Circles in dotted lines show the migration of the four canonical nucleotides used as UV markers.
The T.thermophilus TrmI protein functions as a tRNA (m1A58) MTase in vitro
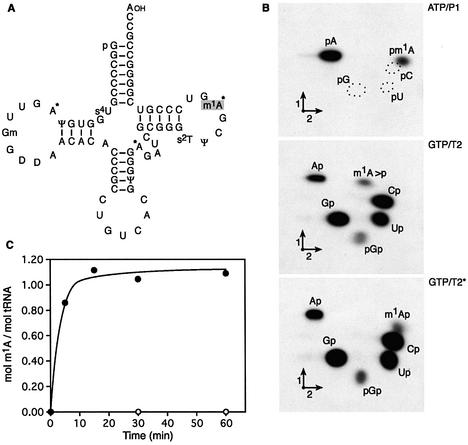
In order to determine whether the TrmI protein displays tRNA MTase activity, the purified recombinant protein was first incubated at 60°C for 30 min with [methyl-14C]AdoMet and unfractionated (bulk) E.coli tRNA. The rationale for using bulk E.coli tRNA arises from the fact that none of the tRNAs from E.coli contains a methyl group on the universally conserved A58 (2); also no GCD14 ortholog has been identified in the whole E.coli genome by bioinformatics analysis (15). After the incubation, the tRNA was recovered by phenol extraction and ethanol precipitation and further completely hydrolyzed by nuclease P1. The resulting hydrolysate was then analyzed by 2-dimensional cellulose TLC followed by autoradiography. The result shown in Figure 2B revealed the presence on the TLC plate of a single radioactive compound with migration characteristics identical to that of N1-methyladenosine 5′-phosphate (pm1A).
In a second series of experiments, the purified enzyme was incubated under identical experimental conditions as above but with either an [α-32P]ATP- or an [α-32P]GTP-labeled precursor tRNA substrate obtained after in vitro transcription by T7 RNA polymerase of a synthetic T.thermophilus tRNAAsp gene. Examination of the tRNA sequence database reveals indeed that m1A is found at position 58 (in the T-loop) of this particular T.thermophilus tRNA species (33) (Fig. 3A) as well as in several other tRNAs from this organism (2). After the incubation, the formation of N1-methyladenosine in the T.thermophilus tRNAAsp was analyzed as above, except that the [α-32P]ATP-labeled tRNA was completely hydrolyzed with nuclease P1 to generate 5′-phosphate nucleosides with the [32P]phosphate present only in AMP (5′P) and AMP (5′P) derivatives (Fig. 3B, ATP/P1), while the [α-32P]GTP-labeled tRNA was digested with RNase T2, thus generating the different 3′-phosphate nucleosides of which only those that were 5′-adjacent to GMP harbored a 32P-radiolabeled phosphate (nearest neighbours analysis; Fig. 3B, GTP/T2 and GTP/T2*). Figure 3B (ATP/P1) again demonstrates the unambiguous presence of pm1A, while Figure 3A (GTP/T2 and GTP/T2*) attests to the presence of m1Ap 5′-adjacent to a GMP in T.thermophilus tRNAAsp. The difference of m1Ap migration in the two chromatograms results from the accumulation of the 2′-3′ cyclic form of m1Ap (m1A>p) when the ratio of tRNA to RNase T2 is high and of exclusively the non-cyclic 3′-phosphate nucleoside when the ratio of tRNA to RNase T2 is low (excess of RNase T2). Of note, the dinucleotide A*G (where * indicates 32P) exists only in three positions in T.thermophilus tRNAAsp. Based on the sequencing data (33), only the A*G dinucleotide at positions 58–59 could correspond to G*m1A (Fig. 3A). Quantification of the relative amount of 32P in the different radioactive spots on the TLC plates reveals that about 1 mole of m1A (*m1A as in Fig. 3B, ATP/P1, or m1A* as in Fig. 3B, GTP/T2*) is formed per mole of tRNA after 30 min incubation at 60°C. Figure 3C shows the corresponding kinetics.
Figure 3.
Affinity-purified T.thermophilus TrmI protein methylates A58 of in vitro transcribed T.thermophilus tRNAAsp. (A) Nucleotide sequence of T.thermophilus tRNAAsp (33). m1A58 is shown in a gray box. The adenosines 5′-adjacent to a guanosine are indicated by an asterisk. (B) Autoradiograms of 2-dimensional chromatograms of P1 or T2 hydrolysates of [α-32P]ATP- or [α-32P]GTP-labeled T.thermophilus tRNAAsp transcripts incubated for 1 h at 60°C in the presence of the purified TrmI protein (see Materials and Methods for details). In the case of the chromatogram designated GTP/T2*, the T2 hydrolysis was carried out in the absence of carrier yeast tRNA. Circles of dotted lines show the migration of the pG, pC and pU nucleotides used as UV markers. (C) Kinetics of in vitro formation of m1A58 in T.thermophilus tRNAAsp in the presence (closed circles) or absence (open circles) of the purified TrmI protein.
Altogether, these data demonstrate that the TrmI protein from T.thermophilus is the enzyme that catalyzes the formation of m1A at position 58 in the T-loop of tRNAs.
Thermus thermophilus TrmI protein is a tetramer
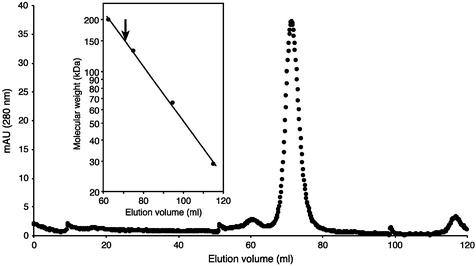
The theoretical molecular weight of the T.thermophilus TrmI sequence identified in the present work is 28 582 Da. Taking into account that the closely related Rv2118c protein was reported to be a tetramer according to the X-ray crystallographic analysis and gel filtration experiments (16), we investigated the quaternary structure of T.thermophilus TrmI by gel filtration experiments. Figure 4 shows the result of gel filtration chromatography of the purified T.thermophilus TrmI protein. The apparent molecular weight of the protein is ∼150 kDa (see inset of Fig. 4), consistent with the protein being tetrameric.
Figure 4.
Estimation of the apparent molecular weight of the purified His-tagged T.thermophilus TrmI protein by gel filtration chromatography on a Superdex 200 prep grade 16/60 column (Pharmacia Biotech). The sample consisted of 2.5 mg protein in 50 mM Tris–HCl pH 8.5, 500 mM KCl, 200 mM imidazole. Elution was performed with the same buffer. (Inset) Calculation of the apparent molecular weight of the T.thermophilus TrmI protein. The standard consisted of carbonic anhydrase from bovine erythrocytes (29 kDa), bovine serum albumin (66 kDa), bovine serum albumin dimer (132 kDa) and β-amylase from sweet potato (200 kDa). The elution volume corresponding to the T.thermophilus TrmI protein is indicated by an arrow.
Inactivation of the T.thermophilus trmI gene results in a thermosensitive phenotype
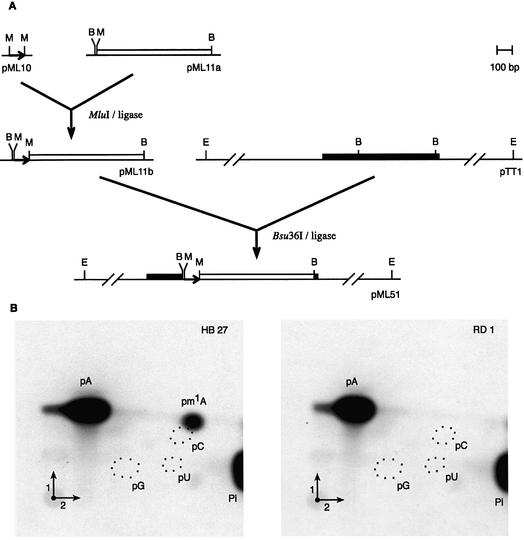
To generate a T.thermophilus strain lacking tRNA (m1A58) MTase activity, a kanamycin resistance cassette was introduced into the chromosomal trmI gene by homologous recombination. The mutant strain was obtained by a double homologous recombination event between the wild-type chromosomal allele and a non-replicating plasmid-borne allele in which a part of the trmI coding sequence was replaced by the kanamycin nucleotidyltransferase (knt) gene. Indeed, homologous recombination is a highly efficient mechanism in T.thermophilus (31) and insertional inactivation of a target gene with an antibiotic resistance cassette has already been reported (34). To facilitate selection, the knt gene was transcribed from the 16S rRNA promoter of T.thermophilus. Figure 5A outlines the different steps leading to the plasmid construct used for homologous recombination. The resulting disrupted strain (RD1) was resistant to 50 µg/ml kanamycin. The presence of the antibiotic resistance cassette within the trmI gene was checked by PCR (result not shown). Figure 5B shows that the RD1 strain lacks TrmI activity.
Figure 5.
Generation of a T.thermophilus mutant lacking TrmI activity, by homologous recombination. (A) Plasmid construction in E.coli for T.thermophilus trmI gene inactivation (see Materials and Methods for details). The trmI gene is represented by a solid bar, the thermostable knt gene (Kmr) by an open bar and the 16S promoter by a horizontal arrow. Restriction sites are indicated as follows: B, Bsu36I; E, EcoRI; M, MluI. (B) Autoradiography of 2-dimensional chromatograms of P1 hydrolysates of [α-32P]ATP-labeled T.thermophilus tRNAAsp transcripts incubated for 1 h at 60°C in the presence of a crude extract of the T.thermophilus wild-type strain (HB27) or trmI mutant (RD1). Circles of dotted lines show the migration of the pG, pC and pU nucleotides used as UV markers.
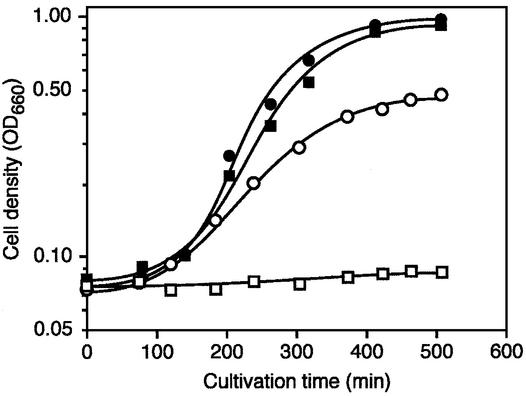
The growth rates of T.thermophilus HB27 (wild-type) and RD1 strains were compared at different temperatures. Figure 6 shows that at 70°C, mutant and wild-type strains had the same growth rate. However, cells lacking tRNA (m1A58) MTase activity failed to grow at 80°C. This is a clear indication that the methylation machinery (either the presence of the enzyme and/or the presence of a methylated A58 in tRNA) is (or are) required for T.thermophilus growth at high temperature.
Figure 6.
The T.thermophilus mutant lacking TrmI activity shows a temperature-sensitive phenotype. Growth curves of the T.thermophilus wild-type strain (HB27) (circles) and of the trmI mutant (RD1) (squares). Growth temperature was 70°C (filled symbols) or 80°C (open symbols).
DISCUSSION
In this work we have characterized the tRNA (m1A58) MTase of T.thermophilus encoded by the gene now designated trmI. This gene was identified by amino acid sequence similarity with the Gcd14p subunit of genuine eukaryotic tRNA (m1A58) MTase from S.cerevisiae and the putative RNA (m1A) MTase Rv2118c from M.tuberculosis. The interest in studying the bacterial enzyme is due to the fact that none of the bacterial genomes sequenced so far contains an ortholog of S.cerevisiae GCD10, encoding a subunit which is absolutely required for the MTase activity of the yeast enzyme. Indeed, the purified recombinant Gcd14p subunit, which binds AdoMet, cannot bind tRNA in vitro in the absence of the Gcd10p subunit (12). Also, both GCD14 and GCD10 are essential S.cerevisiae genes, attesting to the importance of each subunit for the methylation of tRNA in vivo (11). Together with the fact that Gcd14p and Gcd10p are homologous, these observations led one of us to suggest that the heteromultimeric eukaryotic tRNA (m1A58) MTase could have arisen from gene duplication and divergent evolution of a possibly homomultimeric prokaryotic enzyme (15). Here we have demonstrated that the purified recombinant protein encoded by the T.thermophilus trmI gene catalyzes in vitro the formation of m1A58 in T.thermophilus tRNAAsp. This enzyme acts in the absence of a Gcd10p-like protein and behaves as a homotetramer in vitro. The same multimeric state was demonstrated for the closely related Rv2118c protein of M.tuberculosis both in the crystal and in solution (16). Moreover, close inspection of the crystal structure of this MTase revealed that this tetramer is formed by two pairs of extensively interacting subunits stabilized by relatively small numbers of contacts between the two dimers. A preliminary homology modeling study of the T.thermophilus TrmI protein structure based on the Rv2118c structure suggests that the dimer–dimer interactions in these two proteins are nearly identical (data not shown). However, to what extent the binding of the tRNA substrate and the catalysis depend on the spatial organization of TrmI into a tetramer has still to be determined.
Of note, beside tRNA (m1A58) MTase, there are two other examples of multimeric tRNA modifying enzymes in yeast: the Tad2p/Tad3p complex and the newly identified Trm8p/Trm82p complex, which catalyze, respectively, the site-specific formation of inosine at position 34 of the anticodon in several tRNAs (35) and N7-methylguanosine at position 46 in the variable loop of several tRNAs (36). Interestingly the corresponding E.coli enzymes (TadA and YggH) have been recently identified (37,38) and shown to catalyze the expected corresponding reactions in vitro without the need of any other protein, attesting again that the structural information needed by the bacterial counterpart is present in only one type of polypeptide chain.
Thermus thermophilus is an extreme thermophilic bacterium that grows over a wide range of temperature (48–85°C). Cell components of this organism, including tRNA, are found to be much more thermostable than those of mesophiles. Nucleotide modification can provide a significant physicochemical contribution to tRNA stabilization at high temperatures (39,40). Moreover, for T.thermophilus, a positive correlation was observed between the growth temperature of the cells and the presence of 2-thioribothymidine in position 54 of tRNA (s2T54) (41). The presence of a thiol group on ribothymidine leads to an increase in the melting temperature of T.thermophilus tRNA1Ile (42). However, other modifications, in particular m1A58 and 2′-O-methylguanosine 18 (Gm18), were also proposed to stabilize the T.thermophilus tRNA at high temperatures (42). In this work we have shown that a trmI mutant of T.thermophilus lacking tRNA (m1A58) MTase activity shows a temperature-sensitive phenotype (growth defect at 80°C). The presence of a methyl group on N1 of A58 not only reinforces the hydrophobicity around the methyl group but also introduces a positive charge on the tRNA which may also contribute to the overall stability of the tRNA molecule. However, a role of the TrmI protein, independent of its MTase activity, cannot be ruled out.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to J.-P. Waller for critical reading of the manuscript and to J.-P. ten Have for the art work. We thank Y. Koyama (Tsukuba, Japan) for the gift of the pYK189 plasmid. L.D. is a Research Associate of the FNRS (Fonds National de la Recherche Scientifique). J.M.B. is supported by an EMBO/HHMI Young Investigator award. This work was supported in Belgium by grants from the FRFC (Fonds pour la Recherche Fondamentale Collective), from the French Community of Belgium (Actions de Recherches Concertées) and from the Université Libre de Bruxelles (Fonds E. Defay). In France, H.G. was supported by grants from the CNRS (Centre National de la Recherche Scientifique) and from the French Ministry of Scientific Research (Geomex Program). L.D. and H.G. benefited from a travel grant from the Ministère des Affaires Etrangères (France) and the Commissariat Général aux Relations Internationales de la Communauté Française de Belgique: Actions Intégrées Franco-Belges (Programme Tournesol no. 99035).
DDBJ/EMBL/GenBank accession no. AJ516007
REFERENCES
- 1.McCloskey J.A. and Crain,P.F. (1998) The RNA modification database–1998. Nucleic Acids Res., 26, 196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sprinzl M., Horn,C., Brown,M., Ioudovitch,A. and Steinberg,S. (1998) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res., 26, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baguley B.C. and Staehelin,M. (1968) Substrate specificity of adenine-specific transfer RNA methylase in normal and leukemic tissues. Eur. J. Biochem., 6, 1–7. [DOI] [PubMed] [Google Scholar]
- 4.Kuchino Y. and Nishimura,S. (1974) Methylation of Escherichia coli transfer ribonucleic acids by adenylate residue-specific transfer ribonucleic acid methylase from rat liver. Biochemistry, 13, 3683–3688. [DOI] [PubMed] [Google Scholar]
- 5.Glick J.M. and Leboy,P.S. (1977) Purification and properties of tRNA (adenine-1)-methyltransferase from rat liver. J. Biol. Chem., 252, 4790–4795. [PubMed] [Google Scholar]
- 6.Morozov I.A., Gambaryan,A.S., Lvova,T.N., Nedospasov,A.A. and Venkstern,T.V. (1982) Purification and characterization of tRNA (adenine-1-)-methyltransferase from Thermus flavus strain 71. Eur. J. Biochem., 129, 429–436. [DOI] [PubMed] [Google Scholar]
- 7.Mutzel R., Malchow,D., Meyer,D. and Kersten,H. (1986) tRNA (adenine-N1)-methyltransferase from Dictyostelium discoideum. Purification, characterization and development changes in activity. Eur. J. Biochem., 160, 101–108. [DOI] [PubMed] [Google Scholar]
- 8.Yamazaki N., Hori,H., Ozawa,K., Nakanishi,S., Ueda,T., Kumagai,I., Watanabe,K. and Nishikawa,K. (1992) Purification and characterization of tRNA (adenosine-1-)-methyltransferase from Thermus thermophilus HB27. Nucleic Acids Symp. Ser., 27, 141–142. [PubMed] [Google Scholar]
- 9.Yamazaki N., Hori,H., Ozawa,K., Nakanishi,S., Ueda,T., Kumagai,I., Watanabe,K. and Nishikawa,K. (1994) Substrate specificity of tRNA (adenine-1)-methyltransferase from Thermus thermophilus HB27. Biosci. Biotech. Biochem., 58, 1128–1133. [DOI] [PubMed] [Google Scholar]
- 10.Garcia A.G. and Goodenough-Lashua,D.M. (1998) Mechanisms of tRNA-modifying and editing enzymes. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 135–168.
- 11.Anderson J., Phan,L., Cuesta,R., Carlson,B.A., Pak,M., Asano,K., Björk,G.R., Tamame,M. and Hinnebusch,A.G. (1998) The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev., 12, 3650–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson J., Phan,L. and Hinnebusch,A.G. (2000) The Gcd10p/Gcd14p complex is the essential two-subunit tRNA (1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 97, 5173–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calvo O., Cuesta,R., Anderson,J., Gutierrez,N., Garcia-Barrio,M.T., Hinnebusch,A.G. and Tamame,M. (1999) Gcd14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 4167–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fauman E.B., Blumenthal,R.M. and Cheng,X. (1999) Structure and evolution of AdoMet-dependent MTases. In Cheng,X. and Blumenthal,R.M. (eds), S-Adenosylmethionine-dependent Methyltransferases: Structure and Functions. World Scientific Inc., Singapore, pp. 1–38.
- 15.Bujnicki J.M. (2001) In silico analysis of the tRNA:m1A58 methyltransferase family: homology-based fold prediction and identification of new members from Eubacteria and Archaea. FEBS Lett., 507, 123–127 [DOI] [PubMed] [Google Scholar]
- 16.Gupta A., Kumar,H., Dineshkumar,T.K., Varshney,U. and Subramanya,H.S. (2001) Crystal structure of Rv2118c: an AdoMet-dependent methyltransferase from Mycobacterium tuberculosis H37Rv. J. Mol. Biol., 312, 381–391. [DOI] [PubMed] [Google Scholar]
- 17.Caldas T., Binet,E., Bouloc,P., Costa,A., Desgres,J. and Richarme,G. (2000) The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23S ribosomal RNA methyltransferase. J. Biol. Chem., 275, 16414–16419. [DOI] [PubMed] [Google Scholar]
- 18.Pintard L., Lecointe,F., Bujnicki,J.M., Bonnerot,C., Grosjean,H. and Lapeyre,B. (2002) Trm7p catalyses the formation of two 2′-O-methylriboses in yeast tRNA anticodon loop. EMBO J., 21, 1811–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Motorin Y., Keith,G., Simon,C., Foiret,D., Simos,G., Hurt,E. and Grosjean,H. (1998) The yeast tRNA:pseudouridine synthase Pus1p displays a multisite substrate specificity. RNA, 4, 856–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lasa I., Caston,J.R., Fernandez-Herrero,L.A., de Pedro,M.A. and Berenguer,J. (1992) Insertional mutagenesis in the extreme thermophilic eubacteria Thermus thermophilus HB8. Mol. Microbiol., 6, 1555–1564. [DOI] [PubMed] [Google Scholar]
- 21.Glansdorf N. (1965) Topography of cotransducible arginine mutations in Escherichia coli K12. Genetics, 51, 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramseier T.M., Nègre,D., Cortay,J.-C., Scarabel,M., Cozzone,A.J. and Saier,M.H.,Jr (1983) In vitro binding of the pleiotropic transcriptional regulatory protein, FurR, to the fru, pps, ace, pts and icd operons of Escherichia coli and Salmonella typhimurium. J. Mol. Biol., 234, 28–44. [DOI] [PubMed] [Google Scholar]
- 23.Buck M., Connick,M. and Ames,B.N. (1983) Complete analysis of tRNA-modified nucleosides by high-performance liquid chromatography: the 29 modified nucleosides of Salmonella typhimurium and Escherichia coli tRNA. Anal. Biochem., 129, 1–13. [DOI] [PubMed] [Google Scholar]
- 24.Sampson J.R. and Uhlenbeck,O.C. (1988) Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl Acad. Sci. USA, 85, 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reyes V.M. and Abelson,J. (1987) A synthetic substrate for tRNA splicing. Anal. Biochem., 166, 90–106. [DOI] [PubMed] [Google Scholar]
- 26.Silberklang M., Gillum,A.M. and RajBhandary,U.L. (1979) Use of 32P labeling in the sequence analysis of nonradioactive tRNAs. Methods Enzymol., 59, 58–109. [DOI] [PubMed] [Google Scholar]
- 27.Keith G. (1995) Mobilities of modified ribonucleotides on two-dimensional cellulose thin layer chromatography. Biochimie, 77, 142–144. [DOI] [PubMed] [Google Scholar]
- 28.Droogmans L. and Grosjean,H. (1991) 2′-O-methylation and inosine formation in the wobble position of anticodon-substituted tRNAPhe in a homologous yeast in vitro system. Biochimie, 73, 1021–1025. [DOI] [PubMed] [Google Scholar]
- 29.Hartmann R.K. and Erdmann,V.A. (1989) Thermus thermophilus 16S rRNA is transcribed from an isolated transcription unit. J. Bacteriol., 171, 2933–2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao H., McKenzie,T. and Hageman,R. (1986) Isolation of a thermostable enzyme variant by cloning and selection in a thermophile. Proc. Natl Acad. Sci. USA, 83, 576–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koyama Y., Hoshino,T., Tomizuka,N. and Furukawa,K. (1986) Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J. Bacteriol., 166, 338–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keith G., Yusupov,M., Brion,C., Moras,D. and Kern,D. (1993) Sequence of tRNAAsp from Thermus thermophilus HB8. Nucleic Acids Res., 21, 4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simitsopoulou M., Avila,H. and Franceschi,F. (1999) Ribosomal gene disruption in the extreme thermophile Thermus thermophilus HB8. Generation of a mutant lacking ribosomal protein S17. Eur. J. Biochem., 266, 524–532. [DOI] [PubMed] [Google Scholar]
- 35.Gerber A.P. and Keller,W. (1999) An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science, 286, 1146–1149. [DOI] [PubMed] [Google Scholar]
- 36.Alexandrov A., Martzen,M.R. and Phizicky,E. (2002) Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA, 8, 1253–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolf J., Gerber,A.P. and Keller,W. (2002) TadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J., 21, 3841–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Bie L.G.S., Roovers,M., Oudjama,Y., Wattiez,R., Tricot,C., Stalon,V., Droogmans,L. and Bujnicki,J.M. (2003) The yggtt gene of Escherichia coli encodes a tRNA (m7G46) methyltransferase.. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yokoyama S., Watanabe,K. and Miyazama,T. (1987) Dynamic structures and functions of transfer ribonucleic acids from extreme thermophiles. Adv. Biophys., 23, 115–147. [DOI] [PubMed] [Google Scholar]
- 40.Kowalak J.A., Dalluge,J.J., McCloskey,J.A. and Stetter,K.O. (1994) The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry, 33, 7869–7876. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe K., Shinma,M., Oshima,T. and Nishimura,S. (1976) Heat-induced stability of tRNA from an extreme thermophile, Thermus thermophilus. Biochem. Biophys. Res. Commun., 72, 1137–1144. [DOI] [PubMed] [Google Scholar]
- 42.Horie N., Hara-Yokoyama,M., Yokoyama,S., Watanabe,K., Kuchino,Y., Nishimura,S. and Miyazawa,T. (1985) Two tRNA1Ile species from an extreme thermophile, Thermus thermophilus HB8: effect of 2-thiolation of ribothymidine on the thermostability of tRNA. Biochemistry, 24, 5711–5715. [DOI] [PubMed] [Google Scholar]