Abstract
Single-stranded guanosine-rich oligodeoxyribonucleotides (GROs) have a propensity to form quadruplex structures that are stabilized by G-quartets. In addition to intense speculation about the role of G-quartet formation in vivo, there is considerable interest in the therapeutic potential of quadruplex oligonucleotides as aptamers or non-antisense antiproliferative agents. We previously have described several GROs that inhibit proliferation and induce apoptosis in cancer cell lines. The activity of these GROs was related to their ability to bind to a specific cellular protein (GRO-binding protein, which has been tentatively identified as nucleolin). In this report, we describe the physical properties and biological activity of a group of 12 quadruplex oligonucleotides whose structures have been characterized previously. This group includes the thrombin-binding aptamer, an anti-HIV oligonucleotide, and several quadruplexes derived from telomere sequences. Thermal denaturation and circular dichroism (CD) spectropolarimetry were utilized to investigate the stability, reversibility and ion dependence of G-quartet formation. The ability of each oligonucleotide to inhibit the proliferation of cancer cells and to compete for binding to the GRO-binding protein was also examined. Our results confirm that G-quartet formation is essential for biological activity of GROs and show that, in some cases, quadruplex structures formed in the presence of potassium ions are significantly more active than those formed in the presence of sodium ions. However, not all quadruplex structures exhibit antiproliferative effects, and the most accurate factor in predicting biological activity was the ability to bind to the GRO-binding protein. Our data also indicate that the CD spectra of quadruplex oligonucleotides may be more complex than previously thought.
INTRODUCTION
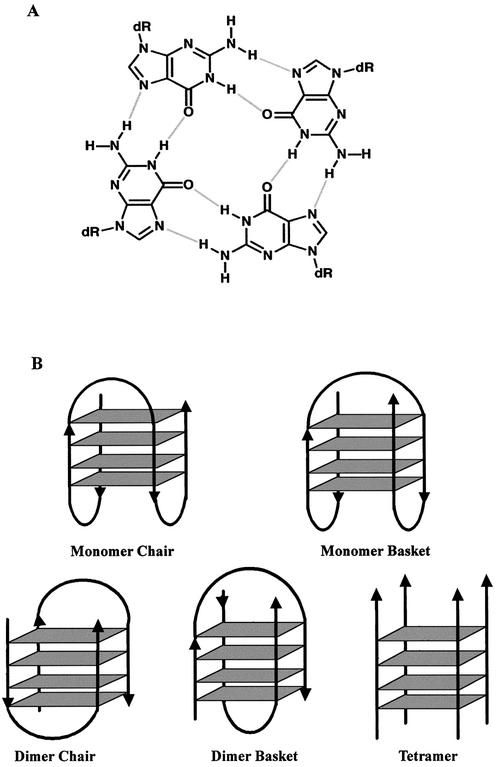
It has long been recognized that G-rich nucleic acid sequences can adopt intermolecular or intramolecular quadruplex structures that are stabilized by the presence of G-quartets (Fig. 1A). There is currently substantial interest in the potential roles of quadruplex formation in vivo because many biologically important G-rich sequences are capable of forming G-quartets under physiological conditions in vitro (1–7). In addition, the number of reports describing specific G-quartet-binding proteins is now considerable [reviewed in Shafer and Smirnov (8)]. Telomerase, a promising molecular target for cancer therapy because it is expressed in almost all tumor cells but absent in most somatic tissues (9–11), has sparked additional interest in quadruplexes because stabilization of telomeres by G-quartet-interactive compounds may inhibit the activity of this enzyme (12–15).
Figure 1.
Schematic presentation of G-quartet structures. (A) G-quartet. (B) Molecularity and loop orientation of quadruplexes
G-quartet formation has also been implicated in the non-antisense antiproliferative effects of G-rich oligonucleotides (GROs). In several cases, the biological effects of oligonucleotides designed as antisense agents were found to be unrelated to inhibition of target protein expression, but instead were associated with the formation of G-quartet structures (16–19). Although disadvantageous for antisense activity, these quadruplex effects may have therapeutic utility. We recently have reported on 3′-protected G-rich phosphodiester oligodeoxyribonucleotides, specifically GRO29A, that have impressive antiproliferative activity when added to cancer cell lines (20–22). Active GROs were shown to be stabilized by G-quartet structures that are nuclease resistant, so that even a 3′-unmodified phosphodiester analog of GRO29A was undegraded in serum-containing cell culture medium for several days (22). The precise molecular mechanism of GRO action is not yet known, but appears to be related to the ability of oligonucleotides to bind to a specific protein, which was identified as nucleolin or a nucleolin-like protein (20). Binding of nucleolin to other G-quartet-forming sequences such as telomeric DNA, immunoglobulin switch regions and ribosomal genes has also been reported (2,5,23,24). Treatment of tumor cells with GROs was found to inhibit cell cycle progression by specifically interfering with DNA replication, whereas GRO-treated normal skin cells exhibited minimal perturbation of the cell cycle (21).
GROs can form a variety of possible quadruplex structures, depending on both thermodynamic and kinetic considerations. The structures formed can be influenced by oligonucleotide base sequence and concentration, as well as the conditions (temperature and buffer) used for annealing, especially the presence of monovalent cations such as K+ and Na+. Quadruplexes can be formed by one, two or four molecules of oligonucleotide, which are referred to as monomer, dimer and tetramer structures, respectively (Fig. 1B). Monomer and dimer quadruplexes have been classified further based on the positioning of their loop regions into chair (lateral loop) or basket (diagonal loop) forms. The relative strand orientation (5′ to 3′ polarity) of the four strands of the quadruplex may be parallel (e.g. Fig. 1B, tetramer), antiparallel (Fig. 1B, monomer chair or dimer chair) or mixed (Fig. 1B, monomer basket or dimer basket).
The major aim of this study was to identify quadruplex features that are associated with antiproliferative activity in order to facilitate the design of GROs with optimal antitumor effects. A second goal was to investigate the physical properties of quadruplex oligonucleotides and their dependence on sodium and potassium cations. Therefore, we chose to study 12 quadruplex oligodeoxynucleotides whose structural characteristics had been determined previously by nuclear magnetic resonance (NMR) and/or X-ray crystallographic (XRC) techniques. The sequences and attributes of these oligonucleotides are shown in Table 1, and are described in more detail in the following section. The structures of many of these sequences have also been discussed in a recent excellent review (25). In many of the cases where both NMR and XRC structures are available, there is disagreement between the two techniques regarding the loop structure (basket or chair) or conformation (syn or anti) of the quartet guanosines (25), although the molecularities of the structures formed (tetramer, dimer or monomer) are in accord. These apparent discrepancies have been resolved to some degree by more recent structural evaluations (26,27), but it is possible that, in some cases, both structures may be correct and any disparities reflect the different experimental conditions used for quadruplex annealing.
Table 1. Sequences and structural properties of synthetic oligonucleotides.
| Oligonucleotide | Sequence | Description | References |
|---|---|---|---|
| KS-A (6mer) | d(TGGGGT) | Tetramer, telomere | (28–30,59) |
| KS-B (15mer) | d(GGTTGGTGTGGTTGG) | Monomer, thrombin aptamer | (27,31–33) |
| KS-C (12mer) | d(GGGGTTTTGGGG) | Dimer, telomere | (26,34–36,43) |
| KS-D (7mer) | d(GCATGCT) | Dimer, non-GQ | (37,60) |
| KS-E (11mer) | d(GCGGTTTGCGG) | Dimer, fragile X | (3) |
| KS-F (4mer) | d(TAGG) | Tetramer, telomere | (38) |
| KS-G (10mer) | d(GGGTTTTGGG) | Dimer, telomere | (39–41,61) |
| KS-H (28mer) | d(GGGGTTTTGGGGTTTTGGGGTTTTGGGG) | Monomer, telomere | (42,43,62) |
| KS-I (24mer) | d(TTGGGGTTGGGGTTGGGGTTGGGG) | Monomer, telomere | (44) |
| T30695 (16mer) | d(GGGTGGGTGGGTGGGT) | Monomer, anti-HIV | (45) |
| GRO20A (20mer) | d(GGTTTTGGTTTTGGTTTTGG) | Monomer, basket, experimental | (46) |
| GRO23A (23mer) | d(GGGGTTGGGGTGTGGGGTTGGGG) | Mixed basket/chair, experimental | (46) |
| GRO29A (29mer) | d(TTTGGTGGTGGTGGTTGTGGTGGTGGTGG) | Active GRO (proposed dimer) | (20–22) |
| GRO15B (15mer) | d(TTGGGGGGGGTGGGT) | Control GRO | (20–22) |
Underlined bases form loop regions.
Oligonucleotide KS-A, d(TGGGGT), is derived from the Oxytricha nova telomeric sequence, and previous work (both XRC and NMR) has shown that it forms a parallel tetrameric quadruplex in the presence of sodium ions (28–30). Oligo nucleotide KS-B, d(GGTTGGTGTGGTTGG), is known as the thrombin-binding aptamer and was developed by an in vitro selection approach. It forms a monomeric chair structure that is strongly stabilized by potassium ions (31–33). The NMR and crystal structures are in accord concerning the chair conformation, but differ in the folding pattern (although it should be noted that the NMR analysis was carried out in the presence of potassium, whereas in the crystal structure the aptamer is in the presence of thrombin). However, re-evaluation of higher resolution XRC data (27) has also indicated that the diffraction data could fit equally well to the NMR solution structure. Oligonucleotide KS-C, d(G4T4G4), is derived from the O.nova telomere sequence and forms a dimeric hairpin quadruplex in the presence of K+, Na+ or NH4+ (34). This dimer adopts a chair conformation by XRC analysis (35), but exists in the basket conformation according to the NMR structure (36). A more recent XRC study indicates that a similar basket conformation can also be formed in the crystalline state (26). The KS-D sequence is a heptamer, d(GCATGCT), that forms a four-stranded structure that does not involve G-quartets. Rather, it forms a novel dimer structure stabilized by the non-classical base-pairing interactions of two folded molecules (37). Oligonucleotide KS-E, d(GCGGTTTGCGG), represents the fragile X gene repeat sequence, and has been shown to form a dimeric chair structure in the presence of sodium ions that contains two G-quartets and two G·C·G·C tetrads (3). Oligonucleotide KS-F, d(TAGG), is a short analog of the Bombyx mori telomere repeat and has been analyzed by a combined NMR–molecular dynamics approach. It forms an unusual structure containing two G-quartets, in which each strand has a parallel and an antiparallel neighbor (38). The KS-G sequence, d(G3T4G3), is another O.nova telomere sequence and forms a dimeric quadruplex structure in the presence of either K+ or Na+ (39,40), but is stabilized preferentially by potassium (41). NMR studies show that this quadruplex is in the basket form, but that the glycosidic conformations are different from the similar KS-C sequence. Both KS-H, d(G4T4G4T4G4T4G4), and KS-I, d(G4T2G4T2G4T2G4), which are derived from, respectively, O.nova and Tetrahymena thermophila telomere sequences, form folded monomeric quadruplexes, but they have considerably different structures. The KS-H monomer is stabilized by four stacked G-quartets with two lateral T4 loops and a diagonal central loop (42,43) so that each strand is adjacent to one parallel and one antiparallel strand (basket monomer). Due to the shorter sequence between the G4 repeats, KS-I can only form three stacked G-quartets, and the folding of this structure is also quite different from that of KS-H. For KS-I, the quadruplex is formed with a lateral GTTG loop, a central lateral TTG loop and an unusual TT loop that spans from the top of the quadruplex to the bottom such that the fourth strand polarity is parallel to the adjacent first and third strands (44). T30695 is a potent anti-HIV oligonucleotide that has been analyzed by a combined NMR–molecular modeling approach and shown to form a potassium-stabilized chair monomer containing three stacked G-quartets (45). The sequences of GRO20A and GRO23A were based on a structure–activity study carried out by Marathias and Bolton (46). Using NMR analysis, these authors showed that the sequence similar to GRO20A (in the NMR study, some of the loop thymines were substituted by uracil) formed a well defined monomer basket quadruplex in the presence of sodium ions and did not change significantly upon addition of K+. On the other hand, the spectrum of GRO23A was affected by the addition of potassium and was not consistent with a single well defined structure, so it was proposed that this oligonucleotide formed a complex mixture of chair and basket conformations.
MATERIALS AND METHODS
Oligodeoxyribonucleotides
All oligonucleotides had phosphodiester backbones and were synthesized with a 3′-propylamine modification. Oligonucle otides were purchased from Oligos Etc. (Wilsonville, OR) or were synthesized on a Beckman Oligo1000M synthesizer using reagents from Beckman, and 3′-propylamine CPG columns from Glen Research (Sterling, VA). We have shown previously that oligonucleotide activity is independent of the source of synthetic oligonucleotides. Oligonucleotides were resuspended in water, precipitated with butan-1-ol, washed with 70% ethanol, dried and resuspended in 10 mM Tris–HCl pH 7.4. They were then sterilized by filtration through a 0.2 µm filter, and the concentration was determined by UV spectroscopy. Extinction coefficients were calculated as the sum of individual nucleotide ε260 (11 700 M–1cm–1 for deoxyguanosine, 8800 M–1cm–1 for thymidine, 15 400 M–1cm–1 for deoxyadenosine, and 7300 M–1cm–1 for deoxycytidine). Oligonucleotides were diluted to 250 µM with sterile 10 mM Tris–HCl and stored in aliquots at –20°C. Each oligonucleotide was checked for integrity by 5′-radiolabeling followed by denaturing polyacrylamide gel electrophoresis. The 3′-propylamine was added to ensure that all oligonucleotides had similar stability in serum-containing medium, although we have shown that this modification does not greatly affect the activity, thermal stability (22) or circular dichroism (CD) spectrum (P.J.Bates and A.Rodger, unpublished observations) of active GROs.
Antiproliferative activity of G-rich oligonucleotides in buffers containing potassium and sodium
Oligonucleotides were diluted in 10 mM Tris–HCl pH 7.4 to give a final concentration of 150 µM. Samples were boiled for 5 min and placed on ice. Subsequently, sterile 2 M KCl or 2 M NaCl was added to produce a final 50 mM concentration of NaCl or KCl, and samples were incubated for 40 h at 60°C. These lengthy annealing conditions were used to ensure dissociation of any existing structures formed during oligonucleotide synthesis and resuspension, so that the final solutions contained the most thermodynamically stable forms in either potassium or sodium. This method therefore does not take into account the kinetic factors that may influence conformation in solution, and further studies will be required to establish which structural forms are prevalent under biological conditions.
HeLa cervical carcinoma cells were plated at a density of 103 cells/well in a 96-well plate in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, which had been heat inactivated for 30 min at 55°C. Annealed oligonucleotides (10 µl of 150 µM) were added to cells to give a final concentration of 10 µM oligonucleotide per well. Cells were placed in an incubator at 37°C in an atmosphere of 10% CO2, and culture medium was not replaced for the duration of the experiment. Seven days after the addition of oligonucleotides, cell viability was determined using the MTT assay (47). Experiments were performed in triplicate, and bars represent the standard error of the data. Results were found to be reproducible, and treatment of cells with buffers containing amounts of NaCl or KCl equivalent to experimental samples was not significantly toxic (data not shown).
Thermal denaturation–renaturation using UV-visible spectroscopy
Oligonucleotides were resuspended in Tm buffer (140 mM KCl, 2.5 mM MgCl2 and 20 mM Tris–HCl pH 8.0) at a final concentration ranging from 2 to 10 µM, depending on oligonucleotide length. Oligonucleotides were annealed in Tm buffer by boiling for 5 min, slow cooling to room temperature and overnight incubation at 4°C. Samples (1.6 ml, which filled the cuvette) were placed in a stoppered quartz cuvette of 1 cm path length and were allowed to reach ambient temperature before beginning each experiment. Thermal denaturation–renaturation was carried out using an Ultrospec 2000 UV/visible spectrophotometer equipped with a Peltier effect heated cuvette holder and temperature controller (Amersham Pharmacia Biotech). A temperature range of 25–95 or 20–90°C was used to monitor absorbance at 295 nm at a heating/cooling rate of 0.5°C/min. The melting and annealing temperatures were calculated as the temperature where the absorbance at 295 nm was halfway between the absorbance of the annealed species and the absorbance of the denatured species.
Circular dichroism spectroscopy
Oligonucleotides, at a final concentration of 5 µM, were resuspended in 10 mM sodium phosphate buffer pH 7.0 containing either 0.1 M NaCl or 0.1 M KCl. Samples were boiled for 5 min, placed on ice and annealed at 60°C for 56 h. CD spectra were collected on a Jasco J-715 spectropolarimeter at 320–200 nm, using 16 scans at 100 nm/min, 1 s response time, 1 nm bandwidth. Cuvettes of 4 mm width with black quartz sides to mask the light beam were used for the measurements. A buffer baseline was collected in the same cuvette and subtracted from sample spectra. Final spectra were normalized to have zero ellipticity at 320 nm.
Protein-binding assay
An electrophoretic mobility shift assay (EMSA) to determine the ability of GROs to compete for binding to a specific GRO-binding protein (tentatively identified as nucleolin) was described previously (20). Briefly, a G-quartet-forming oligonucleotide representing the human telomere sequence, d(TTAGGGTTAGGGTTAGGGTTAGGG), referred to as ‘TEL’, was labeled with 32P using T4 kinase. Labeled TEL (2 × 104 c.p.m. per reaction, ∼1 nM final concentration) was pre-incubated alone or in the presence of an unlabeled competitor oligonucleotide (40 nM final concentration) for 30 min at 37°C. HeLa nuclear extracts (2.5 µg bandshift grade, Promega, Inc., Madison, WI) were added and samples were incubated for an additional 30 min at 37°C. Pre-incubation and binding reactions were carried out in buffer A [20 mM Tris–HCl pH 7.4, 140 mM KCl, 2.5 mM MgCl2, 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride and 8% (v/v) glycerol]. Electrophoresis was carried out using 5% polyacrylamide gels in TBE buffer (90 mM Tris borate, 2 mM EDTA). Densitometry of autoradiograms was performed on the Personal Densitometer SI (Molecular Dynamics, Inc.) and image analyzed with ImageQuaNT™ software.
Non-denaturing polyacrylamide gel electrophoresis
Radiolabeled GRO29A (40 000 c.p.m.) was added to unlabeled oligonucleotide to give a final concentration of 300 µM. Samples were boiled for 5 min and placed on ice. An equal volume of buffer containing 10 mM Tris–HCl pH 7.4, supplemented with 100 mM KCl or 100 mM NaCl, or no salt was added to the sample. Therefore, samples contained final concentrations of 150 µM GRO29A, 10 mM Tris–HCl and 50 mM KCl or NaCl (or no additional salt), which is identical to samples used for the antiproliferative activity assay. Samples were annealed at 60°C for 48 h, and electrophoretic analysis was carried out using a 20% non-denaturing polyacrylamide gel in TBE buffer. Similar experiments were carried out with 1, 10 and 100 µM GRO29A (data not shown).
RESULTS
Cation-dependent antiproliferative activity of quadruplex oligonucleotides
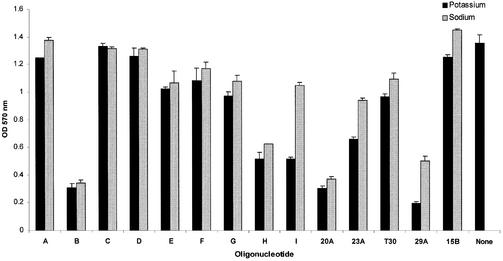
To investigate how the antiproliferative effects of GROs depend on cations, and to determine whether other quadruplex oligonucleotides have similar activity, we annealed oligonucleotides in the presence of either potassium or sodium cations and tested their ability to inhibit the proliferation of HeLa cervical carcinoma cells.
The results of these experiments are shown in Figure 2, and the sequences and descriptions of all oligonucleotides used are outlined in Table 1. Oligonucleotides KS-A, KS-C, KS-D and GRO15B showed minimal antiproliferative activity in both NaCl and KCl buffers. Oligonucleotides KS-E, KS-F, KS-G and T30695 had intermediate activity that was not very different between NaCl and KCl buffers. The group of oligonucleotides that inhibited cellular proliferation by >50% was KS-B, KS-H, KS-I (in KCl), GRO20A, GRO23A (in KCl) and GRO29A. For some of these oligonucleotides (e.g. KS-I, GRO23A and GRO29A), there was a strong dependence on the annealing conditions, with K+-annealed oligonucleotides having markedly enhanced activity compared with the Na+-annealed sample.
Figure 2.
Cation dependence of antiproliferative activity. HeLa cervical carcinoma cells were either treated with a single dose of oligonucleotide (10 µM final concentration) annealed in buffer containing either 50 mM KCl or 50 mM NaCl, or received no oligonucleotide (None). Cell viability was assayed 7 days after addition of oligonucleotide using the MTT assay. All experiments were performed in triplicate, and bars represent the standard error of the data. In this figure, GRO20A, GRO23A, GRO29A, GRO15B and T30695 are abbreviated to 20A, 23A, 29A, 15B and T30 respectively. Dark bars represent oligonucleotides annealed in KCl and light bars represent oligonucleotides annealed in NaCl.
With respect to developing a structure–activity relationship, these data do not appear to provide any definitive answers. Significant activity was observed for most, but not all, monomer quadruplexes, in both basket (e.g. GRO20A) and chair (e.g. KS-B) conformations. Perhaps most significant is the finding that not all quadruplex structures exhibit strong activity, suggesting that recognition of these molecules may be based on subtle features of the quadruplex, in accord with our previous studies (22). The present results also support our hypothesis (22) that the loop region is not a major determinant in quadruplex antiproliferative effects. In other words, although all active oligonucleotides in this study have loop structures, the number of loops, their base sequence and their conformation vary widely among the active molecules, making it unlikely that they contain a consensus domain for recognition.
UV thermal denaturation–renaturation
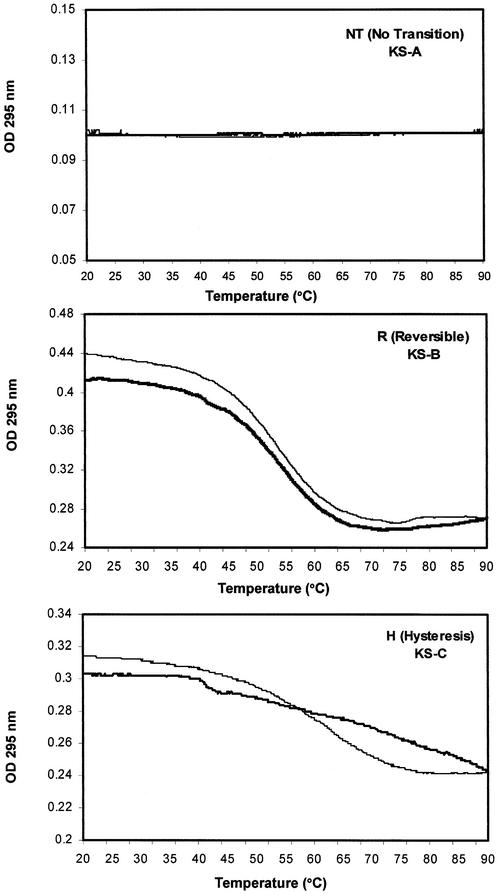
Our next aim was to determine whether biophysical signatures, such as the melting profile or CD spectrum, could predict the antiproliferative activity of quadruplex oligonucleotides. Thermal denaturation–renaturation studies can provide information about both the thermodynamic stability and kinetics of formation of secondary structures. We have used a spectroscopic method to detect G-quartet formation (20,22) that was first described by Mergny et al. (48). This technique relies on the observation that the absorbance of guanosines at 295 nm is higher when in a G-quartet structure than when denatured. In this study, we have determined the denaturation–renaturation profiles of quadruplex oligonucleotides and categorized them into three groups, as shown in Figure 3. The first category (NT) is exemplified by KS-A and showed no clear transition at 295 nm, indicating the absence of stable G-quartets (or possibly the presence of a structure with a melting temperature >90°C). The second category (R) was characterized by a clear and reversible transition where melting and annealing curves were almost superimposable, as in the case of KS-B. We interpreted this profile as reversible G-quartet formation with relatively fast kinetics. The third category (H) was typified by KS-C, and showed reversible melting with considerable hysteresis between melting and annealing curves, which is thought to reflect the slower kinetics (relative to the ‘R’ class) of quadruplex formation. The midpoints of the melting transition (Tmelt) and the annealing transition (Tanneal) were calculated for all oligonucleotides and are shown in Table 2. Several trends can be observed by comparing Table 2 and Figure 2. First, with the exception of T30695 (which is discussed below), all oligonucleotides with the NT profile lack antiproliferative activity, supporting our previous observations (20) that G-quartet formation is necessary for activity. Secondly, not all oligonucleotides with profiles that indicate quadruplex formation are active, confirming that G-quartet formation is not sufficient for activity (22). Finally, although all active oligonucleotides show thermal stability, there is no correlation between melting temperature and activity. For example, KS-B with a Tmelt of 53°C has higher activity than GRO23A, which melts at ∼85°C. Conversely, GRO29A with a Tmelt of ∼62°C has higher activity than GRO20A, which melts at 40°C.
Figure 3.
UV thermal denaturation–renaturation studies. Profiles were classified as NT, R or H, as shown. Oligonucleotides (2–10 µM final concentration) were annealed in Tm buffer (20 mM Tris–HCl pH 8.0, 140 mM KCl, and 2.5 mM MgCl2) and absorbance was measured at 295 nm with a 1 cm path length. The melting curve is the lighter line; the annealing curve is the darker line. Melting and annealing temperatures are reported in Table 2.
Table 2. Melting and annealing temperatures of oligonucleotides.
| Oligonucleotide | Tmelting | Tannealing | Profilea | Molecularity | Activityb |
|---|---|---|---|---|---|
| KS-A | – | – | NTa | Tetramer | – |
| KS-B | 52.5 | 53.0 | R | Monomer | + |
| KS-C | 68.5 | 62.8 | H | Dimer | – |
| KS-D | – | – | NT | Dimer | – |
| KS-E | – | – | NT | Dimer | – |
| KS-F | – | – | NT | Tetramer | – |
| KS-G | 45.6 | 41.4 | R | Dimer | – |
| KS-H | 65.3 | 66.8 | R | Monomer | + |
| KS-I | 86.1 | 82.8 | H | Monomer | + |
| GRO20A | 39.7 | 40.3 | R | Monomer | + |
| GRO23A | 85.6 | 83.8 | R | Monomer | + |
| T30695 | – | – | NT | Monomer | – |
| GRO29A | 61.0 | 63.5 | R | Proposed dimer | + |
| GRO15B | – | – | NT | No structure detected | – |
aNT = no transition; R = reversible; H = hysteresis.
bDefined as >50% inhibition of HeLa cell proliferation at 10 µM in buffer containing 50 mM KCl.
It is not completely clear why some of the quadruplex-forming oligonucleotides in this study do not exhibit transitions in this assay. The utility of this approach in detecting the dissociation of parallel type quadruplexes such as KS-A has not been investigated extensively but, in the case of KS-D, KS-E and KS-F, it is most likely that these sequences do not form stable quadruplexes under the conditions used here. This hypothesis is supported by the fact that these oligonucleotides exhibit small ellipticities in their CD spectra (Fig. 4), which suggests the absence of any strong base stacking interactions. For oligonucleotide T30695, which has been shown previously to form an extremely stable quadruplex in the presence of potassium ions (45,49), it is more likely that the lack of a clear transition in this assay can be attributed a very high melting temperature. This is also supported by the CD of T30695, which shows a well defined spectrum with a large positive peak, indicative of strong base stacking interactions. In most respects, our results are consistent with other reports (50). Previous evaluations of four-stranded structures have shown that, generally, the kinetics of tetramer quadruplex formation are extremely slow, while monomeric quadruplexes form quickly and reversibly. The association rate for dimeric structures is also relatively fast, but melting profiles may show slight hysteresis (50).
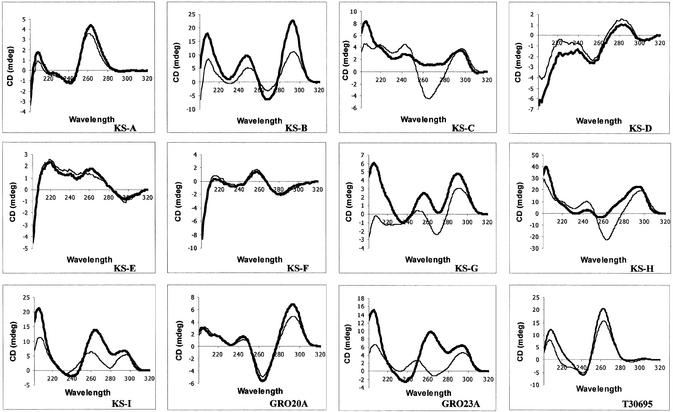
Figure 4.
CD spectroscopy studies. CD spectra of oligonucleotides (5 µM final concentration) were obtained in the presence of either 0.1 M KCl (dark lines) or 0.1 M NaCl (light lines) at 25°C with a path length of 4 mm.
CD spectropolarimetry
CD, which measures the differences between the absorbance of right-handed and left-handed circularly polarized light, has been used extensively to investigate the structure of helical molecules such as nucleic acids (51). The majority of publications have reported that ‘folded’ quadruplexes (monomers and dimers) have a CD spectrum characterized by a positive ellipticity maximum at 295 nm and a negative minimum at 265 nm, while the ‘parallel’ type (tetramers) have a positive maximum at 264 nm and a negative minimum at 240 nm (52–54). This distinction has become accepted by many researchers, and we previously were surprised to discover that the antiproliferative oligonucleotide GRO29A, which is thought to form a folded dimer quadruplex (22), exhibited a spectrum with a positive peak at 264 nm, typical of a parallel quadruplex. Therefore, the purpose of this part of the study was to clarify the relationship between CD spectral characteristics and quadruplex properties, and also to determine if any features in the CD spectra of quadruplexes were associated with biological activity. Figure 4 shows the CD spectra of all oligonucleotides annealed in buffers containing either 0.1 M KCl (thick line) or 0.1 M NaCl (thin line). As in the UV melting analysis, certain trends were observed, but there was no signature CD spectrum that was unequivocally linked with antiproliferative activity. Generally, oligonucleotides with weak activity (KS-A, KS-D, KS-E and KS-F) had small ellipticities with little difference between Na+ and K+ spectra. This suggested that only a small amount of quadruplex formation occurred under these conditions, consistent with the UV data. Those oligonucleotides whose CD signature was most different in form between NaCl and KCl buffers (KS-I and GRO23A) also exhibited clearly different antiproliferative activity in KCl and NaCl buffers. Most of the active oligonucleotides had the classical ‘folded’ CD spectrum (positive peak at 295 nm, negative at 265 nm). Some sequences exhibited this spectrum in both KCl and NaCl buffers, whereas for others the negative peak at 265 nm was only observed in sodium. However, it seems unlikely that the ability to exhibit this specific spectrum is important for biological activity because this spectral characteristic is associated with sodium ions, while activity is promoted by potassium ions. In addition, the spectrum corresponding to the most active oligonucleotide (GRO29A annealed in KCl, Fig. 6) is lacking this feature. In summary, the spectra suggest that there is not one structural reason for activity, but that activity is suggested by the presence of a number of spectral features. These include a large positive CD signal at either 264 or 295 nm, significant ellipticity in the 290–310 nm region (in contrast to duplex DNA, which lacks significant ellipticity at wavelengths >300 nm) and a large positive peak at 210 nm.
Figure 6.
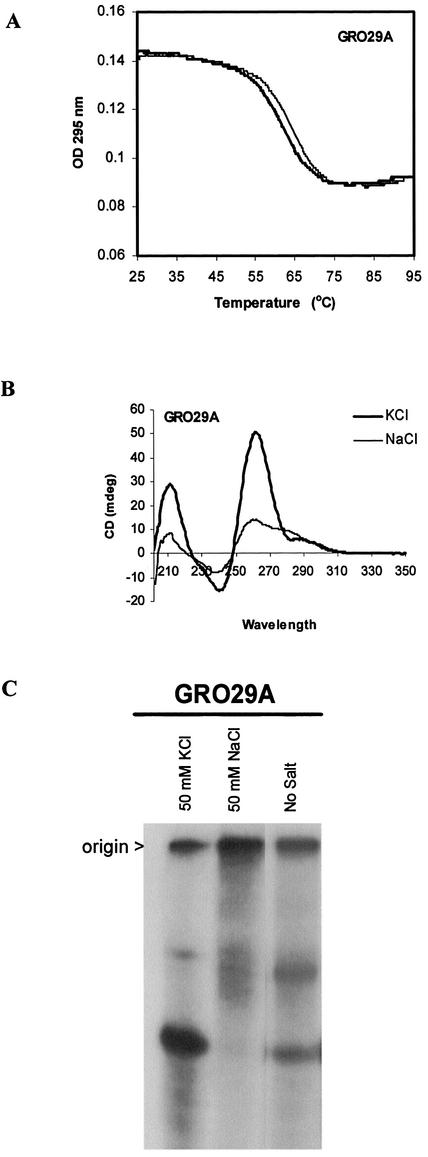
Structural characteristics of GRO29A. (A) UV thermal denaturation–renaturation profile (295 nm) of GRO29A (2 µM concentration, 1 cm path length). (B) CD spectra of GRO29A (5 µM concentration, 4 mm path length) in buffers containing KCl (dark line) or NaCl (light line). (C) Non-denaturing polyacrylamide electrophoresis of GRO29A (150 µM) annealed in 50 mM KCl, 50 mM NaCl or no salt.
What is most striking about these data is that the CD spectra are often highly dependent on ionic conditions and cannot be categorized as simply ‘parallel’ or ‘folded’. Many spectra (e.g. KS-I and GRO23A) contain positive peaks at both 264 and 295 nm. The appearance of these two peaks most probably indicates that more than one molecular species (probably both parallel and folded forms) is present in solution. The formation of multiple quadruplex species is another factor that adds to the difficulty in clearly defining quadruplex molecular structure, and may well contribute to the lack of simple correlation between structure and activity. The situation is made even more complex by the possibility that the predominant species in solution may be determined by kinetic considerations, as well as thermodynamic factors. Furthermore, an oligonucleotide (T30695) that has been well characterized as forming a monomeric folded quadruplex has a CD spectrum with a strong positive peak at 264 nm. In summary, while CD studies are clearly useful in establishing the presence of quadruplex structures, a much better understanding of the various contributions to quadruplex CD spectra will be required before CD data alone can be used to determine quadruplex molecular structure definitively. Until such results are available, researchers in this area will continue to rely on other biophysical methods to complement CD studies.
Binding of quadruplex oligonucleotides to GRO-binding protein
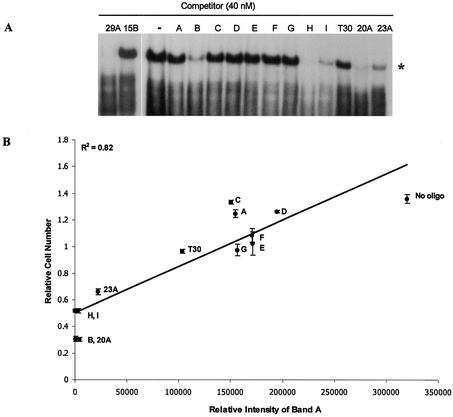
In previous work, we showed that the antiproliferative activity of GROs was related to their ability to compete with a 32P-labeled telomere sequence oligonucleotide for binding to a specific cellular protein in a mobility shift assay (20,22). This specific GRO-binding protein was also recognized by a nucleolin antibody, and therefore we proposed that it was nucleolin or a similar protein (20). Figure 5A shows the relative ability of each quadruplex oligonucleotide to bind to the GRO-binding protein in a physiological buffer containing 140 mM KCl and 2.5 mM MgCl2. In this assay, the ability of an oligonucleotide (which is unlabeled) to compete with labeled TEL for binding to GRO-binding protein (derived from HeLa extracts) is determined by the disappearance of the band indicated by an asterisk. There appears to be a good correlation between antiproliferative activity in KCl buffer and the ability of oligonucleotides to compete for the GRO-binding protein, with the most active oligonucleotides, namely GRO29A, KS-B, KS-H, KS-I and 20A (see Fig. 2), being most effective at competing for binding to GRO-binding protein. Figure 5B shows a plot of the relative intensity of the protein band in Figure 5A (note that in this assay, a high intensity indicates poor binding) versus the relative number of viable cells remaining after treatment with oligonucleotide annealed in KCl buffer (a high number indicates poor antiproliferative activity). The squared correlation coefficient (R2) for the relationship between these variables is 0.82.
Figure 5.
Protein-binding assay. (A) Competitive mobility shift assay showing the ability of unlabeled oligonucleotides to compete with radiolabeled TEL for binding to nuclear proteins. The band marked by an asterisk indicates the specific GRO-binding protein (thought to be nucleolin). The names of the oligonucleotides are abbreviated without the KS or GRO prefixes. (B) Plot of the relative intensity of the protein band in (A) versus the relative number of viable cells remaining after treatment with oligonucleotide annealed in KCl buffer. The squared correlation coefficient (R2) for the relationship between these variables is 0.82. The names of the oligonucleotides are abbreviated without the KS or GRO prefixes.
Implications for the structure of GRO29A
Another reason for carrying out biophysical studies of known quadruplex-forming oligonucleotides was to facilitate interpretation of the biophysical data regarding GRO29A and other active GROs. Previous molecular modeling studies (22) had predicted that GRO29A formed a dimeric quadruplex with lateral loops at opposite ends (chair form). The UV melting profile and CD spectra of GRO29A are shown in Figure 6. This oligonucleotide exhibits a fully reversible transition in Tm buffer (containing 140 mM KCl, 2.5 mM MgCl2) with only slight hysteresis. This is consistent with the relatively fast kinetics of formation seen for both monomer and dimer quadruplexes (Table 2). The CD spectrum of GRO29A in KCl buffer shows a large positive peak at 264 nm with a small shoulder at 295 nm, and in NaCl solution the magnitude of the 264 nm peak is greatly reduced. Although the KCl spectrum (which, as Fig. 2 shows, represents the most active species) may have been interpreted previously as formation of a parallel tetramer quadruplex, our studies have indicated that the interpretation of CD data may not be as unambiguous as we had once thought. Thus, the peak at 264 nm does not necessarily exclude the possibility of a folded antiparallel quadruplex. A further possibility that should be considered is that GRO29A forms a structure that is folded, yet with strands in a parallel orientation. Such unusual structures have been reported recently for the human telomere sequence (55) and d(GGAGGAGGAGGAGGA) (56), and the latter structure does indeed exhibit a CD spectrum with a large positive peak at 264 nm (56).
In an attempt to clarify the molecularity of GRO29A, non-denaturing polyacrylamide electrophoresis was also performed after annealing GRO29A in various buffers (Fig. 6C). In the absence of monovalent cations, GRO29A migrates as two distinct bands on a native gel in TBE buffer, clearly indicating that multiple molecular species are present. Pre-annealing the oligonucleotide in 50 mM NaCl promotes the formation of the slower mobility species, whereas annealing in 50 mM KCl promotes the formation of the faster migrating species. This faster migrating species (which is most probably the active species) migrates parallel with denatured GRO29A and an unstructured oligonucleotide of the same length (data not shown). However, in the KCl sample, this lower band is unlikely to represent unstructured single strand because the melting temperature (in 50 mM KCl) was 49°C, whereas no clear melting transitions were seen in water or 50 mM NaCl. The appearance of bands in KCl buffer was independent of GRO29A concentration, whereas as in NaCl buffer, the intensity of the slower migrating band increased with increasing GRO29A concentration (for 1, 10 and 100 µM, data not shown). Although these electrophoretic data are far from equivocal, the mobility of the lower band suggests that the active species is likely to be a monomer (or, possibly, a very compact dimer). In summary, the biophysical data are not inconsistent with our previous molecular modeling prediction that GRO29A is a dimer (22), but could also be consistent with a monomeric structure. Further experiments are in progress to determine unequivocally the molecularity and structure of the most active form of GRO29A.
DISCUSSION
With growing evidence that G-quartet formation occurs in vivo and that such structures may be important therapeutic targets, there is an urgent need for a better understanding of quadruplex structures. NMR and crystallography are traditionally considered the ‘gold standard’ of structural techniques, but may be limited in their application to quadruplexes by experimental requirements for particular ionic conditions or strand concentrations, and the necessity to prepare a single species for structure determination. Molecular dynamics has also proved to be a powerful tool in examining quadruplex structures, but usually relies on biophysical data to generate starting structures or to confirm predictions. Biophysical methods that can be applied under a variety of experimental conditions to distinguish different G-quartet-containing structures would facilitate studies of quadruplex formation. Previously, biophysical methods such as UV melting, CD, calorimetry and NMR have been applied. These studies have yielded an abundance of thermodynamic and kinetic data that have been summarized comprehensively in a recent review (50). In spite of this, there is still no clear consensus regarding many features of G-quartet formation.
The aim of the present study was not to provide an exhaustive or quantitative analysis of biophysical properties, but to investigate simple relationships between the structure of quadruplexes, their biophysical properties and their antiproliferative activity. The major conclusion is that such simple relationships do not exist; rather the biophysical signatures (especially CD spectra) are exquisitely sensitive to small changes in quadruplex structure. In retrospect, this finding is unsurprising given that the form of the CD spectra will depend on the relative orientation and stacking of bases. These are not simply dependent on sequence, strand polarity or molecularity of the quadruplex, but affected by more complex factors such as glycosidic bond conformation and sugar pucker. The complexity of the spectra is enhanced further by the possibility that more than one quadruplex species may be present. The observation that a monomeric quadruplex (in this case, the anti-HIV oligonucleotide T30695) displays a classical ‘parallel’ CD spectrum with a positive peak at 264 nm is not novel (49,50,57), but should be emphasized because oversimplification of structure–spectrum relationships may lead to misinterpretation of quadruplex CD spectra. Although we cannot absolutely exclude the possibility that T30695 forms a parallel quadruplex under our conditions, this seems unlikely in light of previous studies of this sequence (45,49,57,58).
In our previous work, we have shown that G-quartet-forming oligonucleotides such as GRO29A have inhibitory effects on the proliferation of cancer cells (20–22). A previous study of backbone-modified GRO29A analogs (22) suggested that while G-quartet formation is necessary for activity, it is not sufficient, and binding to the GRO-binding protein is the primary determinant of activity. The ultimate goal of the present study was to extend our investigations to quadruplexes with characterized structures in order to identify features, such as molecularity and loop orientation, which are associated with non-antisense antiproliferative activity. We observed biological activity for some, but not all, of the previously characterized quadruplexes. Active structures included quadruplexes with a variety of conformations, suggesting that biological activity depends on subtle features of the quadruplex, rather than simply recognition of the G-quartet motif or a four-stranded structure. Biological activity requires both a reversible UV melting profile at 295 nm and the ability to compete for the GRO-binding protein (nucleolin) in the EMSA. The results presented in this report show that there is no simple recognition element associated with activity. This supports our hypothesis that protein binding (and thus biological activity) is mediated by recognition of the specific shape of the quadruplex groove, which will depend on many factors and could theoretically be similar in quadruplexes with very different sequences.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Dr Andrew N. Lane for helpful discussions during the preparation of this manuscript. This work was supported by the Department of Defense, the National Cancer Institute, the Commonwealth of Kentucky Research Challenge Trust and the Engineering and Physical Sciences Research Council (UK).
REFERENCES
- 1.Williamson J.R. (1994) G-quartet structures in telomeric DNA. Annu. Rev. Biophys Biomol. Struct., 23, 703–730. [DOI] [PubMed] [Google Scholar]
- 2.Dempsey L.A., Sun,H., Hanakahi,L.A. and Maizels,N. (1999) G4 DNA binding by LR1 and its subunits, nucleolin and hnRNP D. A role for G–G pairing in immunoglobulin switch recombination. J. Biol. Chem., 274, 1066–1071. [DOI] [PubMed] [Google Scholar]
- 3.Kettani A., Kumar,R.A. and Patel,D.J. (1995) Solution structure of a DNA quadruplex containing the fragile X syndrome triplet repeat. J. Mol. Biol., 254, 638–656. [DOI] [PubMed] [Google Scholar]
- 4.Simonsson T., Pecinka,P. and Kubista,M. (1998) DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res., 26, 1167–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanakahi L.A., Sun,H. and Maizels,N. (1999) High affinity interactions of nucleolin with G–G-paired rDNA. J. Biol. Chem., 274, 15908–15912. [DOI] [PubMed] [Google Scholar]
- 6.Catasti P., Chen,X., Moyzis,R.K., Bradbury,E.M. and Gupta,G. (1996) Structure–function correlations of the insulin-linked polymorphic region. J. Mol. Biol., 264, 534–545. [DOI] [PubMed] [Google Scholar]
- 7.Siddiqui-Jain A., Grand,C.L., Bearss,D.J. and Hurley,L.H. (2002) Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA, 99, 11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shafer R.H. and Smirnov,I. (2001) Biological aspects of DNA/RNA quadruplexes. Biopolymers, 56, 209–227. [DOI] [PubMed] [Google Scholar]
- 9.Newbold R.F. (1999) Telomerase as an anti-cancer drug target: will it fulfil its early promise? Anticancer Drug Des., 14, 349–354. [PubMed] [Google Scholar]
- 10.Lavelle F., Riou,J.F., Laoui,A. and Mailliet,P. (2000) Telomerase: a therapeutic target for the third millennium? Crit. Rev. Oncol. Hematol., 34, 111–126. [DOI] [PubMed] [Google Scholar]
- 11.Neidle S. and Kelland,L.R. (1999) Telomerase as an anti-cancer target: current status and future prospects. Anticancer Drug Des., 14, 341–347. [PubMed] [Google Scholar]
- 12.Perry P.J. and Jenkins,T.C. (1999) Recent advances in the development of telomerase inhibitors for the treatment of cancer. Expert Opin. Invest. Drugs, 8, 1981–2008. [DOI] [PubMed] [Google Scholar]
- 13.White L.K., Wright,W.E. and Shay,J.W. (2001) Telomerase inhibitors. Trends Biotechnol., 19, 114–120. [DOI] [PubMed] [Google Scholar]
- 14.Mergny J.L., Mailliet,P., Lavelle,F., Riou,J.F., Laoui,A. and Helene,C. (1999) The development of telomerase inhibitors: the G-quartet approach. Anticancer Drug Des., 14, 327–339. [PubMed] [Google Scholar]
- 15.Bearss D.J., Hurley,L.H. and Von Hoff,D.D. (2000) Telomere maintenance mechanisms as a target for drug development. Oncogene, 19, 6632–6641. [DOI] [PubMed] [Google Scholar]
- 16.Burgess T.L., Fisher,E.F., Ross,S.L., Bready,J.V., Qian,Y.X., Bayewitch,L.A., Cohen,A.M., Herrera,C.J., Hu,S.S., Kramer,T.B. et al. (1995) The antiproliferative activity of c-myb and c-myc antisense oligonucleotides in smooth muscle cells is caused by a nonantisense mechanism. Proc. Natl Acad. Sci. USA, 92, 4051–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anselmet A., Mayat,E., Wietek,S., Layer,P.G., Payrastre,B. and Massoulie,J. (2002) Non-antisense cellular responses to oligonucleotides. FEBS Lett., 510, 175–180. [DOI] [PubMed] [Google Scholar]
- 18.Benimetskaya L., Berton,M., Kolbanovsky,A., Benimetsky,S. and Stein,C.A. (1997) Formation of a G-tetrad and higher order structures correlates with biological activity of the RelA (NF-κB p65) ‘antisense’ oligodeoxynucleotide. Nucleic Acids Res., 25, 2648–2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saijo Y., Uchiyama,B., Abe,T., Satoh,K. and Nukiwa,T. (1997) Contiguous four-guanosine sequence in c-myc antisense phosphorothioate oligonucleotides inhibits cell growth on human lung cancer cells: possible involvement of cell adhesion inhibition. Jpn J. Cancer Res., 88, 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bates P.J., Kahlon,J.B., Thomas,S.D., Trent,J.O. and Miller,D.M. (1999) Antiproliferative activity of G-rich oligonucleotides correlates with protein binding. J. Biol. Chem., 274, 26369–26377. [DOI] [PubMed] [Google Scholar]
- 21.Xu X., Hamhouyia,F., Thomas,S.D., Burke,T.J., Girvan,A.C., McGregor,W.G., Trent,J.O., Miller,D.M. and Bates,P.J. (2001) Inhibition of DNA replication and induction of S phase cell cycle arrest by G-rich oligonucleotides. J. Biol. Chem., 276, 43221–43230. [DOI] [PubMed] [Google Scholar]
- 22.Dapic V., Bates,P.J., Trent,J.O., Rodger,A., Thomas,S.D. and Miller,D.M. (2002) Antiproliferative activity of G-quartet-forming oligonucleotides with backbone and sugar modifications. Biochemistry, 41, 3676–3685. [DOI] [PubMed] [Google Scholar]
- 23.Ishikawa F., Matunis,M.J., Dreyfuss,G. and Cech,T.R. (1993) Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol. Cell. Biol., 13, 4301–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dickinson L.A. and Kohwi-Shigematsu,T. (1995) Nucleolin is a matrix attachment region DNA-binding protein that specifically recognizes a region with high base-unpairing potential. Mol. Cell. Biol., 15, 456–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keniry M.A. (2001) Quadruplex structures in nucleic acids. Biopolymers, 56, 123–146. [DOI] [PubMed] [Google Scholar]
- 26.Haider S., Parkinson,G.N. and Neidle,S. (2002) Crystal structure of the potassium form of an Oxytricha nova G-quadruplex. J. Mol. Biol., 320, 189–200. [DOI] [PubMed] [Google Scholar]
- 27.Padmanabhan K. and Tulinsky,A. (1996) An ambiguous structure of a DNA 15-mer thrombin complex. Acta Crystallogr. D, 52, 272–282. [DOI] [PubMed] [Google Scholar]
- 28.Phillips K., Dauter,Z., Murchie,A.I., Lilley,D.M. and Luisi,B. (1997) The crystal structure of a parallel-stranded guanine tetraplex at 0.95 Å resolution. J. Mol. Biol., 273, 171–182. [DOI] [PubMed] [Google Scholar]
- 29.Aboul-ela F., Murchie,A.I., Norman,D.G. and Lilley,D.M. (1994) Solution structure of a parallel-stranded tetraplex formed by d(TG4T) in the presence of sodium ions by nuclear magnetic resonance spectroscopy. J. Mol. Biol., 243, 458–471. [DOI] [PubMed] [Google Scholar]
- 30.Aboul-ela F., Murchie,A.I. and Lilley,D.M. (1992) NMR study of parallel-stranded tetraplex formation by the hexadeoxynucleotide d(TG4T). Nature, 360, 280–282. [DOI] [PubMed] [Google Scholar]
- 31.Kelly J.A., Feigon,J. and Yeates,T.O. (1996) Reconciliation of the X-ray and NMR structures of the thrombin-binding aptamer d(GGTTGGTGTGGTTGG). J. Mol. Biol., 256, 417–422. [DOI] [PubMed] [Google Scholar]
- 32.Macaya R.F., Schultze,P., Smith,F.W., Roe,J.A. and Feigon,J. (1993) Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl Acad. Sci. USA, 90, 3745–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Padmanabhan K., Padmanabhan,K.P., Ferrara,J.D., Sadler,J.E. and Tulinsky,A. (1993) The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J. Biol. Chem., 268, 17651–17654. [DOI] [PubMed] [Google Scholar]
- 34.Schultze P., Hud,N.V., Smith,F.W. and Feigon,J. (1999) The effect of sodium, potassium and ammonium ions on the conformation of the dimeric quadruplex formed by the Oxytricha nova telomere repeat oligonucleotide d(G(4)T(4)G(4)). Nucleic Acids Res., 27, 3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang C., Zhang,X., Ratliff,R., Moyzis,R. and Rich,A. (1992) Crystal structure of four-stranded Oxytricha telomeric DNA. Nature, 356, 126–131. [DOI] [PubMed] [Google Scholar]
- 36.Smith F.W. and Feigon,J. (1993) Strand orientation in the DNA quadruplex formed from the Oxytricha telomere repeat oligonucleotide d(G4T4G4) in solution. Biochemistry, 32, 8682–8692. [DOI] [PubMed] [Google Scholar]
- 37.Leonard G.A., Zhang,S., Peterson,M.R., Harrop,S.J., Helliwell,J.R., Cruse,W.B., d’Estaintot,B.L., Kennard,O., Brown,T. and Hunter,W.N. (1995) Self-association of a DNA loop creates a quadruplex: crystal structure of d(GCATGCT) at 1.8 Å resolution. Structure, 3, 335–340. [DOI] [PubMed] [Google Scholar]
- 38.Kettani A., Bouaziz,S., Wang,W., Jones,R.A. and Patel,D.J. (1997) Bombyx mori single repeat telomeric DNA sequence forms a G-quadruplex capped by base triads. Nature Struct. Biol., 4, 382–389. [DOI] [PubMed] [Google Scholar]
- 39.Scaria P.V., Shire,S.J. and Shafer,R.H. (1992) Quadruplex structure of d(G3T4G3) stabilized by K+ or Na+ is an asymmetric hairpin dimer. Proc. Natl Acad. Sci. USA, 89, 10336–10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keniry M.A., Strahan,G.D., Owen,E.A. and Shafer,R.H. (1995) Solution structure of the Na+ form of the dimeric guanine quadruplex [d(G3T4G3)]2. Eur. J. Biochem., 233, 631–643. [DOI] [PubMed] [Google Scholar]
- 41.Hud N.V., Smith,F.W., Anet,F.A. and Feigon,J. (1996) The selectivity for K+ versus Na+ in DNA quadruplexes is dominated by relative free energies of hydration: a thermodynamic analysis by 1H NMR. Biochemistry, 35, 15383–15390. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y. and Patel,D.J. (1995) Solution structure of the Oxytricha telomeric repeat d[G4(T4G4)3] G- tetraplex. J. Mol. Biol., 251, 76–94. [DOI] [PubMed] [Google Scholar]
- 43.Smith F.W. and Feigon,J. (1992) Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature, 356, 164–168. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y. and Patel,D.J. (1994) Solution structure of the Tetrahymena telomeric repeat d(T2G4)4 G-tetraplex. Structure, 2, 1141–1156. [DOI] [PubMed] [Google Scholar]
- 45.Jing N. and Hogan,M.E. (1998) Structure–activity of tetrad-forming oligonucleotides as a potent anti-HIV therapeutic drug. J. Biol. Chem., 273, 34992–34999. [DOI] [PubMed] [Google Scholar]
- 46.Marathias V.M. and Bolton,P.H. (1999) Determinants of DNA quadruplex structural type: sequence and potassium binding. Biochemistry, 38, 4355–4364. [DOI] [PubMed] [Google Scholar]
- 47.Morgan D.M. (1998) Tetrazolium (MTT) assay for cellular viability and activity. Methods Mol. Biol., 79, 179–183. [DOI] [PubMed] [Google Scholar]
- 48.Mergny J.L., Phan,A.T. and Lacroix,L. (1998) Following G-quartet formation by UV-spectroscopy. FEBS Lett., 435, 74–78. [DOI] [PubMed] [Google Scholar]
- 49.Jing N., Rando,R.F., Pommier,Y. and Hogan,M.E. (1997) Ion selective folding of loop domains in a potent anti-HIV oligonucleotide. Biochemistry, 36, 12498–12505. [DOI] [PubMed] [Google Scholar]
- 50.Hardin C.C., Perry,A.G. and White,K. (2001) Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers, 56, 147–194. [DOI] [PubMed] [Google Scholar]
- 51.Rodger A. and Nordén,B. (1997) Circular Dichroism and Linear Dichroism. Oxford Chemistry Masters; 1. Oxford University Press, Oxford.
- 52.Balagurumoorthy P. and Brahmachari,S.K. (1994) Structure and stability of human telomeric sequence. J. Biol. Chem., 269, 21858–21869. [PubMed] [Google Scholar]
- 53.Hardin C.C., Henderson,E., Watson,T. and Prosser,J.K. (1991) Monovalent cation induced structural transitions in telomeric DNAs: G-DNA folding intermediates. Biochemistry, 30, 4460–4472. [DOI] [PubMed] [Google Scholar]
- 54.Balagurumoorthy P., Brahmachari,S.K., Mohanty,D., Bansal,M. and Sasisekharan,V. (1992) Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res., 20, 4061–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parkinson G.N., Lee,M.P. and Neidle,S. (2002) Crystal structure of parallel quadruplexes from human telomeric DNA. Nature, 417, 876–880. [DOI] [PubMed] [Google Scholar]
- 56.Matsugami A., Ouhashi,K., Kanagawa,M., Liu,H., Kanagawa,S., Uesugi,S. and Katahira,M. (2001) An intramolecular quadruplex of (GGA)(4) triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction. J. Mol. Biol., 313, 255–269. [DOI] [PubMed] [Google Scholar]
- 57.Porumb H., Monnot,M. and Fermandjian,S. (2002) Circular dichroism signatures of features simultaneously present in structured guanine-rich oligonucleotides: a combined spectroscopic and electrophoretic approach. Electrophoresis, 23, 1013–1020. [DOI] [PubMed] [Google Scholar]
- 58.Jing N., Marchand,C., Liu,J., Mitra,R., Hogan,M.E. and Pommier,Y. (2000) Mechanism of inhibition of HIV-1 integrase by G-tetrad-forming oligonucleotides in vitro. J. Biol. Chem., 275, 21460–21467. [DOI] [PubMed] [Google Scholar]
- 59.Laughlan G., Murchie,A.I., Norman,D.G., Moore,M.H., Moody,P.C., Lilley,D.M. and Luisi,B. (1994) The high-resolution crystal structure of a parallel-stranded guanine tetraplex. Science, 265, 520–524. [DOI] [PubMed] [Google Scholar]
- 60.Salisbury S.A., Wilson,S.E., Powell,H.R., Kennard,O., Lubini,P., Sheldrick,G.M., Escaja,N., Alazzouzi,E., Grandas,A. and Pedroso,E. (1997) The bi-loop, a new general four-stranded DNA motif. Proc. Natl Acad. Sci. USA, 94, 5515–5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Strahan G.D., Keniry,M.A. and Shafer,R.H. (1998) NMR structure refinement and dynamics of the K+-[d(G3T4G3)]2 quadruplex via particle mesh Ewald molecular dynamics simulations. Biophys. J., 75, 968–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith F.W., Schultze,P. and Feigon,J. (1995) Solution structures of unimolecular quadruplexes formed by oligonucleotides containing Oxytricha telomere repeats. Structure, 3, 997–1008. [DOI] [PubMed] [Google Scholar]